Abstract
The greenbug, Schizaphis graminum (Rondani), is an important aphid pest of small grain crops especially wheat (Triticum aestivum L., 2n = 6x = 42, genomes AABBDD) in many parts of the world. The greenbug-resistance gene Gb3 originated from Aegilops tauschii Coss. (2n = 2x = 14, genome DtDt) has shown consistent and durable resistance against prevailing greenbug biotypes in wheat fields. We previously mapped Gb3 in a recombination-rich, telomeric bin of wheat chromosome arm 7DL. In this study, high-resolution genetic mapping was carried out using an F2:3 segregating population derived from two Ae. tauschii accessions, the resistant PI 268210 (original donor of Gb3 in the hexaploid wheat germplasm line ‘Largo’) and susceptible AL8/78. Molecular markers were developed by exploring bin-mapped wheat RFLPs, SSRs, ESTs and the Ae. tauschii physical map (BAC contigs). Wheat EST and Ae. tauschii BAC end sequences located in the deletion bin 7DL3-0.82–1.00 were used to design STS (sequence tagged site) or CAPS (Cleaved Amplified Polymorphic Sequence) markers. Forty-five PCR-based markers were developed and mapped to the chromosomal region spanning the Gb3 locus. The greenbug-resistance gene Gb3 now was delimited in an interval of 1.1 cM by two molecular markers (HI067J6-R and HI009B3-R). This localized high-resolution genetic map with markers closely linked to Gb3 lays a solid foundation for map based cloning of Gb3 and marker-assisted selection of this gene in wheat breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The greenbug (Schizaphis graminum Rondani), is an economically important aphid pest of small grain crops. This pest is especially notorious in the Southern high plains of the US with estimated annual yield loss over $100 million in wheat alone (Webster and Kenkel 1999). A number of greenbug-resistance genes have been identified from different sources including Gb1 from durum wheat (Triticum durum), Gb2 and Gb6 from rye (Secale cereale L.), and Gb5 from Aegilops speltoides. A series of resistance genes (Gbx1, Gba, Gbb, Gbc, Gbd, Gbz, Gb3 and Gbx2) from Ae. tauschii has been identified and mapped to the distal region of wheat chromosome arm 7DL (Zhu et al. 2004, 2005; Weng et al. 2005). Germplasm incorporating some of these Ae. tauschii-derived greenbug-resistance genes have been developed such as Largo with Gb3 (Hollenhorst and Joppa 1983), CI 17959 with Gb4 (Martin et al. 1982), KSU97-85-3 with Gbz (Zhu et al. 2004), KS89WGRC4 with Gbx (Weng and Lazar 2002) and Gb7 in W7984 (Weng et al. 2005). Among these resistance genes, Gb3 in wheat cultivars TAM 110 (Lazar et al. 1997) and TAM 112 is currently the only greenbug-resistance gene widely used in wheat production to confer resistance against prevailing greenbug biotypes (E, I and K).
The wheat-greenbug provides a useful system to study the plant–aphid interactions in the grass genome. However, although aphids are important insect pests, no host-resistance gene has been cloned from any cereal crops. Due to the very large and complex genome of common wheat (~16,000 Mb), fine mapping and map-based gene cloning remains a challenge. Thus, some alternate strategies have been taken to clone genes in hexaploid wheat. Sub-genome chromosome walking using diploid progenitor of wheat provides an efficient method to establish physical contigs and also helps to saturate the target region with molecular markers in wheat. The early successful map-based gene isolation in wheat invariably used the sub-genome mapping strategy (e.g., Stein et al. 2000; Yahiaoui et al. 2003). As an allopolyploid, wheat has complete sets of cytogenetic stocks including deletion lines (Endo and Gill 1996) to reduce the complexity of genetic mapping. In addition, in the recent years, many new genomics resources have been developed in wheat and its relatives, which greatly facilitate gene cloning work. For example, more than one million wheat ESTs are publicly available (http://www.ncbi.nlm.nih.gov/). BAC libraries with 302,976 clones representing 8.5× genome coverage were constructed from the Ae. tauschii accession AL8/78 (Xu et al. 2002), from which physical contigs have been developed (http://avena.pw.usda.gov/wheatD/). Currently, there are 13,647 contigs and 4,730 singletons associated with 267,451 BAC/BIBAC clones of the AL8/78 BAC libraries, and 727 markers that were anchored in the contigs. All these genetic and genomic resources should play important and sometimes indispensable roles in genetic mapping and gene cloning studies in wheat.
Previous studies have located the greenbug-resistance gene Gb3 into the distal 18% region of 7DL (7DL-3 0.82–1.0) of wheat and anchored with microsatellite (SSR) markers (Weng et al. 2005). We developed a mapping population with 558 F2:3 families from a cross between two diploid Ae. tauschii accessions, PI 268210 and AL8/78. The objective of this study was to develop a localized high-resolution genetic map for the Gb3 region in the long arm of Ae. tauschii chromosome 7D. Our goal is to clone this aphid-resistance gene Gb3 with a sub-genome chromosome walking and fine genetic mapping strategy.
Materials and methods
Plant materials
Fine genetic mapping was conducted with 558 F2:3 lines derived from two Ae. tauschii accessions, the greenbug-resistant PI 268210 and the greenbug-susceptible line AL8/78. PI 268210 is the donor of Gb3 (Joppa and Williams 1982) and AL8/78 is the accession used for BAC library construction by the NSF wheat D genome physical mapping project (http://avena.pw.usda.gov/wheatD/). Two hexaploid wheat near-isogenic lines of Gb3 (Lazar et al. 1996), the greenbug-resistant TXGBE273 and susceptible TXGBE281, were used for conversion of two Gb3-linked AFLP markers into STS markers. To determine chromosome arm locations of molecular markers, a set of homoeologous Group-7 aneuploid stocks was used including the ditelosomic 7DS (DT7DS), six nulli-tetrasomic stocks (N7AT7B, N7AT7D, N7BT7A, N7BT7D, N7DT7A and N7DT7B), and six 7DL deletion lines of Chinese Spring (Endo and Gill 1996; Weng et al. 2005).
Evaluation of responses to biotype E greenbug infestation
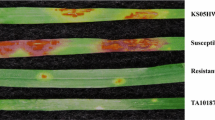
Biotype E greenbugs were used in all screening tests, which is currently the prevailing biotype in wheat fields of Southern high plains of the US. Biotype E greenbug colonies were reared in the growth chamber of an isolated room at the Texas AgriLife Research, Bushland, Texas, USA with continuous evaluation of responses on known-resistant (TAM 110) and susceptible wheat lines (TAM 105 and TAM 107). Greenbug infestation was performed on three-leaf stage seedlings of individual F2 plants and F2-derived F3 progeny following the procedure of Weng and Lazar (2002). Since most F2 plants grew vigorously with many tillers, the tillers were split from each plant to make multiple clones for both self-pollination to produce F3 and greenbug screening. At least 15 plants were screened for each F3 family. The greenbug-resistant PI 268210 and TAM 110, and susceptible AL8/78 and TAM 105 were included in all infestation experiments as controls. Greenbug infestation was evaluated starting from 15 days after infestation when susceptible control plants were almost dead. Each plant was scored qualitatively as either resistant or susceptible. For a subset of 32 F2 plants which were identified to be recombinants between flanking markers WG380 and KSUD2, their genotypes at the Gb3 locus were further verified by one additional screening of their respective F3 progeny that included at least 30 plants for each family. Deviations of observed data from theoretically expected segregation ratios were tested using Chi-squared (χ 2) tests for goodness-of-fit.
AFLP marker conversion
We previously identified two AFLP markers, XM-AGG/P-ATG and XM-GCC/P-AGG that cosegregated with the Gb3 locus in a relatively small hexaploid wheat mapping population (Weng and Lazar 2002). In the present study, we initiated our fine mapping effort by converting the two AFLP markers into STS markers. Template DNAs from two near-isogenic lines (NILs) of Gb3, TXGBE273 and TXGBE281, as well as two Ae. tauschii parental lines, PI 268210 and AL8/78 were PCR amplified using the original AFLP primer pairs (M-GCC/P-AGG and M-AGG/P-ATG). The PCR products were resolved in 6% polyacrylamide sequencing gel, and displayed with silver staining. Polymorphic, target AFLP fragments were excised from the gel and purified using Qiagen’s QIAquick Gel Extraction Kit (Valencia, CA, USA). The purified DNA was cloned into the pGEM®-T Easy vector (Promega, Madison, WI, USA) and transformed in E. coli cells (H5 alpha strain). Plasmid DNA was isolated from white colonies using the QIAprep® Spin Miniprep kit (Qiagen, Valencia, USA) and the cloned fragments were sequenced. For each plant material, at least eight clones were sequenced. The sequences were aligned to identify polymorphism.
Marker development strategy
Since relatively few markers have been mapped in wheat chromosome 7DL, we started high-resolution genetic mapping for Gb3 in Ae. tauschii with all available marker resources in the distal end of homoeologous Group-7 chromosomes of wheat. Previously mapped RFLP and EST markers placed in the distal 7DL bin FL0.82–1.00 (Weng et al. 2005) were selected from databases of GrainGenes (http://wheat.pw.usda.gov/), and KOMUGI (http://www.shigen.nig.ac.jp/wheat/komugi), as well as Hossain et al. (2004). The marker sequences, when available were downloaded from the GenBank (http://www.ncbi.nlm.nih.gov/). SSR or SCAR markers linked with disease- or insect-resistance genes in distal 7DL (Groenewald et al. 2003; Francki et al. 2004; Somers et al. 2004; Weng et al. 2005; Gupta et al. 2006; Hayden et al. 2006; Giovanini et al. 2007; Shen and OhmH 2007) were also employed for polymorphism screening between the two Ae. tauschii parental lines. A number of bin-mapped RFLPs did not have sequence information. In this case, RFLP clones were requested from respective laboratories and end sequenced (see supplemental materials for sequence information). The EST or RFLP sequences were BLASTed against the TREP-Repeat databases (http://blast.jcvi.org/). EST/RFLP sequences showing unique or low hits of similarity values less than e-25 were considered for designing primers. PCR primers were designed with Primer3 software (http://frodo.wi.mit.edu/primer3/) and synthesized commercially. The two parental lines (PI 268210 and AL8/78) were amplified and the primers showing discrete fragments were selected.
In a few cases, polymorphisms (insertion/deletion, ‘Indel’ hereinafter) were detected in the PCR products between the two parental lines, which could be used directly in the mapping population. Those monomorphic STS markers from EST, RFLP and AFLP sequences were the first subjected to deletion mapping with homoeologous Group-7 aneuploid stocks and 7DL deletion lines to assure that these markers were located in the expected 7DL distal bin. These markers were then considered for CAPS (cleaved amplified polymorphic sequence) marker development. The parental lines PI 268210 and AL8/78 were amplified and the PCR products were purified for cycle sequencing. The sequences from the parental lines were aligned using SegMan of Lasergene package (DNASTAR Inc, Madison, WI). Polymorphic restriction enzyme recognition sites were identified with restriction maps (http://arbl.cvmbs.colostate.edu/molkit/mapper/). All markers except two were genotyped by PCR amplification followed by restriction enzyme digestion and agarose gel electrophoresis. The markers BG604843 and CJ579846 were genotyped by direct sequencing of PCR products.
Molecular marker analysis
Genomic DNAs from all plant materials were extracted from young leaf tissues following Weng et al. (2000). The DNA was quantified and diluted to 20 ng/μl. Each PCR reaction contained 20 ng of template DNA, 0.3 μM of each of two primers, 5 μl of 2× master mix (Promega, Madison, WI) in a total volume of 10 μl, and the PCR was performed in an ABI 2720 thermocycler (Applied Biosystems, Carlsbad, CA) with a touchdown PCR program for all primer sets (Weng et al. 2000) which consisted of 3 min initial denaturation at 95°C, six cycles of 45 s at 94°C, 5 min at 68°C, 1 min at 72°C. The annealing temperature was reduced by 2°C per cycle, eight cycles of 45 s at 94°C, 2 min at 58°C, 1 min at 72°C, and the annealing temperature reduced by 1°C per cycle, a final 25 cycles of 45 s at 94°C, 2 min at 50°C and 1 min at 72°C. PCR products were separated in either regular or high-resolution agarose gels, depending on amplicon size, and were visualized by ethidium bromide staining. The 7DL bin locations of all markers were verified with a set of wheat chromosome Group-7 aneuploid stocks and 7DL deletion lines as described in Weng et al. (2005).
BAC library screening
BAC end sequences were also explored for developing more closely linked markers for Gb3. The Ae. tauschii accession AL8/78 BAC library used for initial screening contained 73,728 clones in four high-density filters which were obtained from Dr. Hongbin Zhang’ Lab in the Texas A and M University at College Station, TX. The AL8/78 BAC libraries deposited in the University of California at Davis were also employed in late stages of library screening. Two robust markers linked with Gb3, AFLP-STS11 and BF293421 were used to initiate BAC library screening. AFLP-STS11 was converted from AFLP marker XMaggPatg (Weng and Lazar 2002), and BF293421 was a wheat EST-derived CAPS marker. The PCR products of the two markers from the parental line AL8/78 were purified and radioactively labeled with 32P dATP by NEBlot kit following manufacturer’s protocol (New England Biolab, Ipswich, MA). The labeled probe was purified with a Sephadex G-50 column and denatured at 100°C for 10 min. The filters were pre-hybridized overnight at 65°C in hybridization solution [2% SSPE, 0.5% SDS, 5× Denhart’s buffer (10% of PVP-360 + Ficoll, bovine albumin in 500 ml), 20 mg Herring sperm DNA)] in a rotary glass tube. The labeled probe was mixed with 5 mL hybridization solution and incubated at 65°C overnight. Then the filters were washed in the wash solution containing 2% SSPE and 0.5% SDS and rinsed with 1× SSC. The washed filters were exposed to X-ray film for 1–3 days. Positive clones were used to search the Wheat D Genome Physical Mapping Database to identify Ae. tauschii BAC contigs (http://avena.pw.usda.gov/wheatD/). Those positive clones that were belonged to a BAC physical contig were then selected and grown overnight in LB media with chloroamphenicol. BAC DNA was extracted with standard alkaline lysis procedure (Sambrook et al. 1989) for finger printing and PCR amplification. Five micrograms of pure BAC DNA of all positive clones in each screen was extracted using QIAGEN Large-Construct Kit (Qiagen, Valencia, CA, USA) and used for direct cycle sequencing of BAC ends. The position of BAC clones in the contig was confirmed by PCR amplification with all mapped BAC-end-sequence markers using BAC clone DNA.
Genetic mapping
Mapping of 7DL markers was conducted in three phases. First, a set of anchored SSR markers were mapped with all 558 F2 plants. Then, based on the recombination between the markers WMC94 and CFD175, a subset of 93 recombinant F2 plants was selected for genotyping all newly developed EST and RFLP markers from this study. Further, the subset of 93 F2 plants were screened with two flanking markers WG380 and KSUD2 and 32 F2 plants with recombination events between these markers were identified. In the third phase, all BAC-end-sequence-based markers were mapped with these 32 F2 recombinant plants. Linkage analysis was performed initially with MAPMAKER/EXP version 3.0 (Lincoln et al. 1993) and later with JoinMap 3.0 (Van Ooijen and Voorrips 2001). Kosambi mapping function was used to estimate the map distance (Kosambi 1944).
Results
Inheritance of greenbug resistance in Ae. tauschii
The F2 individuals derived from PI 268210 and AL8/78 and their F2:3 families were evaluated for responses to greenbug biotype E infestation. Of 532 F2 plants tested, 395 were resistant and 137 susceptible, which did not deviate significantly from the expected 3:1 resistant-to-susceptible ratio (χ 2 = 0.16, P = 0.69). Among the 431 F3 families screened, 74 were homozygous resistant, 234 were heterozygous and 123 were susceptible. A χ 2 test revealed that the 431 corresponding F2 families genotypically segregated in a ratio of 1Gb3Gb3:2Gb3gb3:1gb3gb3 (χ 2 = 14.31, P = 0.007) with less than expected homozygous-resistant plants. The results were largely consistent with earlier studies on the inheritance of the greenbug resistance in PI 268210 which is controlled by a single dominant-resistance gene (Gb3) (Joppa and Williams 1982; Lazar et al. 1996; Weng and Lazar 2002; Weng et al. 2005).
Conversion of AFLP markers into STS
Previously we identified two AFLP markers, M-GCC/P-AGG and M-AGG/P-ATG that were cosegregating with the Gb3 locus in a hexaploid-mapping population (Weng and Lazar 2002). The two AFLP fragments, 218 bp for M-GCC/P-AGG and 107 bp for M-AGG/P-ATG, were amplified with PCR, recovered from the gels and cloned. For each fragment, eight clones were randomly selected and sequenced. The eight sequences of the M-GCC/P-AGG 218 bp fragment contained three different groups of sequences. All these sequences were subjected to a homology non-redundant BLASTn search in the GenBank, but no significant homology sequence was found. New internal primers were designed from each of the three type sequences of the M-GCC/P-AGG 218 bp fragment and used to amplify genomic DNAs from PI 268210 and AL8/78. The PCR products were sequenced. A single nucleotide polymorphism (SNP) within an MseI recognition site was identified in PI 268210. Thus, the AFLP of M-GCC/P-AGG was due to a point mutation in the internal region of the AFLP fragment rather than a mutation in the restriction site or in the selective base regions. When treated with MseI restriction enzyme, the 210 bp PCR product yielded two bands in AL8/78 which were 115 and 95 bp in size, respectively; whereas in PI 268210, the fragment size remained intact (210 bp). This internally amplified STS marker was named AFLP-STS11.
All eight sequences obtained for the M-AGG/P-ATG 107 bp fragment were identical in nucleotide composition. No polymorphism was found among these sequences, or among sequences amplified from newly designed internal primers. Probably the sequence polymorphism of MseI or PstI was located outside the amplified internal region or all the sequences cloned were not the target sequence associated with the AFLP marker.
SSR and EST-derived CAPS markers for Gb3
Our high-resolution mapping effort for Gb3 started with markers previously mapped in the distal region of the long arms of wheat homoeologous Group-7 chromosomes. Polymorphism screening test was performed with 21 SSRs located within the region delimited by SSR markers WMC94 and CDF175 (Somers et al. 2004), the SSR marker, Ust2001-7DL, linked with eyespot-resistance genes pch1 (Groenewald et al. 2003; Chapman et al. 2008) and 102 RFLPs. Among the markers tested, nine SSRs (BARC111, BARC1046, CFD69, CFD175, WMC094, WMC273, WMC634 and WMC824) and five RFLP-derived STS or CAPS markers (FBB79R, WG380, KSUD2, G278 and PSR680) were polymorphic between PI 268210 and AL8/78 and were mapped (Fig. 1; Table 1).
A linkage map of Ae. tauschii 7DL distal region encompassing Gb3 locus. Numbers to the left of the chromosome are the genetic distances of adjacent markers in centiMorgans. The arrows show the orientation of chromosome 7DL. The right is a diagram of wheat 7D deletion map showing the physical location of the markers mapped on the high-resolution genetic map
There were four RAPD-converted SCAR markers reported to be linked with the leaf rust-resistance gene Lr19 in 7DL (Gupta et al. 2006). None of them showed direct length polymorphism between the two Ae. tauschii parental lines. However, comparison of the sequences of PCR products between the two parents identified a SNP in the restriction enzyme Taq I recognition site of SCS253 (Table 1) which enabled us to develop a CAPS marker that was mapped distal to SSR marker WMC94 (Fig. 1).
Meanwhile, wheat ESTs physically placed in the distal bins of wheat 7AL, 7BL and 7DL arms were targeted for the marker development. A total of 101 primer pairs were designed from those EST sequences, of which 15 showed sequence polymorphism between PI 268210 and AL8/78. Consequently, 15 EST-derived markers were developed. Of them, only CJ833370 showed a length polymorphism with a 58-bp deletion in AL8/78. For EST marker BG604843 and CJ579846, a SNP (G/A) was detected between the two parental lines, but no restriction enzyme was available to detect this SNP. Therefore genotyping was performed by direct sequencing. All the remaining 12 EST markers were converted to CAPS markers (Table 1). In addition, the wheat EST DQ462308 is part of the gene Hfr-3 that encodes a lectin-like protein during defensive responses upon Hessian fly feeding (Giovanini et al. 2007). This EST was also converted into a CAPS marker and mapped between SCS253 and FBB79R. For the EST marker BF293421 an improved version of the respective CAPS marker was developed based on the extended sequence obtained from the BAC internal sequence of HI083E15.
To summarize, by exploring previously mapped SSR, AFLP and RFLP markers, as well as deletion bin-mapped EST sequences, we were able to develop 30 PCR-based markers in the Gb3 region in Ae. tauschii. Among these markers, AFLP-STS11 was the closest; BF293421 and BJ309404 were the other two closely linked markers (Fig. 1).
Marker enrichment based on Ae. tauschii BAC contigs
Our initial mapping effort identified 19 new PCR-based markers for the Gb3 locus. At that time, the Ae. tauschii D genome physical mapping project developed many physical contigs of Ae. tauschii BAC clones (http://avena.pw.usda.gov/wheatD/). We explored this genomics resource for fine mapping of Gb3. The initial search of the database with already mapped Gb3-linked marker WG380 identified a BAC contig ctg6081 that consisted of 79 BAC clones. Four clones in this contig, HI113I16, RI109K22, HB117O20 and RI046F23, were end sequenced and two markers (RI109K22-R and RI046F23-F) were developed. However, although both BAC-end sequence based markers were mapped in 7DL, none of them was linked with WG380 in this mapping population. Probably WG380 had multiple paralogous copies in the Ae. tauschii genome and the copy we sequenced and the corresponding markers we developed were not the one represented by the Gb3-linked copy.
We initiated the chromosome walking by screening the Ae. tauschii BAC library with AFLP-STS11, which was the closest marker to Gb3 by then. This screening yielded three positive BAC clones, HB001G18, BB011A23, and HB110F7 (Fig. 2). Using the BAC clone ID, database search identified Ae. tauschii BAC contig ctg3176 which contained 76 clones spanning 538 kb. Fourteen BAC clones from this contig were end sequenced resulting six new markers including five CAPS (HD095A22-F, HB007K7-F, HI085O14-R, HI067J6-F, and HI067J6-R) and one Indel (BB001M22-R) markers. Of them, HI067J6-F and HI067J6-R were from the two ends of the same BAC clone (Table 1; Fig. 2). Genetic mapping of the six new markers covered a genetic distance of 0.9 cM with eight recombination events between CJ579846 and AFLP-STS11, with three markers cosegregating with AFLP-STS11 (Fig. 2).
On the other side of the Gb3 locus, we started BAC library screening with the distally flanking marker BF293421. Eleven positive BAC clones, HB098O03, HD003I01, HD005A18, HD005H01, HB067D18, HI021M14, HI035F15, HI071D04, HI071D13, HI071E09, RI108O08 were identified, of which HD067D18, HI021M14, HI035F15 and HI071E09 were found to be members of same contig, ctg6340. The contig ctg6340 contained 22 clones spanning a total physical length of 231 kb. All 22 BAC clones in this contig were end sequenced resulting in six new markers including HI009B3-F, HI083E15-F, HI009B3-R, HI021M14-F, HD134A2-R and HI133A22-R that covered seven recombination events between HI009B3-F and HI133A22-R which corresponded to 0.6 cM (Fig. 1). From the BAC clone HI009B3, one marker from each end was developed that covered four recombination events between Gb3 and HI021M14-F. All 112 BAC end sequences from this study have been deposited in the GenBank (accession numbers JM174961 to JM175072) for public access.
High density genetic map of Gb3
Twenty-two SSR markers previously mapped in 7DL were tested for polymorphism between PI 268210 and AL8/78, and 9 were eventually mapped. All of them were co-dominant markers. From the 45 markers that were mapped in the Gb3 region of 7DL with the F2 mapping population derived from PI 268210 and AL8/78, 30 of them were newly developed in this study (Fig. 1; Table 1). The Gb3 locus was flanked by HI067J6-R and HI009B3-F (Fig. 2) with a genetic distance of 1.1 cM. Among the 32 recombinant plants between WG380 and KSUD2, only one plant was homozygous recombinant.
Discussion
In our previous studies, using hexaploid common wheat mapping populations, we identified several SSR and AFLP markers closely linked with the greenbug-resistance locus Gb3 (Weng and Lazar 2002; Weng et al. 2005). The complexity of the hexaploid wheat genome presents a significant challenge for fine genetic mapping and ultimate map-based cloning of this aphid-resistance gene. In the present study, we focused our effort on high-resolution mapping of the Gb3 gene with a population developed from two diploid Ae. tauschii accessions. Unfortunately, on historical genetic maps (for example, Somers et al. 2004), the Gb3 region in 7DL is sparsely saturated with a limited number of molecular markers due to overall low polymorphism in the D genome chromosomes. Consequently, we explored all available resources to develop molecular markers for the Gb3 region. We were able to place 45 markers in this region, 30 of which were newly developed (Fig. 1). This is the first high-density linkage map of Ae. tauschii in this region. Marker order on this map is largely collinear with previously published wheat or Ae. tauschii 7DL maps (Somers et al. 2004; Weng et al. 2005). This map provides a solid foundation for map-based cloning of Gb3. In addition, this map should be useful in whole genome assembly of this genome region for the ongoing hexaploid wheat or Ae. tauschii genome sequencing or physical mapping projects.
In this study, we attempted to convert two AFLP markers cosegregating with Gb3 (Weng and Lazar 2002) into STS markers. In large genomes such as wheat and Ae. tauschii, AFLP conversion is in general difficult because the target fragment is often contaminated by similar-size, co-migrating non-target DNA sequences (Guo et al. 2003). To identify the target sequence from the cloned fragment, we sequenced multiple clones, and we took the extension-AFLP strategy (Xu and Ban 2004) to minimize non-target sequence contaminations. Indeed, for the 218 bp AFLP fragment (marker M-GCC/P-AGG), we sequenced eight clones, which were found to contain three different sequence reads. Extension PCR enabled to identify the target sequence and we successfully converted the 218 bp M-GCC/P-AGG fragment into a co-dominant STS marker AFLP-STS11, which proved to be the critical and the closest marker for Gb3 (0.7 cM from Gb3 at the proximal side (Fig. 1).
The conversion of the other AFLP fragment (M-AGG/P-ATG 107 bp) into STS marker was not successful due to the fact that no internal sequence polymorphism was found. To exploit sequence polymorphism that caused the AFLP, the extended sequence from both ends must be obtained either by inverse PCR (Bradeen and Simon 1998) or by utilizing the BAC sequence (Azhaguvel et al. 2006). However, further attempt of any of these above-mentioned methods is needed to convert this marker.
For fine genetic mapping in the Gb3 region, we explored all available SSR markers and exhausted the bin-mapped wheat ESTs. A low rate of success in generating markers was observed. Only 6.8% (7/102) of RFLPs and 15% of EST (14/101) were mapped on this genetic map. This prompted us to explore new ways of marker development. We made full use of the Ae. tauschii BAC library and physical contigs developed by the wheat community, which was proved to be fruitful. Based on information from three molecular marker-anchored BAC contigs (ctg6081, ctg3176 and ctg6340), 40 BAC clones were end sequenced from both ends. While most of these BAC end sequences (68%) were repetitive in nature, we were able to design new markers from these sequences. From the 80 end sequences, 81 primer pairs were designed and 14 new markers were developed. This result indicated that BAC end sequencing was a useful method for developing new molecular markers in target regions with a luxury of having options to select BAC clones in the BAC contig even though Ae. tauschii had a high percentage of repetitive DNA sequences in its genome.
In Fig. 1, with the exception of two dominant markers (PSR680 and BM134450), all other markers were co-dominant. Among the 45 markers placed on the map (Fig. 1), only 15 showed length polymorphism in the agarose gel, and 28 were CAPS markers and the remaining two markers were SNP markers. The cost of developing the CAPS marker is directly related to the cost of restriction enzymes. However, the reduced sequencing costs allowed for SNP genotyping by direct sequencing of PCR products. The genotyping of markers CJ579846 and BG604843 was conducted in 32 recombinant plants by direct sequencing of 32 PCR products from one end with relatively low cost ($48.00, $1.5 per reaction). Although there are many SNP genotyping platforms available, most of them require a very high start-up cost that is not realistic for small to medium labs with molecular breeding work. In this case, direct sequencing should be a cost-effective method of choice if the small set of recombinants is used in fine mapping studies.
The initial results of BAC screening confirmed the resolving power of the developed high-resolution map. The genetic and physical mapping in the 7DL distal region allowed us to analyze the physical to the genetic distance ratio in two physical contigs of about 768 kb physical length. With a genetic distance of 0.9 cM between the two markers HD095A22-R and HI067J6-R (Figs. 1, 2), the overall ratio of physical to genetic distance on the proximal side of Gb3 was about 558 kb/cM (502 kb/0.9 cM); whereas in the region between HI009B3-F and HI133A22-R, a relatively low ratio of 360 kb/cM (216 kb/0.6 cM) was observed. Interestingly the two markers developed from BAC end sequences of BAC clone HI009B3 (139 kb) covered four recombination events with 0.3 cM genetic length providing additional evidence of high recombination on the distal side of Gb3 locus (463 kb/cM). Erayman et al. (2004) estimated a relatively high genetic to physical ratio in this region (863 kb/cM) from FBB79 to KSUD2. Our estimate was probably more accurate because we used exact physical length and a large mapping population to calculate the genetic distance in this region.
In this study, we developed a high-resolution genetic map in Ae. tauschii for the Gb3 gene and placed 45 markers on the genetic map. Clearly, we are still some distance away for fine mapping and candidate gene identification of the Gb3 aphid-resistance gene. Since we have exhausted SSR markers and the EST resources in this region, we are taking two additional approaches to saturate the Gb3 region. First, since wheat chromosome 7 and rice chromosome 6 are largely collinear, marker enrichment could be achieved using wheat EST resources and wheat–rice colinearity (Hossain et al. 2004). A high colinearity of 7BL and rice 6L in the Bo1 region (boron tolerance) was reported (Schnurbusch et al. 2007). Comparing the common markers, the Gb3 is proximal to Bo1 and further research is necessary to determine the extent of colinearity of 7DL with rice 6L. Second, chromosome walking cannot rely solely on single Ae. tauschii BAC library AL8/78 which is susceptible to greenbug biotype E, and the Gb3 homologs may not be present in this library. Wheat sub-genome libraries have been constructed for many chromosomes or arms (Šafář et al. 2004; 2010; Janda et al. 2004; Janda et al. 2006; Šimková et al. 2008), including the 7DL-spefic BAC library (Šimková et al. 2011). The 7DL-specific BAC library is likely to be very useful in our effort of map-based cloning of the Gb3 gene. Our initial screening of this library using Gb3-linked markers has shown that this 7DL-specific BAC library was robust and less complex (Šimková et al. 2011) as compared with the Ae. tasuchii BAC libraries we used in the present study, which will be an excellent alternate resource for the positional gene isolation of Gb3.
Aphids are important insect pests of most crop plants. In several dicot crops, aphid-resistance genes or gene candidates have been cloned. For example, in tomato, the Mi-resistance gene for root knot nematode (Meloidogyne incognita) and potato aphid (Macrosiphum euphorbiae) was isolated (Milligan et al. 1998; Rossi et al. 1998). An aphid (Aphis gossypii)-resistance gene Vat in melon has been recently isolated and patented (Dogimont et al. 2009). A candidate gene for soybean aphid-resistance gene Rag1 was identified in soybean (Kim et al. 2010). All of the cloned aphid-resistance genes belong to (or predicted) the nucleotide binding leucine-rich repeats (NB-LRR) R gene family. No aphid-resistance gene has been cloned in cereal crops. The NB-LRR is a large class gene family and will be difficult to land on the right NB-LRR in a large genome such as wheat. Using this genetic map of Ae. Tauschii, we delimited Gb3 to a 1.1-cM interval, which may present a significant step toward positional cloning of this gene. In addition, the genetic marker resources, map and the recombinant plants developed in this study will be helpful to clone the gene that eventually will contribute to a better understanding of the molecular mechanism of greenbug resistance.
References
Azhaguvel P, Vidya Saraswathi D, Komatsuda T (2006) High-resolution linkage mapping for the non-brittle rachis locus btr1 in cultivated x wild barley (Hordeum vulgare). Plant Sci 170:1087–1094
Bradeen JM, Simon PW (1998) Conversion of an AFLP fragment linked to the carrot Y2 locus to a simple, codominant, PCR-based marker form. Theor Appl Genet 97:960–967
Chapman NH, Burt C, Dong H, Nicholson P (2008) The development of PCR-based markers for the selection of eyespot resistance genes Pch1 and Pch2. Theor Appl Genet 117:425–433
Dogimont C, Bendahmane A, Pitrat M, Burget-Bigeard E,Hagen L, Le Menn A, Pauquet J, Rouselle P, Caboche M, Chovelon V (2009) Gene resistant to Aphis gossypii. US Patent Application US Patent 7576264
Endo TR, Gill BS (1996) The deletion stocks of common wheat. J Hered 87:295–307
Erayman M, Sandhu D, Sidhu D, Dilbirligi M, Baenziger PS, Gill KS (2004) Demarcating the gene-rich regions of the wheat Genome. Nucleic Acids Res 32:3546–3565
Francki M, Carter M, Ryan K, Hunter A, Bellgard M, Appels R (2004) Comparative organization of wheat homoeologous group 3S and 7L using wheat-rice synteny and identification of potential markers for genes controlling xanthophyll content in wheat. Funct Integr Genomics 4:118–130
Giovanini MP, Saltzmann KD, Puthoff DP, Gonzalo M, Ohm HW, Williams CE (2007) A novel wheat gene encoding a putative chitin-binding lectin is associated with resistance against Hessian fly. Mol Plant Pathol 8:69–82
Groenewald JZ, Marais AS, Marais GF (2003) Amplified fragment length polymorphism-derived microsatellite sequence linked to the Pch1 and Ep-D1 loci in common wheat. Plant Breed 122:83–85
Guo PG, Bai GH, Shaner GE (2003) AFLP and STS tagging of a major QTL for Fusarium head blight resistance in wheat. Theor Appl Genet 106:1011–1017
Gupta SK, Charpe A, Prabhu KV, Haque QM (2006) Identification and validation of molecular markers linked to the leaf rust resistance gene Lr19 in wheat. Theor Appl Genet 113:1027–1036
Hayden MJ, Stephenson P, Logojan AM, Khatkar D, Rogers C, Elsden J, Koebner RMD, Snape JW, Sharp PJ (2006) Development and genetic mapping of sequence-tagged microsatellites (STMs) in bread wheat (Triticum aestivum L.). Theor Appl Genet 113:1271–1281
Hollenhorst MM, Joppa LR (1983) Chromosomal locations of genes for resistance to greenbug in ‘Largo’ and ‘Amigo’ wheats. Crop Sci 23:91–93
Hossain KG, Kalavacharla V, Lazo GR, Hegstad J, Wentz MJ, Kianian PM, Simons K, Gehlhar S, Rust JL, Syamala RR, Obeori K, Bhamidimarri S, Karunadharma P, Chao S, Anderson OD, Qi LL, Echalier B, Gill BS, Linkiewicz AM, Ratnasiri A, Dubcovsky J, Akhunov ED, Dvorák J, Miftahudin Ross K, Gustafson JP, Radhawa HS, Dilbirligi M, Gill KS, Peng JH, Lapitan NL, Greene RA, Bermudez-Kandianis CE, Sorrells ME, Feril O, Pathan MS, Nguyen HT, Gonzalez-Hernandez JL, Conley EJ, Anderson JA, Choi DW, Fenton D, Close TJ, McGuire PE, Qualset CO, Kianian SF (2004) A chromosome bin map of 2148 expressed sequence tag loci of wheat homoeologous group 7. Genetics 168:687–699
Janda J, Bartoš J, Šafář J, Kubaláková M, Valárik M, Číhalíková J, Šimková H, Caboche M, Sourdille P, Bernard M, Chalhoub B, Doležel J (2004) Construction of a subgenomic BAC library specific for chromosomes 1D, 4D and 6D of hexaploid wheat. Theor Appl Genet 109:1337–1345
Janda J, Šafář J, Kubaláková M, Bartoš J, Kovářová P, Suchánková P, Pateyron S, Číhalíková J, Sourdille P, Šimková H, Fairaivre-Rampant P, Hřibová E, Bernard M, Lukaszewski A, Doležel J, Chalhoub B (2006) Advanced resources for plant genomics: BAC library specific for the short arm of wheat chromosome 1B. Plant J 47:977–986
Joppa LR, Williams ND (1982) Registration of Largo, a greenbug resistant hexaploid wheat. Crop Sci 22:901–902
Kim K-S, Bellendir S, Hudson KA, Hill CB, Hartman GL, Hyten DL, Hudson ME, Diers BW (2010) Fine mapping the soybean aphid resistance gene Rag1 in soybean. Theor Appl Genet 120:1063–1071
Kosambi D (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Lazar MD, Worrall WD, Porter KB, Tuleen NA (1996) Registration of eight closely related wheat germplasm lines differing in biotype E greenbug resistance. Crop Sci 36:1419
Lazar MD, Worrall WD, Peterson GL, Porter KB, Rooney LW, Tuleen NA, Marshall DS, McDaniel ME, Nelson LR (1997) Registration of TAM 110. Crop Sci 37:1978–1979
Lincoln SE, Daly MJ, Lander ES (1993) Constructing genetic linkage maps with MAPMAKER/EXP 3.0. Whitehead Institute for Biomedical Research Technical Report, Cambridge, 1993
Martin TJ, Harvey TL, Hatchett JH (1982) Registration of greenbug and Hessian fly resistant wheat germplasm. Crop Sci 22:1089
Milligan SB, Bodeau J, Yaghoobi J, Kaloshian I, Zabel P, Williamson VM (1998) The root-knot nematode gene Mi from tomato is a member of the leucine zipper-nucleotide binding leucine-rich repeat family of plant genes. Plant Cell Rep 10:1037–1319
Rossi M, Goggin FL, Milligan SB, Kaloshian I, Ullman DE, Williamson VM (1998) The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc Natl Acad Sci USA 95:9750–9754
Šafář J, Bartoš J, Janda J, Bellec A, Kubaláková M, Valárik M, Pateyron S, Weiserová J, Tušková R, Číhalíková J, Vrána J, Šimková H, Faivre-Rampant P, Sourdille P, Caboche M, Bernard M, Doležel J, Chalhoub B (2004) Dissecting large and complex genomes: flow sorting and BAC cloning of individual chromosomes from bread wheat. Plant J 39:960–968
Šafář J, Šimková H, Kubaláková M, Číhaliková J, Suchánková P, Bartoš J, Doležel J (2010) Development of chromosome-specific BAC resources for genomics of bread wheat. Cytogenet Genome Res 129:211–223
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Schnurbusch T, Collins NC, Eastwood RF, Sutton T, Jefferies SP, Langridge P (2007) Fine mapping and targeted SNP survey using rice-wheat gene colinearity in the region of the Bo1 boron toxicity tolerance locus of bread wheat. Theor Appl Genet 115:451–461
Shen X, Ohm H (2007) Molecular mapping of Thinopyrum-derived Fusarium head blight resistance in common wheat. Mol Breed 20:131–140
Šimková H, Šafář J, Suchánková P, Kovarova P, Bartoš J, Kubaláková M, Janda J, Cihalikova J, Mago R, Lelley T, Doležel J (2008) A novel resource for genomics of Triticeae: BAC library specific for the short arm of rye (Secale cereale L.). BMC Genomics 9:237
Šimková H, Šafář J, Kubaláková M, Suchánková P, Číhalíková J, Robert-Quatre H, Azhaguvel P, Weng Y, Peng J, Lapitan NLV, Ma Y, You FM, Luo M-C, Bartoš J, Doležel J (2011) BAC libraries from wheat chromosome 7D: efficient tool for positional cloning of aphid resistance genes. J Biomed Biotechnol Volume 2011 (2011), Article ID 302543
Somers DJ, Isaac P, Edwards K (2004) A high density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114
Stein N, Feuillet C, Wicker T, Schlagenhauf E, Keller B (2000) Subgenome chromosome walking in wheat: a 450-kb physical contig in Triticum monococcum L. spans the Lr10 resistance locus in hexaploid wheat (Triticum aestivum L.). Proc Natl Acad Sci USA 97:13436–13441
Van Ooijen JW, Voorrips RE (2001) JoinMap® version 3.0: software for the calculation of genetic linkage maps. Plant Research International, Wageningen
Webster JA, Kenkel P (1999) Benefits of managing small-grain pests with plant resistance. In: Wiseman BR, Webster JA (eds) Economic, environmental, and social benefits of resistance in field crops. Entomol Soc Am, Lanham, pp 87–114
Weng Y, Lazar MD (2002) Amplified fragment length polymorphism- and simple sequence repeat-based molecular tagging and mapping of greenbug resistance gene Gb3 in wheat. Plant Breed 121:218–223
Weng Y, Tuleen NA, Hart GE (2000) Extended physical maps and a consensus physical map of the homoeologous group-6 chromosomes of wheat. Theor Appl Genet 100:519–527
Weng Y, Li W, Devkota RN, Rudd JC (2005) Microsatellite markers associated with two Aegilops tauschii-derived greenbug resistance loci in wheat. Theor Appl Genet 110:462–469
Xu DH, Ban T (2004) Conversion of AFLP markers associated with FHB resistance in wheat into STS markers with an extension-AFLP method. Genome 47:660–665
Xu Z, Deal KR, Li W, Covaleda L, Chang Y-L, Dvorak J, Luo MC, Gill BS, Anderson OD, Zhang HB (2002) Construction and characterization of five large-insert BAC and BIBAC libraries of Aegilops tauschii, the diploid donor of the wheat D genome. In: 10th international plant and animal genome conference, San Diego
Yahiaoui N, Srichumpa P, Dudler R, Keller B (2003) Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J 37:528–538
Zhu LC, Smith CM, Fritz A, Boyko EV, Flinn MB (2004) Genetic analysis and molecular mapping of a wheat gene conferring tolerance to the greenbug (Schizaphis graminum Rondani). Theor Appl Genet 109:289–293
Zhu LC, Smith CM, Fritz A, Boyko E, Voothuluru P, Gill BS (2005) Inheritance and molecular mapping of new greenbug resistance genes in wheat germplasms derived from Aegilops tauschii. Theor Appl Genet 111:831–837
Acknowledgments
This study was supported by the USDA-NRI grant (CSREES# 2006-35301-16892) to Y. W. and partly supported by the Wheat MAS Project (CAP# 2006-55606-16629) to J. C. R. The authors are grateful to Peihuan Yan, Yuanhong Du, Jony Simmons and Gina Rudd for technical support. The authors also thank Takao Komatsuda (National Institute of Agrobiological Sciences, Japan) for useful discussions and critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by C. Feuillet.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Azhaguvel, P., Rudd, J.C., Ma, Y. et al. Fine genetic mapping of greenbug aphid-resistance gene Gb3 in Aegilops tauschii . Theor Appl Genet 124, 555–564 (2012). https://doi.org/10.1007/s00122-011-1728-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-011-1728-z