Abstract
Key message
Making use of wheat chromosomal resources, we developed 11 gene-associated markers for the region of interest, which allowed reducing gene interval and spanning it by four BAC clones.
Abstract
Positional gene cloning and targeted marker development in bread wheat are hampered by high complexity and polyploidy of its nuclear genome. Aiming to clone a Russian wheat aphid resistance gene Dn2401 located on wheat chromosome arm 7DS, we have developed a strategy overcoming problems due to polyploidy and enabling efficient development of gene-associated markers from the region of interest. We employed information gathered by GenomeZipper, a synteny-based tool combining sequence data of rice, Brachypodium, sorghum and barley, and took advantage of a high-density linkage map of Aegilops tauschii. To ensure genome- and locus-specificity of markers, we made use of survey sequence assemblies of isolated wheat chromosomes 7A, 7B and 7D. Despite the low level of polymorphism of the wheat D subgenome, our approach allowed us to add in an efficient and cost-effective manner 11 new gene-associated markers in the Dn2401 region and narrow down the target interval to 0.83 cM. Screening 7DS-specific BAC library with the flanking markers revealed a contig of four BAC clones that span the Dn2401 region in wheat cultivar ‘Chinese Spring’. With the availability of sequence assemblies and GenomeZippers for each of the wheat chromosome arms, the proposed strategy can be applied for focused marker development in any region of the wheat genome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bread wheat (Triticum aestivum L.) is one of the most important crops providing staple food for 35 % of the world’s population. Its annual production of 713 million tons (FAOSTAT 2013) is topped only by rice and maize. Wheat yields are markedly reduced due to attack of a large variety of pests and diseases causing significant economic losses.
Russian wheat aphid, RWA (Diuraphis noxia Kurdjumov) is globally a major invasive pest affecting wheat, as well as barley (Morrison and Peairs 1998). Numerous D. noxia strains (biotypes) varying in virulence have spread in all wheat and barley growing areas with the exception of Australia (Lapitan et al. 2007). Besides causing chlorosis and reducing plant biomass and consequently grain yield (Burd and Burton 1992; Girma et al. 1993), aphid infestation introduces leaf rolling, which reduces the effectiveness of contact insecticides by sheltering the insect colonies. Thus, development of new varieties carrying genes controlling resistance to the pest is the most efficient, economical and environmentally safe approach to reducing the damage. Supplementation of conventional wheat breeding approaches for pest resistance with marker-assisted selection and, potentially, direct gene transfer by molecular methods, promises to enhance the efficiency of the breeding process. Prerequisites of this approach are saturation of genetic maps in the region of interest and, ideally, identification of the gene underlying the resistance.
Numerous RWA resistance genes have been identified in various cultivars and lines of wheat, rye and Aegilops tauschii (Nkongolo et al. 1991; Marais et al. 1998; Martin et al. 2001; Liu et al. 2001, 2002; Peng et al. 2009), but only a few of them, namely Dn7, Dn626580 and Dn2401 (Lapitan et al. 2007; Valdez et al. 2012; Qureshi et al. 2006; Fazel-Najafabadi et al. 2014), confer resistance to the highly virulent US RWA biotype 2, the prevailing biotype in the United States. In the present work, we made efforts towards cloning the Dn2401 gene located in the central part of the short arm of chromosome 7D (7DS). A previous study (Fazel-Najafabadi et al. 2014) identified several 7DS-located microsatellite markers linked with the trait, including Xbarc214 and Xgwm473, delimiting an interval of 2.9 cM. In our work, we aimed to narrow down the Dn2401 interval and potentially span it with a contig of BAC clones.
Marker development and mapping in hexaploid bread wheat (2n = 6x = 42) is highly challenging due to the complex nature of its genome, combining large size (~17 Gbp/1 C), high repeat content (>80 % repetitive sequences), and the presence of three homoeologous subgenomes A, B and D. Moreover, the level of polymorphism among elite wheat cultivars, especially within the D subgenome, the youngest of the subgenomes, is limited (Chao et al. 2009; Berkman et al. 2013).
Previously, positional cloning projects took advantage of mapping in diploid wheat ancestors and close relatives to avoid problems due to polyploidy. Nevertheless, mapping in diploid species is often complicated by inconsistencies between genetic maps of the hexaploid and the diploid and/or absence of the gene of interest from the diploid genome. This is also the case with Dn2401. In such situation, mapping in the hexaploid becomes a necessity. In the past decade, the major source of information for targeted marker development aiming to saturate a narrow region was sequence information of wheat expressed sequence tags (ESTs) and colinearity between wheat and sequenced genomes of model grass species like rice and Brachypodium (Keller et al. 2005; Qin et al. 2011; Terracciano et al. 2013). However, the efficiency of designing locus-specific primers based on ESTs was rather low due to insufficient specificity, accentuated in polyploid genomes, and lack of wheat sequence information that would reveal genomic context of the ESTs (Schnurbusch et al. 2007; Terracciano et al. 2013).
In the past two decades, chromosomal genomics has been developed as a strategy to tackle problems associated with genome complexity and polyploidy in wheat (Doležel et al. 2012). The approach is based on dissecting large genomes into particular chromosomes or chromosome arms by flow-cytometric sorting and has also been applied to other Triticeae species like barley (Lysák et al. 1999), rye (Kubaláková et al. 2003) and Aegilops sp. (Molnár et al. 2011). DNA of flow-sorted chromosomes is of high quality, which enabled the construction of a set of wheat chromosome (arm)-specific BAC libraries (Šafář et al. 2010; http://olomouc.ueb.cas.cz/). The International Wheat Genome Sequencing Consortium (IWGSC, http://www.wheatgenome.org/) adapted the chromosomal approach employing chromosome-specific physical maps as the major strategy towards obtaining a complete reference sequence of the bread wheat genome. Moreover, combining next-generation sequencing technologies with the flow sorting enabled to obtain survey sequences of individual chromosomes (arms) of barley (Mayer et al. 2009, 2011), wheat (IWGSC 2014) as well as rye (Martis et al. 2013). The generated barley chromosomal sequences were exemplarily used to establish so-called ‘GenomeZipper’, a colinearity-based tool comprising of an ordered set of Brachypodium, rice and sorghum genes anchored to a backbone of linkage-mapped barley ESTs (Close et al. 2009), the corresponding barley 454 sequence reads, non-mapped barley ESTs and barley full-length cDNAs (Mayer et al. 2011). Implying virtual gene order in barley, GenomeZipper promises to be a useful tool for targeted development of gene-derived markers from desired regions of the genome.
Application of these chromosomal resources, including a 7DS-specific BAC library and physical map (Šimková et al. 2011), survey sequence assemblies of the 7AS, 7BS and 7DS chromosome arms (Berkman et al. 2013; IWGSC 2014), and barley GenomeZipper (Mayer et al. 2011), together with a new high-density linkage map of Ae. tauschii (Luo et al. 2013), the progenitor of the D subgenome, enabled us to develop an efficient approach for targeted identification of gene-associated markers from the region of interest and to place 11 new markers around the Dn2401 gene. This aided reducing interval delimited by flanking markers and identifying BAC contig spanning the region of interest.
Materials and methods
Plant material
Genetic mapping was performed in 184 F2 plants derived from a cross between RWA resistant winter wheat line CI2401 and susceptible cultivar ‘Glupro’. To identify BAC clones and contigs bearing markers linked to the Dn2401 gene, a BAC library specific for the 7DS chromosome arm of cv. ‘Chinese Spring’ (Šimková et al. 2011) and a 7DS-specific physical contig map (https://urgi.versailles.inra.fr/gb2/gbrowse/wheat_phys_7DS_v1/) were utilized.
7AS, 7BS and 7DS chromosome arms were isolated from double-ditelosomic lines of wheat cv. ‘Chinese Spring’ carrying arms of chromosomes 7A, 7B and 7D as telocentric chromosomes. Seeds for these lines were kindly provided by Prof. Bikram Gill (Kansas State University, Manhattan, USA) and Prof. Adam Lukaszewski (University of California, Riverside, USA).
Preparation of chromosome-specific DNA
7AS, 7BS and 7DS chromosome arms were isolated from the double-ditelosomic lines by flow-cytometric sorting as described in Kubaláková et al. (2002). Chromosomal DNA was amplified by isothermal multiple displacement amplification (MDA) as described in Šimková et al. (2008).
Scoring RWA response
RWA screening was done at the Colorado State University Insectary greenhouse under ambient conditions (14 h, ~25.5 °C days with light intensities between 1100 and 1400 μM m2 s−1 and 10 h, ~20 °C nights). A randomized complete block design with three replications was used. Twelve to fifteen seeds of each F2 derived F3 family (F2:3) were planted. Each tray contained the parents as well as ‘Gamtoos- R’ (containing the Dn7 resistance gene) and ‘Yuma’ which served as resistant and susceptible controls, respectively. 7-day-old seedlings were infested with RWA biotype 2 by scattering aphids evenly across the trays. Each plant was scored for leaf rolling and chlorosis at 7 and 10 days post infestation, as previously described (Collins et al. 2005). Chlorosis scores ranged from 1 (=healthy plants with small hypersensitive lesions) to 9 (=dead or unrecoverable), while leaf rolling scores were on a scale of 1 (=completely flat leaves) to 4 (=tightly rolled leaves with leaf trapping) (Collins et al. 2005). Plants with chlorosis scores of ≤4 and leaf rolling scores ≤2 were considered resistant and those exhibiting chlorosis ≥5 and leaf rolling ≥3 were considered susceptible. The F2 individuals were assigned a phenotypic class (homozygous resistant, heterozygous, or homozygous susceptible) based on the number of resistant and susceptible individuals observed in the F2:3 families, under RWA infestation. Homozygous resistant and susceptible designations were only given to F2 individuals when all (or all but one) of the plants in the F2:3 family were scored with the same corresponding designation.
Mapping of SSR markers
All 184 plants of the mapping population were scored for SSR markers Xcfd14, Xcfd68 and Xgwm473 using primer sequences and PCR conditions given in GrainGenes 2.0 (http://wheat.pw.usda.gov/). Marker Xcfd14 was separated in 6 % non-denaturing polyacrylamide gel (19:1 acrylamide:bisacrylamide) and visualized by ethidium bromide staining. Genotyping of markers Xcfd68 and Xgwm473 was performed by PCR amplification using fluorescently labeled primers and subsequent separation of the PCR products by ABI 3730xl DNA analyzer (Applied Biosystems). Data were analyzed by GeneMarker® software (SoftGenetics).
Marker development strategy
Markers specific for the Dn2401 gene region were developed by two approaches, both employing colinearity with related grass genomes (scheme of the procedure shown in Supplementary File 1).
Since the available flanking markers were microsatellites, the procedure started with identification of genes delimiting the Dn2401 region. BAC pools of the 7DS-specific BAC library were PCR screened with flanking microsatellite markers Xcfd68 and Xgwm473 and corresponding BAC clones as well as BAC contigs were identified (Šimková et al. 2011). The Xcfd68 marker was used in our study instead of the closest marker Xbarc214, located 0.3 cM proximal to Xcfd68 (Fazel-Najafabadi et al. 2014), because of easier and more reliable scoring. Two BAC clones from each of the contigs identified by the above markers were sequenced using Roche/454 technology and the sequence reads were assembled using Newbler software package. Genes adjacent to both Xcfd68 and Xgwm473 were identified and used to initially delimit syntenic regions in sequenced grass genomes.
The first approach to marker development was based on information provided by the barley GenomeZipper (Mayer et al. 2011). After identifying the syntenic region, sequences of relevant barley ESTs were retrieved from the ‘HarvEST: Barley’ database (Close et al. 2007), version 1.77. These were compared with assembled 7AS, 7BS and 7DS sequences of cv. ‘Chinese Spring’ (Berkman et al. 2013; http://www.wheatgenome.info/) using BLASTn algorithm. Genetically mapped barley ESTs located at syntenic positions in Brachypodium, rice and sorghum were selected initially, followed by ESTs only inferred by synteny. Wheat genomic sequences corresponding to 7AS, 7BS and 7DS contigs were identified based on sequence identity with barley ESTs. Contigs from each subgenome were aligned and annotated using GeniousPro software, version 5.4.4 (Drummond et al. 2010) using the Genious or Muscle alignment software with default parameters. Sequences annotated with marked exons and introns were the basis for designing a first set of 7DS-specific primers.
An alternative approach employing synteny between wheat and Ae. tauschii was applied to design a second set of markers using a gene-based linkage map of Ae. tauschii that included information of colinear rice, Brachypodium and sorghum genes (Luo et al. 2013). Sequences of markers from orthologous region of Ae. tauschii were compared with the 7AS, 7BS and 7DS assemblies using BLASTn, and the sequence contigs thus identified were aligned as described above. Exons and introns were determined by comparison with rice genes using BLASTn.
Primer design and identification of polymorphisms
Aligned and annotated sequence contigs from 7AS, 7BS and 7DS were used to design subgenome-specific primers for the region of interest. To avoid amplification from conserved gene domains, primers were preferentially designed in non-coding regions. Primer3 v.0.4.0 software was used for the primer design with default parameters (Rozen and Skaletsky 2000). To verify the specificity of the PCR primers, DNA of flow-sorted chromosome arms 7AS, 7BS and 7DS was used as PCR template. PCR reactions were carried out in 20-μl volume comprising 10 ng of template DNA, 1× PCR buffer (10 mM Tris-HCl, pH 8.8 at 25 °C, 1.5 mM MgCl2, 50 mM KCl, 0.1 % Triton X-100), 1 μM primers, 200 μM dNTPs and 1 U of DyNAzyme DNA Polymerase (Finnzymes, Espoo, Finland). The amplification was performed under the following conditions: initial denaturation at 94 °C for 5 min followed by 35 cycles of 94 °C for 30 s, 58 °C for 30 s, 72 °C for 1 min and final extension at 72 °C for 10 min. PCR products were separated in 4 % non-denaturing polyacrylamide gel and visualised by ethidium bromide staining. PCR conditions (annealing temperature and cycle number) were optimized to obtain a single 7DS-specific product. Primer pairs that proved to be 7DS-specific were applied to amplify DNA of the mapping population parents, CI2401 and ‘Glupro’. PCR was performed under optimized conditions with 20 ng of template DNA. The amplification products were cleaned up by Exonuclease I/FastAP™ Thermosensitive Alkaline Phosphatase (Fermentas, Burlington, Canada) treatment and cycle sequencing was performed using a BigDye® Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, USA) following manufacturer’s instructions. Sequencing products were purified using an Agencourt Clean SEQ kit (Beckman Coulter, Beverly, MA) and analyzed on an ABI 3730xl DNA analyzer (Applied Biosystems). Sequences were aligned to identify polymorphisms between the parents using MEGA 4 software and ClustalW with default parameters. To verify the locus specificity of the primers, the parental sequences were aligned to the ‘Chinese Spring’ 7DS survey sequence (Berkman et al. 2011).
Linkage mapping and integration with the physical map
Mapping of the identified SNP markers was done by amplicon sequencing on 184 plants of the CI2401/‘Glupro’ F2 mapping population. PCR products from individual F2 plants were purified, sequenced and analyzed as described above. Marker Xowm711 showing length polymorphism in the amplicon was scored after separation in 4 % polyacrylamide gel. Genotyping data were analyzed using JoinMap 4.0 (Van Ooijen and Voorrips 2001). Genetic distances between markers were calculated applying the Kosambi function (Kosambi 1944). The markers were then integrated into the existing microsatellite genetic map covering the Dn2401 gene region (Fazel-Najafabadi et al. 2014). Aiming to integrate genetic and physical maps of the region, the mapped markers were used to screen the 7DS-specific BAC library as described in Šimková et al. (2011) and contigs from the 7DS physical map were identified through positive BAC clones.
Results
Delimiting syntenic regions
In order to apply a synteny-based approach to marker development, we had to identify genic sequences adjacent to the available microsatellite markers and through them delimit syntenic regions in genomes of relative species.
Screening of the 7DS-specific BAC library with Dn2401-flanking SSR markers Xcfd68 and Xgwm473 and sequencing of BAC clones from marker-comprising contigs revealed genes corresponding to barley ESTs that enabled identification of the syntenic region in the barley GenomeZipper. This region was delimited by barley EST 40502 and the genes Bradi1g43560.1, Os06g0284200 and Sb10g010460.1 on the distal side, and barley EST 7580 and Bradi3g39810.1, Os08g0503800 and Sb07g028020.1 on the proximal side, and contained 255 predicted genes. In addition, a 2-cM region proximal of Xgwm473 was included in the study. The Dn2401 region comprised a previously described breakpoint of colinearity between the barley, as well as wheat genome and genomes of Brachypodium, rice and sorghum, respectively (Sorrells et al. 2003; Mayer et al. 2011), combining parts of Brachypodium chromosomes 1 and 3, rice chromosomes 6 and 8 and sorghum chromosomes 7 and 10.
Based on genetic positions of the first batch of synteny-derived SNP markers we could more precisely identify the syntenic region in Ae. tauschii. In subsequent experiments we focused on a region distal of the colinearity breakpoint delimited by markers AT7D6468 and AT7D6420 (Luo et al. 2013), which is colinear to a part of rice chromosome 6, Brachypodium chromosome 1 and sorghum chromosome 10.
Development of locus-specific primers
Barley ESTs and Ae. tauschii marker sequences from the selected syntenic regions were used to identify homologous sequences within wheat 7AS, 7BS and 7DS sequence assemblies. In total, 11 ESTs from the GenomeZipper and 17 marker sequences from the Ae. tauschii linkage map were selected for marker development. The respective genic sequences for the three homoeologs, if available, were aligned, annotated and used as a template for designing 7DS-specific primers based on homoeolog sequence variants (Fig. 1a). From the total of 161 primer pairs, 114 primer pairs had at least one primer designed as 7DS-specific based on known sequence information from all three homoeologs (Table 1). For 86 out of these 114 primer pairs, both forward and reverse primers were designed to be 7DS-specific. In the remaining 28 cases, only one primer could be designed as a 7DS homoeolog specific variant (HSV), either due to missing sequence information from 7AS and 7BS (25 primer pairs) or due to a lack of polymorphism between the subgenomes on one side of the locus (3 primer pairs). Out of the 114 7DS-specific primer pairs, 99 produced a single 7DS-specific PCR amplification product, a success rate of 87 %. Further 47 primer pairs were designed with incomplete knowledge of homoeolog sequence. In these cases, 39 (83 %) produced a 7DS-specific PCR amplification product (Table 1). The relatively high success rate in cases of missing homoeolog sequence information reflects the fact that most of the primers were designed in gene-flanking regions which show relatively low level of conservation between A, B and D subgenomes.
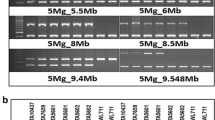
Development of genome-specific markers. a 7DS-specific primers were designed based on HSVs between 7AS, 7BS and 7DS chromosome arms (HSVs are underlined). b PCR was optimized on Phi29-amplified DNA of isolated chromosome arms 7AS, 7BS and 7DS, respectively, aiming to obtain 7DS-specific product only. The increase in annealing temperature (indicated above the gel images) was essential for most loci. c PCR was run on CI2401 and cv. ‘Glupro’ at the optimal annealing temperature. Resulting PCR products were sequenced and aligned to reveal polymorphisms between the parents of the mapping population. The figure shows the procedure for locus STS94
In total, out of 161 primer pairs, 138 (listed in Suppl. File 2) provided 7DS-specific product using DNA from flow-sorted 7AS, 7BS and 7DS arms as a template, but only 15 primer pairs provided the specific product when using the annealing temperature specified by the Primer3 software. In the other 123 cases, PCR conditions had to be optimized by increasing the annealing temperature (Fig. 1b) and/or decreasing number of PCR cycles (data not shown). Eight out of 161 primer pairs amplified the locus from all three subgenomes, while eight primer pairs produced more than one PCR product for each of the 7S arms. In seven cases, no product was obtained. Overall, we obtained a single 7DS-specific PCR product with 86 % (138) of all designed primers. Out of 138 primer pairs successfully tested on the flow-sorted chromosome arms originating from ‘Chinese Spring’, only one failed to provide a product in the parents of the mapping population (CI2401, ‘Glupro’).
Analysis of polymorphisms
All 7DS-specific PCR products from both parental DNAs (CI2401, ‘Glupro’) were sequenced (Suppl. File 3). The sequencing confirmed locus-specificity of the PCR products. A total of 73,070 bp were sequenced for both CI2401 and ‘Glupro’. Comparison of parental sequences revealed 11 single-nucleotide polymorphisms between the parents, of which two were present in one PCR product. One of these two SNPs, representing an indel in a [T]n microsatellite, was excluded from the mapping. The remaining ten SNP markers were designated Xowm701–Xowm710 (Table 2). The last marker (Xowm711) was a 43-bp indel between CI2401 and ‘Glupro’. Based on the sequencing data, we calculated the frequency of polymorphisms between the CI2401 line and ‘Glupro’, as one polymorphism (SNP/indel) per 6,089 bp. Out of the 11 SNPs, four were found in introns and six in gene-flanking regions, just as the long indel. Only one SNP was identified in an exon, reflecting the fact that exon regions show a higher level of sequence conservation compared to introns and intergenic regions.
Mapping new markers
Mapping the Xowm701–Xowm711 markers resulted in six clusters comprising one to three co-segregating markers each (Fig. 2). All markers mapped in the Dn2401 gene region. Genotypes of all markers across the mapping population are stated in Suppl. File 4, segregation ratios in Suppl. File 5. The first two markers, Xowm701 and Xowm702, mapped outside the region delimited by SSR markers Xcfd68 and Xgwm473, 0.28 cM proximal to Xgwm473. In case of the Xowm701, this fitted well with the position of the barley ortholog in the GenomeZipper, whereas a more distal position was expected for marker Xowm702. The remaining nine markers (Xowm703–Xowm711) were located within the region delimited by Xcfd68 and Xgwm473. The order of Ae. tauschii-derived markers Xowm704–Xowm711 in wheat fully corresponded with the order of their orthologs in Ae. tauschii. The data indicate that both the barley GenomeZipper and the Ae. tauschii genetic map are valuable resources for targeted marker development in the region of interest in wheat. Mapping the newly developed markers resulted in the reduction of the Dn2401 interval from 2.47 cM, delimited by Xcfd68 and Xgwm473, to 0.83 cM, delimited by Xowm711 on the distal side of Dn2401, and co-segregating markers Xowm704, Xowm705 and Xowm706 on the proximal side. This also aided a more precise determination of syntenic regions in wheat relatives, positioning the Dn2401 interval distal of the colinearity breakpoint in rice chromosome 6, Brachypodium chromosome 1 and sorghum chromosome 10. The distance from the breakpoint is around 12.5 Mb in Ae. tauschii. In the barley GenomeZipper, the Dn2401 interval comprises 31 predicted genes.
Linkage and physical map of the Dn2401 region and its synteny with Ae. tauschii and barley orthologous regions. Linkage map of the central part of wheat chromosome arm 7DS comprising the Dn2401 locus and newly developed markers (Xowm) in the context of previously mapped SSR markers. Colinearity with orthologous loci in Ae. tauschii linkage map (7DtS) and barley GenomeZipper (7HS) is indicated by solid lines. Dashed lines show colinearity for genes adjacent to flanking SSR markers. The numbers on the left indicate positions of the loci in the Ae. tauschii map, the GenomeZipper, and the wheat 7DS linkage map in cM units. Asterisks point out virtual positioning of the respective barley ESTs on the barley genetic map (HarvEST, version 1.77). Minimum tilling path of BAC contig 783 assigned to the Dn2401 region with approximate positions of flanking markers is shown on the right
Physical map of the Dn2401 region
7DS-specific BAC library of a cv. ‘Chinese Spring’ was screened with all genetically mapped markers aiming to estimate local physical/genetic distance ratio and, potentially, identify a BAC contig spanning the Dn2401 region. Most of the markers located in separate contigs, which prevents the exact calculation of the ratio, but an estimate based on the size of the BAC contigs and the position of the individual markers suggested that on average, the genetic distance of 1 cM exceeded 3 Mbp in the studied region. In contrast to this observation, markers Xowm705 and Xowm711, flanking the gene locus in the genetic distance of 0.83 cM, were found to allocate both to the same BAC contig (ctg783, https://urgi.versailles.inra.fr/gb2/gbrowse/wheat_phys_7DS_v1/) with an estimated distance of ~300 kb from each other represented by four BAC clones of minimum tilling path (Fig. 2). Thus the physical/genetic distance between these markers equaled ~360 kb per 1 cM. The high recombination rate around the gene made possible positioning of the trait in ~300 kb interval with a population of only 184 F2 individuals.
Discussion
Gene-associated markers are important for plant molecular breeding. Their development in hexaploid wheat is hindered by the presence of gene homoeologs and paralogs. Thus, designing genome- and locus-specific primers is an essential step in the development of molecular markers. In our study, we applied a procedure exploiting shotgun-sequence information from isolated chromosome arms, which allowed developing locus-specific primer pairs with an efficiency of 86 %.
In previous studies employing synteny-based approaches for targeted marker development, markers were developed based on genome sequence and gene order in rice or Brachypodium, in combination with sequence information from wheat ESTs (Bossolini et al. 2006; Schnurbusch et al. 2007; Qin et al. 2011; Quraishi et al. 2009). As only conserved transcribed sequences were available, this approach suffered from a lack of specificity in the initial steps of locus-specific marker development. Additional steps had to be incorporated to increase marker specificity, including cloning and sequencing of primary PCR products to identify homoeolog specific sequences and to design locus-specific primers based on homoeolog sequence variants (HSVs). Such approach was used by Schnurbusch et al. (2007) who developed markers linked to the Bo1 gene located on the wheat 7BL chromosome arm. In their work, design of a 7B-specific primer harboring at least one HSV was possible in 24 out of 41 tested primer pairs and a 7B-specific PCR product was obtained for 16 primer pairs. Overall, 7B-specific products could be obtained only for 39 % of the investigated loci.
The success rate achieved in our study was also significantly higher as compared to other studies on development of locus- or genome- specific markers in wheat. Terracciano et al. (2013) derived genome-specific markers to map Lr14 gene on the 7BL chromosome arm of tetraploid durum wheat. Lacking sequence information from 7A and 7B chromosomes, the authors began by sequencing PCR products obtained from exon-derived primers using flow-sorted chromosome arms as a template. The chromosome-specific sequences obtained were used to identify HSVs for the design of genome-specific primers. This approach led to obtaining 7B-specific products for only 30 % of primer pairs, perhaps due to an insufficient level of polymorphism used to secure the primer specificity. The same strategy, including sequencing of PCR products from flow-sorted chromosome arms 1AS, 1AL, 1BS, 1BL, 1DS and 1DL was applied by Michalak de Jimenez et al. (2013) to develop chromosome-specific markers for radiation hybrid mapping on chromosome 1D. They succeeded in developing 1D-specific markers for 39 % of investigated loci harboring at least one HSV in the primer sequence.
These results suggest that the availability of a quality sequence assembly for the desired chromosome arm and its homoeologs can significantly increase the efficiency of marker development. The knowledge of genomic context of genes used for marker development enabled us to efficiently design locus-specific primers, placing them preferentially into introns and gene-flanking regions, which exhibit lower levels of conservation. This markedly reduced non-specific amplification from homoeologous and paralogous loci as well as conserved domains of other genes, which often occurs if the primers are designed on exon sequences (Šimková, unpublished). Comparison of the studies mentioned above indicates that a careful optimization of PCR conditions is essential to obtain locus-specific sequence (also reported by Schnurbusch et al. 2007). Although the initial phase, consisting of precise designing 7DS-specific primers and adjusting PCR conditions, is relatively laborious, it is highly compensated by the reduction of labor and costs that would be expended on sequencing and mapping of non-specific products.
The main challenge affecting the efficiency of our approach was a low level of polymorphism in our mapping population. In the present study, a total of 73,070 bp were sequenced from both ‘Glupro’ and CI2401, yielding 12 polymorphisms (11 SNPs and one indel), which implies a polymorphism frequency of 1 per 6,089 bp. This is ten times lower than the frequency observed by Schnurbusch et al. (2007) who found 1 SNP/613 bp when comparing close to 20 kb of sequence information from two cultivars of hexaploid wheat in the Bo1 region. The low level of polymorphism observed in our region is probably due to the location of the Dn2401 region on the D subgenome of wheat, known to have lower level of polymorphism as compared to the A and B subgenomes (Chao et al. 2009). Low level of polymorphism in the D subgenome was also observed by Berkman et al. (2013) who analyzed the frequency of intervarietal SNPs on 7A, 7B and 7D chromosomes.
The positions of the newly developed markers on the genetic map were compared with those of barley genic sequences in the barley GenomeZipper and markers in the Ae. tauschii linkage map (Fig. 2). Comparison between the mapped Xowm markers developed in the present work and virtually ordered barley ESTs revealed a small discrepancy in case of marker Xowm702, which mapped in our population more proximally as compared to the position predicted by the barley GenomeZipper. As the GenomeZipper represents a hypothetic order of genes, this discrepancy could be due to incorrect positioning of the gene on the virtual barley map. Alternatively, since the GenomeZipper has been constructed based on barley linkage map, the discrepancy may indicate disruption of micro-colinearity between barley and wheat due to a translocation or inversion. Disruptions of colinearity between wheat and barley were reported before (Mayer et al. 2011). When comparing our high-density map of the Dn2401 region with that of Ae. tauschii, no mismatch was detected. The region showed complete colinearity, which is in line with the fact that Ae. tauschii is the ancestor of the wheat D subgenome and became a part of the bread wheat genome only 10,000 years ago. Thus, the linkage map of Ae. tauschii proved to be the most reliable source of markers in our study. On the other hand, the GenomeZipper provides a denser map of genes, including links to annotations in the related genomes (http://mips.helmholtz-muenchen.de/plant/barley/gz/index.jsp), which gives an opportunity to search for candidate genes within the interval of interest. GenomeZippers were recently constructed based on wheat chromosome shotgun sequences and genetic maps (IWGSC 2014). They combine a higher information value of wheat-derived genetic maps with the high density of ordered genes introduced by GenomeZipper, resulting in the most dense virtual gene map for wheat. We expect the wheat chromosome Zippers will serve as an efficient template for wheat marker development.
Integration of genetic and physical maps for the Dn2401 region permitted the analysis of recombination rates around the gene, and the results suggested at least eightfold increase in recombination rate in the proximity of the gene compared to the neighboring regions. Since the flanking markers Xowm705 and Xowm711 have been derived from Ae. tauschii markers AT7D6434 and AT7D6430, respectively, which have been anchored to Ae. tauschii physical map (Luo et al. 2013), we could precisely examine recombination rate in the orthologous region of Ae. tauschii genome. BAC contigs comprising orthologs of the wheat flanking markers showed about threefold increase in recombination rate compared to the average of the Ae. tauschii genome (0.32 cM/Mb, corresponding to 3.13 Mb/cM) and more than tenfold increase when comparing to neighboring BAC contigs. Highly recombining regions of Ae. tauschii were shown to be associated with disease resistance genes and signal transduction genes (GO class “receptor activity”) (Luo et al. 2013), which play important roles in the innate immunity in plants (Vakhrusheva and Nedospasov 2011). In a study of Azhaguvel et al. (2012), another aphid resistance gene—Gb3, which confers resistance to greenbug and locates in the long arm of the 7D (7DL), was also observed to be situated in a highly recombining region (558 kb/cM and 360 kb/cM from the left and the right site, respectively) in Ae. tauschii 7DL. Alternative explanation for the decreased physical/genetic distance ratio around Dn2401 is that the region in cv. ‘Chinese Spring’ has been affected by a deletion, which reduces the physical distance. Comparative analysis methods such as genome mapping in nanochannel arrays (Lam et al. 2012) could shed light on the structure of the region in the concerned cultivars.
Conclusions
We have developed a powerful strategy for targeted marker development in hexaploid wheat, which relies on synteny with close relatives of wheat and on shotgun wheat chromosome sequence information from a complete homoeologous group. The approach was efficient and cost-effective, despite the low level of polymorphism between parents of the available mapping population. It allowed us to increase marker density around the RWA resistance gene Dn2401 by adding 11 new gene-associated markers and reduce interval delimited by flanking markers to 0.83 cM. Screening a 7DS-specific BAC library with the flanking markers identified four BAC clones that span the Dn2401 region in cv. ‘Chinese Spring’. Sequencing of these clones will provide data for development of additional markers and potentially suggest candidates for the resistance gene. With the availability of the shotgun sequence assemblies and GenomeZippers for each of the wheat chromosome arms, the strategy described here can be applied for targeted marker development in any region of the wheat genome.
Author contribution statement
HS implemented marker development, performed linkage mapping and drafted manuscript. HŠ and MV proposed the strategy for marker development and MV aided in bioinformatics analyses. NL performed the RWA scoring. PB, JB and DE sequenced and assembled the wheat group seven chromosomes. ML and JD fingerprinted the BAC library and ML performed integration of wheat and Ae. tauschii physical maps. MK prepared flow-sorted wheat chromosome arms. ZT integrated linkage and physical maps in the region. NS delimited the syntenic region in barley GenomeZipper and contributed to writing the manuscript. HŠ and JD conceived and supervised the project and prepared the final version of the manuscript.
References
Azhaguvel P, Rudd JC, Ma Y, Luo MC, Weng Y (2012) Fine genetic mapping of greenbug aphid-resistance gene Gb3 in Aegilops tauschii. Theor Appl Genet 124:555–564
Berkman PJ, Skarshewski A, Lorenc M, Lai K, Duran C, Ling EYS, Stiller J, Smits L, Imelfort M, Manoli S, McKenzie M, Kubaláková M, Šimková H, Batley J, Fleury D, Doležel J, Edwards D (2011) Sequencing and assembly of low copy and genic regions of isolated Triticum aestivum chromosome arm 7DS. Plant Biotech J 9:768–775
Berkman PJ, Visendi P, Lee HC, Stiller J, Manoli S, Lorenc MT, Lai K, Batley J, Fleury D, Šimková H, Kubaláková M, Weining S, Doležel J, Edwards D (2013) Dispersion and domestication shaped the genome of bread wheat. Plant Biotech J 11:564–571
Bossolini E, Krattinger SG, Keller B (2006) Development of SSR markers specific for the Lr34 resistance region of wheat using sequence information from rice and Aegilops tauschii. Theor Appl Genet 113:1049–1062
Burd JD, Burton RL (1992) Characterization of plant-damage caused by the Russian wheat aphid (Homoptera, Aphididae). J Econ Entomol 85:2017–2022
Chao S, Zhang W, Akhunov E, Sherman J, Ma Y, Luo MC, Dubcovsky J (2009) Analysis of gene-derived SNP marker polymorphism in US wheat (Triticum aestivum L.) cultivars. Mol Breed 23:23–33
Close TJ, Wanamaker S, Roose ML, Lyon M (2007) HarvEST: an EST database and viewing software. Methods Mol Biol 406:161–178
Close TJ, Bhat PR, Lonardi S, Wu Y, Rostoks N, Ramsay L, Druka A, Stein N, Svensson JT, Wanamaker S, Bozdag S, Roose ML, Moscou MJ, Chao S, Varshney RK, Szucs P, Sato K, Hayes PM, Matthews DE, Kleinhofs A, Muehlbauer GJ, DeYoung J, Marshall DF, Madishetty K, Fenton RD, Condamine P, Graner A, Waugh R (2009) Development and implementation of high-throughput SNP genotyping in barely. BMC Genom 10:582
Collins MB, Haley SD, Peairs FB, Rudolph JB (2005) Biotype 2 Russian Wheat Aphid resistance among wheat germplasm accessions. Crop Sci 45:1877–1880
Doležel J, Vrána J, Šafář J, Bartoš J, Kubaláková M, Šimková H (2012) Chromosomes in the flow to simplify genome analysis. Funct Integr Genomics 12:397–416
Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Heled J, Kearse M, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A (2010) Geneious v5.3. http://www.geneious.com
FAOSTAT (2013) FAOSTAT, Food and Agriculture Organization of the United Nations. http://faostat.fao.org/
Fazel-Najafabadi M, Peng J, Peairs FB, Simkova H, Kilian A, Lapitan NLV (2014) Genetic mapping of resistance to Diuraphis noxia (Kurdjumov) Biotype 2 in wheat (Triticum aestivum L.) accession CI2401. Euphytica. doi:10.1007/s10681-014-1284-0
Girma M, Wilde GE, Harvey TL (1993) Russian wheat aphid (Homoptera: Aphididae) affects yield and quality of wheat. J Econ Entomol 86:594–601
Keller B, Feuillet C, Yahiaoui N (2005) Map-based isolation of disease resistance genes from bread wheat: cloning in a supersize geonome. Genet Res 85:93–100
Kosambi D (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Kubaláková M, Vrána J, Číhalíková J, Šimková H, Doležel J (2002) Flow karyotyping and chromosome sorting in bread wheat (Triticum aestivum L.). Theor Appl Genet 104:1362–1372
Kubaláková M, Valárik M, Bartoš J, Vrána J, Číhalíková J, Molnár-Láng M, Doležel J (2003) Analysis and sorting of rye (Secale cereale L.) chromosomes using flow cytometry. Genome 46:893–905
Lam ET, Hastie A, Lin C, Ehrlich D, Das SK, Austin MD, Deshpande P, Cao H, Nagarajan N, Xiao M, Kwok PY (2012) Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat Biotechnol 30:771–776
Lapitan NLV, Peng JH, Sharma V (2007) A high-density map and PCR markers for Russian wheat aphid resistance gene Dn7 on chromosome 1RS/1BL. Crop Sci 47:809–818
Liu XM, Smith CM, Gill BS, Tolmay V (2001) Microsatellite markers linked to six Russian wheat aphid resistance genes in wheat. Theor Appl Genet 102:504–510
Liu XM, Smith CM, Gill BS (2002) Identification of microsatellite markers linked to Russian wheat aphid resistance genes Dn4 and Dn6. Theor Appl Genet 104:1042–1048
Luo MC, Gu YQ, You FM, Deal KR, Ma Y, Hu Y, Huo N, Wang Y, Wang J, Chen S, Jorgensen CM, Zhang Y, McGuire PE, Pasternak S, Stein JC, Ware D, Kramer M, McCombie WR, Kianian SF, Martis MM, Mayer KF, Sehgal SK, Li W, Gill BS, Bevan MW, Šimková H, Doležel J, Weining S, Lazo GR, Anderson OD, Dvorak J (2013) A 4-gigabase physical map unlocks the structure and evolution of the complex genome of Aegilops tauschii, the wheat D-genome progenitor. Proc Natl Acad Sci USA 110:7940–7945
Lysák MA, Číhalíková J, Kubaláková M, Šimková H, Künzel G, Doležel J (1999) Flow karyotyping and sorting of mitotic chromosomes of barley (Hordeum vulgare L.). Chrom Res 7:431–444
Marais GF, Wessels WG, Horn M (1998) Association of a stem rust resistance gene (Sr45) and two Russian wheat aphid resistance genes (Dn5 and Dn7) with mapped structural loci in common wheat. S Afr J Plant Soil 15:67–71
Martin TJ, Sears RG, Seifers DL, Harvey TL, Witt MD, Schlegel AJ, McCluskey PJ, Hatchett JH (2001) Registration of Trego wheat. Crop Sci 41:929–930
Martis MM, Zhou R, Haseneyer G, Schmutzer T, Vrána J, Kubaláková M, König S, Kugler KG, Scholz U, Hackauf B, Korzun V, Schön CC, Doležel J, Bauer E, Mayer KFX, Stein N (2013) Reticulate evolution of the rye genome. Plant Cell 25:3685–3698
Mayer KFX, Taudien S, Martis M, Šimková H, Suchánková P, Gundlach H, Wicker T, Petzold A, Felder M, Steuernagel B, Scholz U, Graner A, Platzer M, Doležel J, Stein N (2009) Gene content and virtual gene order of barley chromosome 1H. Plant Physiol 151:496–505
Mayer KFX, Martis M, Hedley PE, Šimková H, Liu H, Morris JA, Steuernagel B, Taudien S, Roessner S, Gundlach H, Kubaláková M, Suchánková P, Murat F, Felder M, Nussbaumer T, Graner A, Salse J, Endo T, Sakai H, Tanaka T, Itoh T, Sato K, Platzer M, Matsumoto T, Scholz U, Doležel J, Waugh R, Stein N (2011) Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell 23:1249–1263
Michalak de Jimenez MK, Bassi FM, Ghavami F, Simons K, Dizon R, Seetan RI, Alnemer LM, Denton AM, Doğramaci M, Šimková H, Doležel J, Seth K, Luo MC, Dvorak J, Gu YQ, Kianian SF (2013) A radiation hybrid map of chromosome 1D reveals synteny conservation at a wheat speciation locus. Funct Integr Genomics 13:19–32
Molnár I, Kubaláková M, Šimková H, Cseh A, Molnár-Láng M, Doležel J (2011) Chromosome isolation by flow sorting in Aegilops umbellulata and Ae. comosa and their allotetraploid hybrids Ae. biuncialis and Ae. geniculata. PLoS One 6:e27708
Morrison WP, Peairs FB (1998) Response model concept and economic impact. In: Thomas Say Publications in Entomology, Entomological Society of America, Lanham, MD
Nkongolo KK, Quick JS, Limin AE, Fowler DB (1991) Sources and inheritance of resistance to Russian wheat aphid in Triticum species amphiploids and Triticum tauschii. Can J Plant Sci 71:703–708
Peng JH, Bai Y, Haley SD, Lapitan NLV (2009) Microsatellite-based molecular diversity of bread wheat germplasm and association mapping of wheat resistance to the Russian wheat aphid. Genetica 135:95–122
Qin B, Cao A, Wang H, Chen T, You FM, Liu Y, Ji J, Liu D, Chen P, Wang XE (2011) Colinearity-based marker mining for the fine mapping of Pm6, a powdery mildew resistance gene in wheat. Theor Appl Genet 123:207–218
Quraishi UM, Abrouk M, Bolot S, Pont C, Throude M, Guilhot N, Confolent C, Bortolini F, Praud S, Murigneux A, Charmet G, Salse J (2009) Genomics in cereals: from genome-wide conserved orthologous set (COS) sequences to candidate genes for trait dissection. Funct Integr Genomics 9:473–484
Qureshi JA, Michaud JP, Martin TJ (2006) Resistance to biotype 2 Russian wheat aphid (Homoptera: Aphididae) in two wheat lines. J Econ Entomol 99:544–550
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, pp 365–386
Šafář J, Šimková H, Kubaláková M, Číhalíková J, Suchánková P, Bartoš J, Doležel J (2010) Development of chromosome-specific BAC resources for genomics of bread wheat. Cytogenet Genome Res 129:211–223
Schnurbusch T, Collins NC, Eastwood RF, Sutton T, Jefferies SP, Langridge P (2007) Fine mapping and targeted SNP survey using rice–wheat gene colinearity in the region of the Bo1 boron toxicity tolerance locus of bread wheat. Theor Appl Genet 115:451–461
Šimková H, Svensson JT, Condamine P, Hřibová E, Suchánková P, Bhat PR, Bartoš J, Šafář J, Close TJ, Doležel J (2008) Coupling amplified DNA from flow-sorted chromosomes to high-density SNP mapping in barley. BMC Genom 9:294
Šimková H, Šafář J, Kubaláková M, Suchánková P, Číhalíková J, Robert-Quatre H, Azhaguvel P, Weng Y, Peng J, Lapitan NL, Ma Y, You FM, Luo MC, Bartoš J, Doležel J (2011) BAC libraries from wheat chromosome 7D: efficient tool for positional cloning of aphid resistance genes. J Biomed Biotechnol 2011:302543
Sorrells ME, La Rota M, Bermudez-Kandianis CE, Greene RA, Kantety R, Munkvold JD, Miftahudin, Mahmoud A, Ma X, Gustafson PJ, Qi LL, Echalier B, Gill BS, Matthews DE, Lazo GR, Chao S, Anderson OD, Edwards H, Linkiewicz AM, Dubcovsky J, Akhunov ED, Dvorak J, Zhang D, Nguyen HT, Peng J, Lapitan NL, Gonzalez-Hernandez JL, Anderson JA, Hossain K, Kalavacharla V, Kianian SF, Choi DW, Close TJ, Dilbirligi M, Gill KS, Steber C, Walker-Simmons MK, McGuire PE, Qualset CO (2003) Comparative DNA sequence analysis of wheat and rice genomes. Genome Res 13:1818–1827
Terracciano I, Maccaferri M, Bassi F, Mantovani P, Sanguineti MC, Salvi S, Šimková H, Doležel J, Massi A, Ammar K, Kolmer J, Tuberosa R (2013) Development of COS-SNP and HRM markers for haplotyping and marker-assisted selection of Lr14 in durum wheat (Triticum durum Desf.). Theor Appl Genet 26:1077–1101
The International Wheat Genome Sequencing Consortium (IWGSC) (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788
Vakhrusheva OA, Nedospasov SA (2011) System of innate immunity in plants. Mol Biol 45:16–23
Valdez VA, Byrne PF, Lapitan NLV, Peairs FB, Bernardo A, Bai G, Haley SD (2012) Inheritance and genetic mapping of Russian wheat aphid resistance in iranian wheat landrace accession PI 626580. Crop Sci 52:676–682
Van Ooijen JW, Voorrips RE (2001) JoinMap 3.0, software for the calculation of genetic linkage maps. Plant J 3:739–744
Acknowledgments
We thank Prof. B.S. Gill and Prof. Adam Lukaszewski for providing seeds of the wheat ditelosomic lines. We are grateful to Hong Wang and Jeff Rudolph for RWA screening of the mapping population and Jarmila Číhalíková, Romana Šperková, Zdeňka Dubská and Jana Dostálová for the assistance with chromosome sorting and BAC library screening, and Dr. Frank Peairs for the use of the CSU Insectary and for providing aphids. We also thank Andreas Petzold and Stefan Taudien for 454 sequencing and assembling BAC clones. This work was supported by the Czech Science Foundation (Award No. P501/12/2554), Ministry of Education, Youth and Sports of the Czech Republic (National Program of Sustainability I, Grant award LO1204), and Australian Research Council (Projects LP0882095, LP0883462 and DP0985953). Partial support for RWA screening was provided by USDA Cooperative Agreement no. 2010-34205-21350.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H.-Q. Ling.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Staňková, H., Valárik, M., Lapitan, N.L.V. et al. Chromosomal genomics facilitates fine mapping of a Russian wheat aphid resistance gene. Theor Appl Genet 128, 1373–1383 (2015). https://doi.org/10.1007/s00122-015-2512-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-015-2512-2