Abstract
Anti-malarial proguanil (1) and phenoxypropoxy biguanide derivatives (2–9) are prodrugs. Their efficacy is directly proportional to the quantity of active triazine metabolites produced from these prodrugs. Detailed molecular docking analyses for all nine drug candidates in the active site of CYP3A4, CYP2D6, and CYP2C19 were carried out under the influence of induced-fit effect of ligand during molecular dynamic simulations. We have developed a strategy based on docking pose clusters to quantify the production of active metabolites for this class of molecules. For all drugs, site of metabolism based clusters of docking poses were prepared in both phases of the molecular docking analyses and correlated with the percentage of metabolites generated in the pooled human liver microsomes study. The total numbers of docking poses representing active metabolite formation were found to be well correlated with the experimental results in post-induced fit docking analyses. This strategy was first validated using proguanil, PS-15 and JPC-2056. Further, this methodology was employed to correlate the theoretically predicted metabolite formation of 4–9 to the experimentally estimated values which further led to clues on isoenzyme specificity in producing the metabolites. Binding requirements of these leads in the active sites of CYPs were also explored in this study.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Malaria is one of the most prevalent diseases in developing countries claiming about one million lives per year on an average, mostly children and women (World Malaria Report, 2009; Wirth, 1998, 1999; Greenwood et al., 2005, 2008; Rieckmann, 2006). Out of several plasmodium species, Plasmodium falciparum parasite is the most dangerous causative agent which is responsible for most of the morbidities and mortalities due to malaria (World Malaria Report, 2009). Inhibition of the P. falciparum dihydrofolate reductase thymidylate synthase (PfDHFR-TS) enzyme is one of the leading ways to tackle this deadly disease (Dasgupta et al., 2009). Success of two anti-folate drugs like pyrimethamine and cycloguanil (active metabolite of proguanil, Fig. 1) provided wide scope in the development of new leads in this class. However, limitations of current therapy, owing to multi drug resistant species, makes malaria an epidemic disease (Biswas, 2001; Curtis et al., 2002; Delfino et al., 2002; Eskandarian et al., 2002; Le Bras and Durand, 2003; Warhurst, 2002; Yuthavong, 2002; Nyunt and Plowe, 2007; Wiesner et al., 2003; Gemma et al., 2010; Sridaran et al., 2010). Proguanil (Fig. 1) is the biguanide-based drug available in market for the treatment of malaria and is the most commonly used drug for pregnant women until resistance to this drug in developing countries (Nosten et al., 2006; Na-Bangchang et al., 2005). In studies aiming to elucidate the mode of action of proguanil, it was found that it has in vivo anti-malarial activity, however, it became inactive in vitro against malaria infected cells (Tonkin, 1946; Black, 1946; Hawking and Perry, 1948). In an effort to ascertain the mode of action of proguanil, Crowther, and Levi were able to isolate active metabolite of proguanil which proved to be 4,6-diamino-1-p-chlorophenyl-1,2-dihydro-2,2-dimethyl-1,3,5-triazine (cycloguanil) (Crowther and Levi, 1953; Carrington et al., 1951). Although active metabolite, cycloguanil is responsible for anti-malarial activity, direct clinical use of this metabolite was restricted due to its poor bioavailability and local toxicity (Butler et al., 1947; Kaump et al., 1965).
Rieckmann (1973) reported WR99210, an analog of cycloguanil (WR99210) having potent in vitro anti-malarial activity against the resistant strains for chloroquine and pyrimethamine. Crystal structure analysis of bound WR99210 to both the wild- and mutant-type of the PfDHFR-TS enzyme further clarified the effectiveness of WR99210 against the mutant enzymes due to flexible oxygen-bridged side chain which allowed the molecule to achieve a conformation which avoids the steric hindrance (Yuvaniyama et al., 2003). This kind of side chain flexibility is absent in cycloguanil and pyrimethamine making their binding more difficult to specific mutated resistant strain of P. falciparum. Due to poor bioavailability and considerable gastrointestinal intolerance in animal studies further development of WR99210 was stopped (Canfield, 1986; Bajwa et al., 1983). To overcome these issues, prodrug PS-15 (phenoxypropoxy biguanide, Fig. 1) was developed which was metabolized in vivo into WR99210 (Rieckmann et al., 1996; Canfield et al., 1993). The conversion of PS-15 to active metabolite WR99210 was found to be very similar to the conversion of proguanil to cycloguanil (Edstein et al., 1997). However, development of this compound also was terminated for human use mainly due to regulatory issues related to the use of toxic starting materials (Jensen et al., 2001). Jensen et al. (2001) reported another series of phenoxypropoxy biguanide prodrugs showing similar high potency in in vitro DHFR inhibition of both wild- and mutant-type enzyme. These molecules were also proven to be metabolically activated into active triazine derivatives.
Proguanil (1) and its analogous phenoxypropoxy biguanide derivatives (2–9) (Fig. 1) are prodrugs. As discussed earlier, direct clinical use of cycloguanil and dihydrotriazines is restricted due to their poor bioavailability and local toxicity. These prodrugs require metabolic activation under physiological conditions to act as anti-malarial agents. Thus, the quantity of active metabolite formation from these prodrugs becomes the rate limiting step in the bioavailability (effective therapeutic dose) of these drugs. Rate of metabolism (V, expressed as moles per hour per mole of CYP) as functions of the substrate concentration (S) were fitted by nonlinear regression according to the Michaelis–Menton equation. For this purpose, Shearer et al. studied quantitative analysis of the production of active triazine metabolite and other metabolites from compound 1–9 in pooled human liver microsomes (pHLM). LC–MS and/or LC–MS/MS analysis of the metabolic products of the phenoxypropoxy biguanide analogs characterized mainly three metabolites—(i) active dihydrotriazine M1, (ii) multiple possible hydroxylations M2, and (iii) a desalkylation metabolite M3 (Fig. 2). This study showed that most of the analogs were producing the active triazines metabolites (M1) in the range of 60–80%. However, proguanil, 5 and 7 in which the triazine metabolite comprised only 30–45% and multiple possible hydroxylation products were major metabolites (Shearer et al., 2005). Thus, it is very important to quantify the active triazine metabolite production over other unwanted or less important metabolites to put an effort for future development of the lead molecules.
Several in vitro and in vivo metabolic studies have been carried out for the identification of cytochromeP450 (CYP) isoform responsible for activation of proguanil to cycloguanil (Wattanagoon et al., 1987; Birkett et al., 1994; Helsby et al., 1990a, b; Wright et al., 1995). These studies showed that the CYP2C19 isoform is mainly involved in cyclization and activation of proguanil to cycloguanil. There are also some reports discussing about the minor involvement of CYP3A4 and CYP2D6 in metabolic activation of proguanil to cycloguanil (Funck-Brentano et al., 1997; Coller et al., 1999; Lu et al., 2000). More widely it has been accepted that hepatic CYP2C19 plays a major role in the metabolic activation of proguanil to cycloguanil.
Diaz et al. described the role of specific CYP isoforms in the conversion of JPC-2056 to its active triazine metabolite JPC-2067. In this study they have also included proguanil and PS-15 as reference compounds and studied their conversion into the active metabolites cycloguanil and WR99210, respectively. They have carried out various experimental analyses using pHLM, recombinant CYP isoforms, selective CYP chemical inhibitors, and specific anti-CYP inhibitory antibodies to elucidate which CYP isoforms are primarily involved in the production of the active metabolites of JPC-2056, proguanil, and PS-15. The results indicated that JPC-2056 is mainly metabolized by CYP3A4 while PS-15 appears to be metabolized through both CYP3A4 and CYP2D6 (Diaz et al., 2008). The data for proguanil support the earlier observation that CYP2C19 is the preferred isoenzyme involved in its metabolism to active cycloguanil. The major concern for pharmaceutical industry is in the development of anti-malarial prodrugs which can produce maximum amount of active triazine metabolites (over other unwanted metabolites) to achieve maximum efficacy of drug with minimal dose. CYP2C19 and CYP2D6 isoenzymes are reported to show high degree of polymorphism, (particularly in Asian populations) classifying these two isoforms as poor metabolizers for its substrates (Satyanarayana et al., 2009; Jurima et al., 1985; Helsby, 2008; Desta et al., 2002; Ingelman-Sundberg, 2005). Considering the availability of high concentration of CYP3A4 than CYP2C19 in guts and liver, current efforts are focused on the development of anti-malarial prodrugs of this class which can show major metabolic activation through CYP3A4 to achieve maximum bioavailability in minimal dose.
As the efficacy of proguanil (1) and phenoxypropoxy biguanide derivatives (2–9) is largely dependent upon the quantity of active triazine metabolites produced from these prodrugs, it becomes very important to quantify as well as to identify the role of specific CYP isoenzyme in the production of active metabolite. The molecular docking strategies adopted in this study aim to address the intriguing questions like: (i) whether the percentage of docking poses favoring active metabolite formation can be correlated to the percentage of active metabolites formed in the in vitro study and (ii) whether this correlation can be employed to identify the role of specific CYP in the production of active metabolites of drugs/lead candidates. To the best of our knowledge this may be the first report describing the preparation of the clusters of docking poses to identify preferences in the site of metabolism (SOM) and correlate with the percentage of metabolites formed in an in vitro study. An important aspect for the antimalarial activity is to identify the favorable oxidative SOM that directs the active metabolite formation. Molecular docking study was employed to identify the SOM and the result of molecular docking was also validated with pHLM study. The strategies adopted in this study shall assist (i) in the prediction of quantity of the desirable triazine metabolites production over other unwanted or less important metabolites, (ii) a priori prediction of the role of specific CYP in the production of active triazine metabolites from prodrugs at an early discovery stage, and (iii) help in choosing ideal lead candidates, over other structurally related leads, to promote them further for the developmental stage.
Methods
Dataset for analysis
In this study, we have selected nine different biguanide based anti-malarial drugs/leads such as, proguanil (1), PS-15 (2), JPC-2056 (3), PS-26 (4), JPC-2058 (5), JPC-2005 (6), JPC-2028 (7), JPC-2059 (8), and JPC-1084 (9) (Fig. 1) which are reported as substrates for CYPs. For all the substrates (1–9) corresponding experimental quantification of the active metabolite and other metabolites generated from these analogs in human liver microsomes were reported. For the purpose of molecular docking and molecular dynamics (MD) analysis, crystal structures of CYP3A4 (PDB ID: 2V0M), CYP2D6 (PDB ID: 2F9Q) and a homology model of CYP2C19 were used in this study. The homology model was built using CYP2C9 as template and validated by Ramachandran plot.
Ligand preparation for molecular docking
The dataset of ligands consist of biguanide scaffold which are reported to be highly basic and exist in the protonated form under physiological condition. Considering that proguanil is orally bio-available in the protonated state and also considering biguanide based leads are binding to enzyme in the protonated state, 1–9 were considered in the protonated state only. Two sites of protonation N6 (terminal N atom) and N1 (central N atom) are available in the neutral biguanide. Out of the two sites of protonation, we have considered N6 protonated ligands over N1 (central N atom) protonated ligands in this study as N6 site is reported to be favored site for first protonation (Bharatam et al., 2005). The detailed conformational analysis and optimization of proguanil was carried out as a representative example using B3LYP/6-31+G* level of theory using Gaussian03 package (Frisch et al., 2004). The conformation of ligand which favors the cyclization process was chosen as the starting conformer for the molecular docking study. This conformation showed marginally less stability (<2 kcal mol−1) than the most stable conformer. The 3D structures of 2–9 were built taking the chosen conformation of proguanil as template and energy minimization of each ligand was carried out at B3LYP/6-31+G* level of theory using Gaussian03. The well-optimized 3D structures were converted into SD file format for docking in GOLD software (Jones et al., 1997).
Protein preparation for molecular docking
Crystal structures of CYP3A4 (PDB ID: 2V0M), CYP2D6 (PDB ID: 2F9Q), and homology model of CYP2C19 were used for the molecular docking studies of the chosen set of biguanide drugs/leads. In case of the CYP3A4 and CYP2D6, only one unit was used out of the original tetrameric units in PDB. The GLIDE (Friesner et al., 2004; Halgren et al., 2004) protein preparation wizard was used for the preliminary refinement of all the protein structures. The prime module of Schrodinger suite of software is used for refinement of missing side chain residues in chosen crystal structures. In all cases, heme of the protein was modeled by defining correct atom type to each of the atom using the build panel implemented in GLIDE. The localized charge on the Iron was chosen as Fe2+. As a part of protein preparation, all water molecules were deleted from all 3D protein structures. Hydrogens were added to all the protein structures to generate ionization (of the basic and acidic amino acids) and tautomeric states for all hetero groups at pH 7.0. The entire protein was then minimized to a maximum root mean square deviation (rmsd) value of 0.3 Å so as to avoid the steric clashes of added hydrogen atoms. The prepared protein structures are saved in .mol2 format for preliminary GOLD based docking studies to obtain the initial protein–ligand complexes of CYP3A4–PS-15, CYP2D6–PS-15, and CYP2C19–proguanil.
These prepared proteins represent the parent conformation and may require different conformational arrangement to accommodate the biguanide based ligands in the active site. This issue become important since (i) active sites of CYPs are known to be flexible and demonstrate induced-fit effects (Ekroos and Sjögren, 2006; Skopalík et al., 2008), (ii) there is no crystallographic information available for biguanide complexed with any of these CYPs, and (iii) for CYP2D6, it is already observed in literature that the active site of apo CYP2D6 is too small to accommodate known CYP2D6 substrates (Hritz et al., 2008; Ito et al., 2008). Thus, the parent conformation of crystal structures of CYP3A4, CYP2D6, and CYP2C19 (Homology model), which have different size and shape of active sites required to be modified to accommodate the biguanide based dataset of molecules chosen in this study.
To overcome the limitations of single conformation of protein and to understand atomic level information regarding the interactions of these biguanide based drugs in the active sites of CYPs, MD based strategy was employed where active sites of all three enzymes were altered under the influence of induced-fit effect of ligands. In this approach, MD simulations (McCammon and Harvey, 1987; Leach, 2001) for the complex structures of CYP3A4–PS-15, CYP2D6–PS-15, and CYP2C19–proguanil were performed. Complexes of docking poses representing proximity of methine carbon (C–α–H) towards the heme iron centroid within the 6 Å distance, which favors the cyclization process, were selected as the initial geometries for MD simulations. We selected docking poses of PS-15 complexed with CYP3A4 and CYP2D6 for MD analysis based on the report of Diaz et al. (2008), which showed importance of both CYP3A4 and CYP2D6 in its metabolism. Due to structural similarities of the PS-15 with other phenoxypropoxybiguanide derivatives, we expected similar induced-fit effect by other leads in terms of altering the shape and size of the active sites of CYP3A4 and CYP2D6. Similarly, proguanil complexed with CYP2C19 was used to understand induced-fit effect of proguanil. From the MD simulations of all three complexes, five different conformations were selected for each protein at interval of every 200 ps after the 500 ps equilibration (out of total 2 ns run). These five conformations of all three proteins were further used for the molecular docking analysis of the compounds 1–9.
Desmond software (2009) was used to perform the MD simulations of the three complex structures of CYP2C19–proguanil, CYP2D6–PS-15, CYP3A4–PS-15 as the starting structures. Each complex was solvated with TIP3P water molecules with the cubic box size of 10 Å. To maintain charge neutrality of the systems counter ions were added into the systems (total charge on CYP3A4, CYP2D6, and CYP2C19 were +3, −4, and −1, respectively). The default OPLS-AA force field (Jorgensen et al., 1988, 1996) parameters were applied to the whole system. Simulations were performed in the NPT ensemble and with the Martyna–Tobias–Klein algorithm at a constant temperature of 300 K and a pressure of 1 bar (Martyna et al., 1994). All the prepared systems were allowed for the default relaxation protocol which includes two stages of minimization (restrained and unrestrained) followed by four stages of MD runs with gradually reducing restraints. However, a positional restraint of 1 kcal/mol was maintained for the heme residue while keeping other protein residues unrestrained during simulation. For all the systems, the whole simulation time including default relaxation period was set to 2 ns of MD simulations with periodic boundary conditions. The particle mesh Ewald method (Essman et al., 1995) was used to treat the long-range electrostatic interactions. A 9 Å cutoff of non-bonded van der Waals interactions and a 2-fs time step were used. Snapshots of the MD trajectories were collected at every 4.8 ps for analysis.
Molecular docking
Genetic Optimization for Ligand Docking (GOLD) is a flexible ligand docking program (version 4.1.1) for predicting ligand binding to the protein based on a genetic algorithm which explores the full range of ligand conformational flexibility. The prepared ligands (1–9) were exported as SD files for GOLD docking. Active site of 10 Å radius from the point ~1.5 Å above the Fe, for CYP3A4, CYP2D6, and CYP2C19 was considered for all the prepared protein structures. Docking calculations were performed using the default Gold fitness function (VDW = 4.0, H-bonding = 2.5) and default evolutionary parameters: population size = 100; selection pressure = 1.1; number of operations = 100,000; number of islands = 5; niche size = 2; migration = 10; mutation = 95; crossover = 95; number of Genetic Algorithm (GA) runs = 100; number of poses to keep = 30. ChemScore was used to predict the SOM in the given set of ligands. The special heme parameters available with the GOLD, was applied while performing independent docking runs for ChemScore calculations in the prediction of SOM for the given set of ligands.
Methodology
The biguanide based anti-malarial prodrugs 1–9 (Fig. 1) are known to undergo metabolism at three different sites—(i) methine carbon α to N which leads to the production of active metabolite and N-dealkylated metabolite, (ii) methyl carbons β to N which leads to the production of alkyl hydroxylated metabolites, and (iii) ring carbons which leads to the production of aryl hydroxylated metabolites (Fig. 2) (Shearer et al., 2005).
Since the bioavailability of 1–9 is mostly depend upon the metabolic activation of these prodrugs, it would be extremely advantageous if information about the preferences for SOM and role of specific cytochrome for the production of metabolites are known early in the discovery phase. In the first step of this study, we carried out detailed molecular docking analyses of 1–9 in the active site of CYP3A4, CYP2D6, and homology model of CYP2C19 using GOLD software.
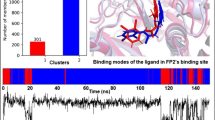
In the current work (Fig. 3), we explored the ligand induced-fit effect based on the MD simulation of selected ligands in the active site of CYP3A4, CYP2D6, and CYP2C19. Under the influence of induced-fit effect of ligands during MD simulation, five different conformations of protein were generated. 30 docking poses were saved for each substrate in the active site of all five conformations of the three CYPs during molecular docking runs based on GOLD algorithm. Generally, only the top ranked pose (or one of the 30 poses which suits) is considered for analysis. Occasionally, top 3 ranked poses are considered where the major focus is to get the insight of the most preferred binding orientation of drugs. In this analysis, all the 30 poses are considered and they are classified to generate clusters as follows (Fig. 4). Correlation was made between the cluster of docking poses for all nine leads in all three CYP isoforms and quantitative production of metabolic products from in vitro metabolism of compounds 1–9 in pHLM study.
-
1.
Cluster-I: poses representing proximity (<6 Å distance from the heme iron centroid) of methine carbon of isopropyl group which leads to the production of active metabolites.
-
2.
Cluster-II: poses representing proximity (<6 Å distance from the heme iron centroid) of methyl carbon of isopropyl group which leads to the production of alkyl hydroxylated metabolites.
-
3.
Cluster-III: poses representing proximity (<6 Å distance from the heme iron centroid) of ring carbon of side chain phenyl group which leads to the production of aryl hydroxylated metabolites.
-
4.
Cluster-IV: All other poses, which are not included for further analysis.
Visual inspection of 30 docking poses and preparation of clusters for different binding poses provided scope to quantify the preference of different SOM with respect to specific substrate. This is because all the 30 chosen poses in each isoform are treated equivalent without assigning any preference to the scores and ranks. Also, the different binding modes (based on SOM) for all drugs may vary depend upon the available space inside the active sites and complementary interactions available to interact inside the active sites of all three CYP isoforms. Thus, the % of poses in each cluster which represents the percentage of docking poses based on SOM, may also vary in each CYP isoform. Sums of percentages of clustered poses belonging to each CYP isoforms are estimated, which can be correlated to the experimental quantitative production of metabolites. For example, if lead-1 shows 30% of docked poses in Cluster-I, in the active site of CYP3A4, 20% in CYP2D6 and 10% in CYP2C19, the probability of formation of cyclization product of lead-1 is 60% which can be correlated to the results from pHLM study. On the other hand, isoform specificity can be estimated from the same data. The percentage of docking poses favoring production of active metabolites by individual CYP isoform may be directly correlated with the ability of that isoform in producing metabolically active products. Thus, this correlation may be useful in identifying specific isoforms responsible for 1–9 which produces maximum quantity of metabolically active products.
Results and discussion
The first portion of this section dealt with the correlation of percentage of docking poses that favored production of active metabolite formation and the pHLM-based quantitative estimation of active metabolites generated from proguanil (1), PS-15 (2), and JPC-2056 (3) was considered as a validation step of docking strategy. Validated protocol was used to predict (i) the quantitative estimation of the production of active metabolites and (ii) the role of specific CYPs in the production of active metabolites, for 4–9. Quantitative estimation of hydroxylated metabolites generated from 1 to 9 was also considered.
Validation of the docking protocol for the quantification and the identification of the role of specific CYP
In this study, compounds 1–3 were considered as reference compounds, because of available experimental knowledge about the role of specific isoform responsible for the production of cyclized active metabolites from all three substrates (Diaz et al., 2008). Also, for these three substrates quantification of metabolic products generated in pHLM-based study was available (Shearer et al., 2005). These three substrates were thus used to establish and validate docking strategy (Fig. 3) by correlating the percentage of docking poses favoring active metabolite formation to the percentage of active metabolites formed in an in vitro study. In establishing the role of specific isoform responsible for the active metabolite production from 1 to 3, several experiments were performed by Diaz et al. (2008) in which they considered five isoforms of CYP, i.e., CYP3A4, CYP2D6, CYP2C19, CYP1A2, and CYP2C9. However, this study revealed that CYP1A2 and CYP2C9 played insignificant role in the conversion of inactive prodrug to active metabolites. Thus, we first carried out molecular docking analysis for protonated form of biguanide derivatives of 1–3 in the active site of crystal structures of three isoforms of CYPs, CYP3A4, CYP2D6, and CYP2C19.
For the purpose of quantitative estimation of percentage of docking poses favoring cyclization process (pose representing Cluster-I), molecular docking analyses of compounds 1–3 were carried out in the active sites of five conformations generated from MD analysis of each of CYP3A4, CYP2D6, and CYP2C19. Docking algorithm GOLD (ChemScore) was used to generate 30 docking poses in each conformation. ChemScore based docking analysis (Supporting information) for protonated proguanil (1) predicted 18% (in case of CYP2C19) and 9% (in case of CYP2D6) (Table 1; Fig. 5) of docking poses to fall in Cluster-I category, which favors the production of active metabolite cycloguanil. The docking analysis, however, was unable to predict any docking pose favoring production of active metabolite for proguanil in CYP3A4 active sites. The total 27% prediction favoring production of active metabolite is slightly lower than 40–45% reported in in vitro pHLM-based quantitative production of cycloguanil (Shearer et al., 2005). However, the order of isoform specificity for the cycloguanil production was found to be in good agreement with the recombinant enzyme analysis reported by Diaz et al. Figure 5 clearly shows that for the cycloguanil production CYP2C19 is the major contributor.
Docking poses belonging to Cluster-I, which favor the production of active metabolite of PS-15 (2), were predicted to be 45, 18, and 11% for the isoforms CYP3A4, CYP2D6, and CYP2C19, respectively (Fig. 5; Table 1). The results demonstrate that the metabolic activation of PS-15 is more favored by CYP3A4 isoform followed by the CYP2D6 and CYP2C19. These docking results are quite comparable to the experimentally observed isoform preference data in the production of active metabolite WR99210 from PS-15 (Fig. 5). The total 74% docking poses favoring the production of active metabolite in the active sites of all three isoforms were slightly larger compared to the ~60% production of WR99210 into the in vitro pHLM-based study (Diaz et al., 2008).
Similar to the metabolic profile of PS-15, the major role of CYP3A4 in the metabolism of JPC-2056 (3) was also envisaged (Fig. 5). ChemScore based docking analysis in the active site of selected five conformers of CYP3A4 favored an average 50% of docking poses belonging to Cluster-I category. These docking analyses also showed that only 13% of docking poses favored the production of active metabolite of JPC-2056 by CYP2D6. This percentage was found to be slightly lower than those predicted for PS-15 in the active site of CYP2D6 for the Cluster-I category. The docking analysis predicted the production of total ~63% active metabolite of JPC-2056 with the major involvement of CYP3A4 followed by little involvement of CYP2D6 and no involvement of CYP2C19. The results are very well correlated with the experimental data which also predicted major metabolism of JPC-2056 through CYP3A4 with minor involvement of CYP2D6 and CYP2C19.
The overall performance of docking runs based on the selection of the average of the five conformers of each CYP2C19, CYP2D6, and CYP3A4 obtained from the MD analyses was found to be satisfactory. The cluster of docking poses representing different sites of metabolism obtained in docking experiment with ChemScore were found to be in good correlation with the experimental quantification of the production of active metabolites from 1 to 3. These docking experiments in conjunction with MD analysis predicted correctly the role of specific CYP450 in the production of active metabolites of 1–3 in the active site of CYP3A4, CYP2D6, and CYP2C19. These results gave us confidence in the above adopted docking protocol (Fig. 3), which reliably quantified and identified the role of specific CYP450, in the production of active metabolites from 1 to 3.
Prediction of active triazine metabolites for 4–9
The validated protocol was adopted in the quantification and the identification of the role of specific CYP, in the production of active metabolites of the other phenoxypropoxy biguanide derivatives (4–9). Quantitative estimation for the production of metabolic products (characterized by LC–MS and/or LC–MS/MS technique followed by metabolite ID analysis) were available from in vitro metabolism of compounds 4–9 by pHLM (Shearer et al., 2005). However, no information is available for the role of specific CYP450 in the production of active metabolites of 4–9. We performed molecular docking analysis for compounds 4–9 in the active sites of previously used five conformations generated from MD analysis of CYP3A4, CYP2D6, and CYP2C19. Cluster of docking poses representing active site was used to establish correlation with experimentally known quantification of the active metabolites from 4 to 9. Based on the above correlation, we predicted the role of specific CYP in the production of active metabolites of 4–9.
As shown in Fig. 6, molecular docking experiment predicted percentage of docking poses of PS-26 (4) representing Cluster-I (favoring the production of active metabolite) were 2, 21, and 43% for the CYP2C19, CYP2D6, and CYP3A4, respectively (Supporting information). The results suggest that CYP3A4 metabolizes PS-26 into the active metabolite much more favorably, followed by CYP2D6 and CYP2C19. These results were similar in order for the involvement of CYPs in the quantitative estimation of docking poses belonging to Cluster-I as in the case of PS-15 but slightly greater contribution from CYP2D6 and very minor contribution from CYP2C19. The indirect confirmation for the correct prediction made by docking protocol in estimating quantitative production of active metabolite of PS-26 may be obtained by comparing the total production of active metabolite in the docking experiment in the active sites of CYP2C19, CYP2D6, and CYP3A4 and in vitro pHLM-based study. In the docking analysis, the total production of active metabolite in the active sites of CYP2C19, CYP2D6, and CYP3A4 was found to be 66% (Figs. 6, 7) which is comparable to the quantity (~70%) of active metabolite obtained in pHLM-based study (Shearer et al., 2005).
LC–MS/MS analysis of the metabolites generated for JPC-2058 (5) and JPC-2028 (7) in the pHLM-based study showed the production of active dihydrotriazine to be only about 30–35% (Shearer et al., 2005). The present ChemScore based docking analysis for protonated form of 5 and 7 predicted 27 and 34%, respectively, for the docking poses, which favor the production of active metabolites in the isoform CYP3A4. CYP2D6 also seems to play a major role in the metabolism of JPC-2058 (5) with a contribution of ~17% production of active metabolite in terms of favored docking poses. A total of ~45% docking poses predicted production of active metabolites for both of 5 and 7, which were estimated slightly higher than the measured value ~30–35% in the in vitro pHLM-based study (Fig. 7). In addition, for these two analogs the hydroxylated metabolites were the most abundant, representing greater than 65% of all the metabolites. Similar to proguanil, these analogs also favored lower percentage of docking poses for the formation of active metabolites. However, 5 and 7 seems to have different isoenzyme specificity (CYP3A4 and CYP2D6) in the production of active metabolites than proguanil which prefer to be metabolized by CYP2C19. This may be attributed to the fact that size of 5 and 7 are larger as compared to proguanil and may require larger pocket size of enzyme for undergoing metabolism.
Metabolic profile prediction of JPC-2005 (6) based on the docking analysis in this work showed that percentage of docking poses which favor the production of active metabolite was 55 and 20% for the CYP3A4 and CYP2D6 isoforms, respectively (Fig. 6). None of the docking poses were successful in case of the Cluster-I category for isoform CYP2C19. This suggested that CYP2C19 does not favor the production of active metabolite for lead candidate 6. The total docking poses predicted to favor production of active metabolite for 6 was ~75%, which was comparable with the measured value ~80% in the in vitro pHLM-based study (Fig. 7) (Shearer et al., 2005). These docking results predicted CYP3A4 as the major isoenzyme to metabolize 6 into the active metabolite. However, the role of CYP2D6 also cannot be under estimated as the docking poses favoring production of active metabolite in the active sites of CYP2D6 was predicted to be 20%. This observation becomes important particularly when we are looking for higher efficacy of drug in minimal dose, since isoform CYP2D6 is reported to have polymorphism in major population in developing countries. From these results, suggestion may be made for the further experimental clarification of 6 before it can be considered in the development stage.
For JPC-2059 (8) and JPC-1084 (9), the percentage of docking poses which favor the production of active metabolites were predicted to be 50 and 57%, respectively, in the active site of CYP3A4 (Fig. 6). These docking results also showed that ~14% of docking poses of 8 and 9 predicted to favor the production of active metabolites by CYP2D6. The total docking poses predicted to favor production of active metabolite for 8 and 9 were ~66 and 73%, which were found to be comparable with the experimentally measured value ~80% in the pHLM-based study (Fig. 7) (Shearer et al., 2005). These values are comparable to the, experimental as well as validated docking cluster based, metabolic profile of production of active metabolite shown by JPC-2056 (3). Relatively less contribution of CYP2D6 in the production of active metabolites of 8 and 9, as compared to other lead molecules such as PS-15, PS-26, and JPC-2005, favored the consideration of 8 and 9 along with JPC-2056 in the development stage.
Prediction of hydroxylated metabolites
For a successful lead candidate in the class of biguanides to be effective anti-malarial drug, it is necessary to show the minimal quantity of unwanted metabolites along with high production of active metabolites. Correlation was also made for docking poses representing Cluster-II and Cluster-III obtained for 1–9 in the active site of all three CYP isoforms with the pHLM-based quantitative estimation of unwanted metabolites such as methyl hydroxylated and ring hydroxylated metabolites (Shearer et al., 2005). Proguanil, JPC-2028, and JPC-2058 showed higher percentage of docking poses that favor the production of such hydroxylated metabolites. Quantitative analysis suggested that for proguanil (1), CYP2C19 favored ~30%, CYP2D6 favored ~8%, and CYP3A4 favored ~5% of the docking poses that leads to ring hydroxylated metabolite formation (Supporting information). These results are slightly higher than the quantity of hydroxylated product obtained in pHLM study which estimated ~30% of ring hydroxylated metabolite formation from proguanil. Minor percentage (~5%) of docking poses in all three enzymes was predicted which favored methyl hydroxylation in proguanil. However, no quantitative estimation is available for methyl hydroxylated product in pHLM-based study of proguanil. The higher percentage of ring hydroxylated product in proguanil was expected due to the availability of more number of Ar–H sites. It was also noted that percentage of docking poses that favored ring hydroxylated products drastically reduced when number of chloro substitution increased in the ring. Docking analyses were not able to predict any pose that favors the production of ring hydroxylated product in case of PS-26 and PS-15 which have two and three chloro substitution, respectively, in the ring. Minor involvement of ring hydroxylated product was noted in case of PS-26 during pHLM-based estimation; however, docking quantification did not favor such involvement. This may be due to some of the discarded docking poses which showed proximity of chloro group over Ar–H towards heme center. Experimental results suggested that similar to the multiple chloro substitution on aromatic ring, trifluoromethyl or trifluoromethoxy substitution at para position of aromatic ring also prevented formation of ring hydroxylated product (Shearer et al., 2005). This also became evident in the molecular docking studies of JPC-2056 (3), JPC-2005 (6), JPC-2028 (7), JPC-2059 (8), and JPC-1084 (9) which showed none or very negligible percentage of docking poses that favors ring hydroxylation in all three isoenzymes. Most of the docking poses in which the aromatic ring head is approaching towards the heme center in the active site were discarded because of electrostatic interaction between CF3 and heme center. Docking pose analysis of JPC-2028, however, was showing ~50% of docking poses favoring the production of methyl hydroxylated product in CYP3A4 and CYP2D6. This follows the trend in metabolite profile of JPC-2028 in pHLM study which estimated >65% of methyl hydroxylated metabolite. Positioning trifluoromethoxy at the ortho-position in JPC-2058 (5), resulted in the formation of >65% hydroxylation products estimated by pHLM study. Molecular docking analysis also shows that ~50% of docking poses favor the production of ring hydroxylated product in CYP3A4 and CYP2D6.
Structural insights of biguanide derivatives into the active sites of CYP isoforms
Crystallographic data for biguanide derivatives interacting with different isoforms of CYPs is unavailable, thus detailed structural analysis was carried out (i) to get insight of changes in protein conformation due to induced-fit effect of ligands in the complex structures of CYP3A4–PS-15, CYP2D6–PS-15, and CYP2C19–proguanil under dynamical conditions, and (ii) to understand the important interactions involved in the binding of proguanil and phenoxypropoxy biguanide (PS-15) to specific CYPs. Superimposition of five conformations of each CYP on the respective initial reference conformation may provide the insights of structural changes occurred in the protein (especially in the active site) during the MD simulations. The entire protein C-alpha rmsd for all five conformations with respect to their initial reference conformation were found to be in the range of, 1.977–2.232 Å in case of CYP2C19, 1.394–1.911 Å in case of CYP2D6, and 1.641–1.912 Å in case of CYP3A4. Similarly, we analyzed the C-alpha rmsd for the active site residues within 10 Å area surrounding the ligands which were found to be in the range of, 2.567–3.256 Å in case of CYP2C19, 1.172–4.458 Å in case of CYP2D6, and 1.786–3.983 Å in case of CYP3A4. This suggests that under dynamical condition over all protein conformation is not much affected and stays stable at less than 2.5 Å rmsd. However, the larger ranges of rmsd for the active site residues in all three proteins suggested induced-fit changes in active sites conformations to accommodate the ligands in the active site during MD simulations.
Analysis of the superimposed 10 Å active sites of all five conformations of CYP3A4 on to the reference conformation showed that there are major movements of active site residues like Phe57, Phe108, Leu211 Phe213, Phe220, Phe241, Phe304, and Leu482 in the process of accommodating PS-15 (Fig. 8). Hydrophobic pocket formed by Phe108, Phe220, Phe57, and Arg106 stabilize the aromatic ring of the PS-15. Hydrophobic side chains of Phe304, Phe241, and Leu482 stabilize the biguanide moiety of PS-15. Furthermore, the side chain of Leu211 was found to move outwards and hence facilitate the opening of the cavity during MD simulations. There were no H-bonding interactions observed between PS-15 and any of the active site residues of CYP3A4. However, hydrophobic interactions provided by Phe108, Phe220, and Phe57 act as an anchor by holding aromatic ring of PS-15. These types of hydrophobic interactions in CYP3A4 available for aromatic ring of PS-15 is possible may be due to length of flexible linker side chain (–O–CH2–CH2–CH2–CH2–O–) between biguanide moiety and aromatic ring which can easily reach the hydrophobic cavity formed by Phe108, Phe220, and Phe57. This may help in positioning the reported SOM in the desirable pose for metabolism of PS-15. But the same are not possible for proguanil, because the absence of such flexible long chain in proguanil. This explains the experimentally observed minor role of CYP3A4 in the metabolism of proguanil and significant role of CYP3A4 in the metabolism of PS-15 and other phenoxypropoxy biguanide derivatives.
Superimposed active sites residues of all five conformations of CYP3A4 chosen from molecular dynamics simulation at regular interval of 200 ps on to the reference conformation. Green color ligands represent the conformations of PS-15 during MD simulations and magenta represents the reference conformation of PS-15. Cyan color amino acid residues represent the reference conformation of CYP3A4 and yellow color amino acid residues represent the conformations of CYP3A4 obtained during MD simulations
Binding analysis of PS-15 in the active site of CYP2D6 during MD showed major movement of Phe120, Leu213, Glu216, Val308, and Met374 which are the residues covering inside layer of binding site and may play important role in the binding of PS-15 and other biguanide derivatives (Fig. 9). Glu216 residue makes hydrogen bond interaction with biguanide unit of PS-15 and holds the PS-15 and other biguanide based substrates in such an orientation that favors metabolism. Hritz et al. (2008) reported flexibility of Phe483 in binding of the substrate, however, no such significant movement of Phe483 was observed during MD simulation of CYP2D6–PS-15 complex. Thus, residue Phe483 may be playing less significant role in the binding and metabolism of this class of molecules in the active site of CYP2D6. There was no aromatic ring stabilization (π stacking) of the PS-15 by the hydrophobic interactions in the active site of CYP2D6.
Superimposed active sites residues of all five conformations of CYP2D6 chosen from molecular dynamics simulation at regular interval of 200 ps. on to the reference conformation. Green color ligands represent the conformations of PS-15 during MD simulations and magenta represents the reference conformation of PS-15. Cyan color amino acid residues represent the reference conformation of CYP2D6 and yellow color amino acid residues represent the conformations of CYP2D6 obtained during MD simulations
Analysis of proguanil in the active site of 5 different conformations of CYP2C19 obtained from MD simulations showed that biguanide moiety was stabilized by active site residues like Asp293, Ala297, Val113, Leu336, and Thr301 (Fig. 10). Asp293 residue makes hydrogen bond interaction with biguanide unit of proguanil. This interaction is analogous to the H-bonding interaction shown by PS-15 in the active site of CYP2D6 with Glu216. Similar to aromatic ring stabilization in the hydrophobic pocket by PS-15 in the active site of CYP3A4, hydrophobic pocket formed by Phe100, Phe114, Phe476, Leu102 and basic residues like Arg108, Asn204 in the active site of CYP2C19 also provided stabilization to aromatic ring of proguanil. The above analyses suggest that aromatic ring stabilization through the hydrophobic interactions in CYP2C19 is available at a shorter range from heme centroid. This suggests that proguanil (with shorter side chain between biguanide moiety and aromatic ring) is suitable for binding into the active site of CYP2C19. Thus, in PS-15 and other phenoxypropoxy biguanides with flexible linker side chain between biguanide moiety and aromatic rings, the aromatic ring may elongate beyond the hydrophobic pocket in CYP2C19 and may not bind in the active site properly. This may be the reason why proguanil showed major metabolism by CYP2C19 while PS-15 and other phenoxypropoxy biguanides showed minimal or negligible metabolism by this isoform.
Superimposed active sites residues of all five conformations of CYP2C19 chosen from molecular dynamics simulation at regular interval of 200 ps. on to the reference conformation. Green color ligands represent the conformations of proguanil during MD simulations and magenta represents the reference conformation of proguanil. Cyan color amino acid residues represent the reference conformation of CYP2C19 and yellow color amino acid residues represent the conformation of CYP2C19 obtained during MD simulations
Conclusions
A novel molecular modeling strategy based on the molecular docking was introduced to quantitatively estimate (i) the preferred metabolite generation, (ii) the amount of metabolites produced, and (iii) CYP isoform specificity in generating the preferred metabolite. This strategy is based on clustering of 30 docking poses, obtained from a molecular docking exercise, into four clusters. All the 30 poses are given equal importance while clustering. Each cluster based on SOM represents experimentally observed metabolites such as—(i) cyclic active metabolite, (ii) alkyl hydroxylated metabolite, and (iii) aryl hydroxylated metabolite. A set of nine important leads (in their protonated state) for anti-malarial activity are considered in the study. The structures of CYP3A4, CYP2D6, and CYP2C19 obtained from crystal structure/homology model were subjected to MD simulations to accommodate induced-fit effects. The protein structures obtained from MD simulations are employed in the molecular docking analysis, using ChemScore as scoring function.
The analysis predicted the major role of CYP2C19 in case of proguanil (1), CYP3A4 and CYP2D6 in case of PS-15 (2), and CYP3A4 in case of JPC-2056 (3) in the production of respective cyclic active metabolites. The percentage of docking poses that favored production of active metabolite formation from these three drugs are well correlated with the pHLM-based quantitative estimation of active metabolites generated from 1 to 3. This study also predicted higher quantity of ring hydroxylated product for proguanil (1) by CYP2C19 which is consistent with the experimental estimation.
Validated protocol has been used to predict the role of specific CYPs in the production of active metabolites of 2–9 (lead molecules). The docking cluster based results are well correlated with the experimental pHLM study, supporting that PS-15 (2), JPC-2056 (3), PS-26 (4), JPC-2005 (6), JPC-2059 (8), and JPC-1084 (9) produce higher amount of cyclic active metabolite formation while JPC-2058 (5) and JPC-2028 (7) are shown to produce more amounts of unwanted hydroxylated metabolites. This study also indicated that CYP2C19 plays negligible role in the metabolism of all these molecules except proguanil. For the lead molecules PS-26, JPC-2005, and JPC-2028, CYP3A4 plays major role in metabolizing and producing active metabolites, however, in these three leads, CYP2D6 also seems to be playing moderate role in the production of active metabolites. For the lead molecule JPC-2058, CYP3A4 and CYP2D6 both were predicted to play major role in metabolizing and producing active metabolites as well as hydroxylated metabolites. Whereas, CYP3A4 was predicted to play major role in metabolizing and producing active metabolites for the lead molecules JPC-2059 and JPC-1084, only minor contribution was found from CYP2D6 in the production of active metabolites for these two leads. From the above analysis, we may recommend the consideration of JPC-2059 and JPC-1084 along with JPC-2056 into the clinical studies and development.
This study also suggested binding requirements of these leads in the active sites of CYPs which supported the reason for major metabolism of proguanil by CYP2C19 while in case of PS-15 and other phenoxypropoxy biguanides CYP3A4 plays major role in metabolism. Structural insight suggest that proguanil (with direct aromatic substitution without linker chain) is suitable for binding into the active site of CYP2C19. Its binding in the cavity of CYP3A4 may not lead to metabolism because of the large distance between the hydrophobic pocket (molecular recognition site) and heme center in CYP3A4. In 2–9, long, flexible chain is present, it facilitates a suitable pose of ligands in CYP3A4 (but not in CYP2C19), which maximizes hydrophobic interactions and place the methine carbon close to the heme center. Such details provided clues for the success in the molecular modeling strategy adopted in this work.
The out-of-the-box solution successfully predicted the percentage of metabolite production and also the isoform specificity in the metabolite production. This study is suggesting JPC-2059 and JPC-1084 compounds towards the developmental stage in addition to the JPC-2056 which is already in the process. The strategy adopted in this work can be executed to many metabolism based drug discovery analysis in determining which compounds should be taken for clinical trials. Thus, such a strategy can find in many practical applications in future.
References
Bajwa BS, Acton N, Brossi A (1983) The chemistry of drugs. III. Acid hydrolysis of antimalarial 5-alkoxy-6,6-dimethyl-5,6-dihydro-s-triazines. Heterocycles 20:839–843
Bharatam PV, Patel DS, Iqbal P (2005) Pharmacophoric features of biguanide derivatives: an electronic and structural analysis. J Med Chem 48:7615–7622
Birkett DJ, Rees D, Andersson T, Gonzalez FJ, Miners JO, Veronese ME (1994) In vitro proguanil activation to cycloguanil by human liver microsomes is mediated by CYP3A isoforms as well as by S-mephenytoin hydroxylase. Br J Clin Pharmacol 37:413–420
Biswas S (2001) Plasmodium falciparum dihydrofolate reductase Val-16 and Thr-108 mutation associated with in vivo resistance to antifolate drug: a case study. Indian J Malariol 38:76–83
Black RH (1946) The effect of anti-malarial drugs on Plasmodium falciparum (New Guinea Strains) developing in vitro. Trans R Soc Trop Med Hyg 40:163–170
Butler R, Davey DG, Spinks A (1947) A preliminary report of the toxicity and the associated blood concentrations of paludrine in laboratory animals. Br J Pharmacol Chemother 2:181–188
Canfield CJ (1986) New antimalarials under development. In: Bruce-Chatt LJ (ed) Chemotherapy of malaria. Monograph series 27, 2nd edn. World Health Organization, Geneva, pp 99–100
Canfield CJ, Milhous WK, Ager AL, Rossan RN, Sweeney TR, Lewis NJ, Jacobus DP (1993) PS-15: A potent, orally active antimalarial from a new class of folic acid antagonists. Am J Trop Med Hyg 49:121–126
Carrington HC, Crowther AF, Davey DG, Levi AA, Rose FL (1951) A metabolite of paludrine with high antimalarial activity. Nature 168:1080
Coller JK, Somogyi AA, Bochner F (1999) Comparison of (S)-mephenytoin and proguanil oxidation in vitro: contribution of several CYP isoforms. Br J Clin Pharmacol 48:158–167
Crowther AF, Levi AA (1953) Proguanil—the isolation of a metabolite with high anti-malarial activity. Br J Pharmacol Chemother 8:93–97
Curtis J, Maxwell CA, Msuya FHM, Mkongewa S, Alloueche A, Warhurst DC (2002) Mutations in DHFR in Plasmodium falciparum infections selected by chlorproguanil-dapsone treatment. J Infect Dis 186:1861–1864
Dasgupta T, Chitnumsub P, Kamchonwongpaisan S, Maneeruttanarungroj C, Nichols SE, Lyons TM, Tirado-Rives J, Jorgensen WL, Yuthavong Y, Anderson KS (2009) Exploiting structural analysis, in silico screening, and serendipity to identify novel inhibitors of drug-resistant falciparum malaria. ACS Chem Biol 4:29–40
Delfino RT, Santos-Filho OA, Figueroa-Villar JD (2002) Molecular modeling of wild-type and antifolate resistant mutant Plasmodium falciparum DHFR. Biophys Chem 98:287–300
Desmond (2009) Version 2.2, Schrödinger, LLC, New York, NY
Desta Z, Zhao X, Shin JG, Flockhart DA (2002) Clinical significance of the cytochrome P450 2C19 genetic polymorphism. Clin Pharmacokinet 41:913–958
Diaz DS, Kozar MP, Smith KS, Asher CO, Sousa JC, Schiehser GA, Jacobus DP, Milhous WK, Skillman DR, Shearer TW (2008) Role of specific P450 isoforms in the conversion of phenoxypropoxybiguanide analogs in human liver microsomes to potent antimalarial dihydrotriazines. Drug Metab Dispos 36:380–385
Edstein MD, Bahr S, Kotecka B, Shanks GD, Rieckmann KH (1997) In vitro activities of the biguanide PS-15 and its metabolite, WR99210, against cycloguanil-resistant Plasmodium falciparum isolates from Thailand. Antimicrob Agents Chemother 41:2300–2301
Ekroos M, Sjögren T (2006) Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc Natl Acad Sci USA 103:13682–13687
Eskandarian AA, Keshavarz H, Basco LK, Mahboudi F (2002) Do mutations in Plasmodium falciparum dihydropteroate synthase and dihydrofolate reductase confer resistance to sulfadoxine-pyrimethamine in Iran? Trans R Soc Trop Med Hyg 96:96–98
Essman U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys 103:8577–8593
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739–1749
Frisch MJ, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Zakrzewski VG, Montgomery JA Jr, Stratmann RE, Burant JC, Dapprich S, Millam JM, Daniels AD, Kudin KN, Strain MC, Farkas O, Tomasi J, Barone V, Cossi M, Cammi R, Mennucci B, Pomelli C, Adamo C, Clifford S, Ochterski J, Petersson GA, Ayala PY, Cui Q, Morokuma K, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Cioslowski J, Ortiz JV, Baboul AG, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Gomperts R, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Gonzalez C, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Andres JL, Gonzalez C, Head-Gordon M, Replogle ES, Pople JA (2004) Gaussian 03 Revision C.02. Gaussian, Inc., Wallingford, CT
Funck-Brentano C, Becquemont L, Lenevu A, Roux A, Jaillon P, Beaune P (1997) Inhibition by omeprazole of proguanil metabolism: mechanism of the interaction in vitro and prediction of in vivo results from the in vitro experiments. J Pharmacol Exp Ther 280:730–738
Gemma S, Travagli V, Savini L, Novellino E, Campiani G, Butini S (2010) Malaria chemotherapy: recent advances in drug development. Recent Pat Antiinfect Drug Discov 5:195–225
Greenwood BM, Bojang K, Whitty CJ, Targett GA (2005) Malaria. Lancet 365:1487–1498
Greenwood BM, Fidock DA, Kyle DE, Kappe SH, Alonso PL, Collins FH, Duffy PE (2008) Malaria: progress, perils, and prospects for eradication. J Clin Invest 118:1266–1276
Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL (2004) Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem 47:1750–1759
Hawking F, Perry WLM (1948) Activation of pauldrine. Br J Pharmacol Chemother 3:320–325
Helsby NA (2008) Pheno- or genotype for the CYP2C19 drug metabolism polymorphism: the influence of disease. Proc West Pharmacol Soc 51:5–10
Helsby NA, Ward SA, Edwards G, Howells RE, Breckenridge AM (1990a) The pharmacokinetics and activation of proguanil in man: consequences of variability in drug metabolism. Br J Clin Pharmacol 30:593–598
Helsby NA, Ward SA, Howells RE, Breckenridge AM (1990b) In vitro metabolism of the biguanide antimalarials in human liver microsomes: evidence for a role of the mephenytoin hydroxylase (P450 MP) enzyme. Br J Clin Pharmacol 30:287–291
Hritz J, de Ruiter A, Oostenbrink C (2008) Impact of plasticity and flexibility on docking results for cytochrome P450 2D6: a combined approach of molecular dynamics and ligand docking. J Med Chem 51:7469–7477
Ingelman-Sundberg M (2005) Genetic polymorphisms of cytochromeP450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J 5:6–13
Ito Y, Kondo H, Goldfarb PS, Lewis DFV (2008) Analysis of CYP2D6 substrate interactions by computational methods. J Mol Graph Model 26:947–956
Jensen NP, Ager AL, Bliss RA, Canfield CJ, Kotecka BM, Rieckmann KH, Terpinski J, Jacobus DP (2001) Phenoxypropoxybiguanides, prodrugs of DHFR inhibiting diaminotriazine antimalarials. J Med Chem 44:3925–3931
Jones G, Willett P, Glen RC, Leach AR, Taylor R (1997) Development and validation of a genetic algorithm for flexible docking. J Mol Biol 267:727–748
Jorgensen WL, Tirado-Rives J (1988) The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc 110:1657–1666
Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 118:11225–11236
Jurima M, Inaba T, Kadar D, Kalow W (1985) Genetic polymorphism of mephenytoin p(4′)-hydroxylation: difference between Orientals and Caucasians. Br J Clin Pharmacol 19:483–487
Kaump DH, Schardein JL, Fisken RA, Sorenson OJ (1965) Toxicity of the repository antimalarial compound cycloguanil pamoate. Toxicol Appl Pharmacol 7:781–793
Le Bras J, Durand R (2003) The mechanisms of resistance to antimalarial drugs in Plasmodium falciparum. Fundam Clin Pharmacol 17:147–153
Leach AR (2001) Molecular modelling: principles and applications, 2nd edn. Prentice Hall, Englewood Cliffs, NJ
Lu AH, Shu Y, Huang SL, Wang W, Ou-Yang DS, Zhou HH (2000) In vitro proguanil activation to cycloguanil is mediated by CYP2C19 and CYP3A4 in adult Chinese liver microsomes. Acta Pharmacol Sin 21:747–752
Martyna GJ, Tobias DJ, Klein ML (1994) Constant pressure molecular dynamics algorithms. J Chem Phys 101:4177–4189
McCammon JA, Harvey SC (1987) Dynamics of proteins and nucleic acids. Cambridge University Press, Cambridge
Na-Bangchang K, Manyando C, Ruengweerayut R, Kioy D, Mulenga M, Miller GB, Konsil J (2005) The pharmacokinetics and pharmacodynamics of atovaquone and proguanil for the treatment of uncomplicated falciparum malaria in third-trimester pregnant women. Eur J Clin Pharmacol 61:573–582
Nosten F, McGready R, d’Alessandro U, Bonell A, Verhoeff F, Menendez C, Mutabingwa T, Brabin B (2006) Antimalarial drugs in pregnancy: a review. Curr Drug Saf 1:1–15
Nyunt MM, Plowe CV (2007) Pharmacologic advances in the global control and treatment of malaria: combination therapy and resistance. Clin Pharmacol Ther 82:601–605
Rieckmann KH (1973) The in vitro activity of experimental antimalarial compounds against strains of Plasmodium falciparum with varying degrees of sensitivity to pyrimethamine and chloroquine, in chemotherapy of malaria and resistance to antimalarials. World Health Organization Technical Report Series. World Health Organization, Geneva, Switzerland
Rieckmann KH (2006) The chequered history of malaria control: are new and better tools the ultimate answer? Ann Trop Med Parasitol 100:647–662
Rieckmann KH, Yeo AE, Edstein MD (1996) Activity of PS-15 and its metabolite, WR99210, against Plasmodium falciparum in an in vivo-in vitro model. Trans R Soc Trop Med Hyg 90:568–571
Satyanarayana CR, Devendran A, Jayaraman M, Mannu J, Mathur PP, Gopal SD, Rajagopal K, Chandrasekaran A (2009) Influence of the genetic polymorphisms in the 5′ flanking and exonic regions of CYP2C19 on proguanil oxidation. Drug Metab Pharmacokinet 24:537–548
Shearer TW, Kozar MP, O’Neil MT, Smith PL, Schiehser GA, Jacobus DP, Diaz DS, Yang YS, Milhous WK, Skillman DR (2005) In vitro metabolism of phenoxypropoxybiguanide analogues in human liver microsomes to potent antimalarial dihydrotriazines. J Med Chem 48:2805–2813
Skopalík J, Anzenbacher P, Otyepka M (2008) Flexibility of human cytochromes P450: molecular dynamics reveals differences between CYPs 3A4, 2C9, and 2A6, which correlate with their substrate preferences. J Phys Chem B 112:8165–8173
Sridaran S, McClintock SK, Syphard LM, Herman KM, Barnwell JW, Udhayakumar V (2010) Anti-folate drug resistance in Africa: meta-analysis of reported dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) mutant genotype frequencies in African Plasmodium falciparum parasite populations. Malar J 9:247
Tonkin IM (1946) The testing of drugs against exoerythrocytic forms of P-gallinaceum in tissue culture. Br J Pharmacol Chemother 1:163–173
Warhurst DC (2002) Resistance to antifolates in Plasmodium falciparum, the causative agent of tropical malaria. Sci Prog 85:89–111
Wattanagoon Y, Taylor RB, Moody RR, Ochekpe NA, Looareesuwan S, White NJ (1987) Single dose pharmacokinetics of proguanil and its metabolites in healthy subjects. Br J Clin Pharmacol 24:775–780
Wiesner J, Ortmann R, Jomaa H, Schlitzer M (2003) New antimalarial drugs. Angew Chem Int Ed Engl 42:5274–5293
Wirth D (1998) Malaria: a 21st century solution for an ancient disease. Nat Med 4:1360–1362
Wirth DF (1999) Malaria: a third world disease in need of first world drug development. Annu Rep Med Chem 34:349–358
World Malaria Report (2009) World Health Organization, Geneva, Switzerland
Wright JD, Helsby NA, Ward SA (1995) The role of S-mephenytoin hydroxylase (CYP2C19) in the metabolism of the antimalarial biguanides. Br J Clin Pharmacol 39:441–444
Yuthavong Y (2002) Basis for antifolate action and resistance in malaria. Microbes Infect 4:175–182
Yuvaniyama J, Chitnumsub P, Kamchonwongpaisan S, Vanichtanankul J, Sirawaraporn W, Taylor P, Walkinshaw MD, Yuthavong Y (2003) Insights into antifolate resistance from malarial DHFR-TS structures. Nat Struct Biol 10:357–365
Acknowledgment
The author thanks Department of Science and Technology (DST), New Delhi for financial support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Patel, D.S., Ramesh, M. & Bharatam, P.V. CytochromeP450 isoenzyme specificity in the metabolism of anti-malarial biguanides: molecular docking and molecular dynamics analyses. Med Chem Res 21, 4274–4289 (2012). https://doi.org/10.1007/s00044-011-9966-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-011-9966-9