Abstract
Almost all clinical oncologists agree that the discovery of reliable, accessible, and non-invasive biomarkers is necessary to decrease cancer mortality. It is possible to employ reliable biomarkers to diagnose cancer in the early stages, predict the patient prognosis, follow up the response to treatment, and estimate the risk of disease recurrence with high sensitivity and specificity. Extracellular vesicles (EVs), especially exosomes, have been the focus of translational research to develop such biomarkers over the past decade. The abundance and distribution of exosomes in bodily fluids, including serum, saliva, and urine, as well as their ability to transport various biomolecules (nucleic acids, proteins, and lipids) derived from their parent cells, make exosomes reliable, accessible, and potent biomarkers for diagnosis and follow-up of solid and hematopoietic tumors. In addition, exosomes play a vital role in various cellular processes, including tumor progression, by participating in intercellular communication. Although these advantages underline the high potential of tumor-derived exosomes as diagnostic biomarkers, the lack of standardized effective methods for their isolation, identification, and precise characterization makes their application challenging in clinical settings. We discuss the importance of non-coding RNAs (ncRNAs) in cellular processes, and the role of tumor-derived exosomes containing ncRNAs as potential biomarkers in several types of cancer. In addition, the advantages and challenges of these studies for translation into clinical applications are covered.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Despite a huge research effort over the past decades, cancer remains a leading cause of death from non-infectious disease and is the second leading cause of death in the USA [1, 2]. Although there have been advances in different therapies over the past two decades, and patient survival has increased markedly, the overall prognosis of patients remains poor. Modern cancer treatment includes surgery radiotherapy, chemotherapy (systemic or regional), targeted therapy, immunotherapy, and anti-cancer vaccines, but the overall success has been limited [3]. Mainly due to delays in diagnosis, many tumors have reached an advanced or metastatic stage and are resistant to treatment. Therefore, prompt non-invasive diagnostic methods are required in the fight against cancer [2, 4]. DNA damage has been known for decades to be a leading cause of carcinogenesis. Although protein products arising from DNA mutations are generated in cancerous cells, only 2% of the entire genome is translated into these functional carcinogenic proteins. Advances in whole genome sequencing have led to rapid and easily accessible methods, such as microarrays and next generation sequencing (NGS), to understand the function and nature of the non-protein-coding portion of the genome. It is now accepted that the 98% non-coding part of the genome is as important as the protein-coding regions [5].
The role of biological molecules, such as circulating RNAs and exosomal proteins, is gaining increased interest from cancer biologists, because of the current emphasis on liquid biopsies for improving cancer diagnosis. If cancer-specific biomarkers can be detected in biological fluids, this removes the requirement for traditional invasive tissue biopsy procedures. Moreover, the heterogeneity of cancer cells, often leads to problems in diagnosis and treatment [6]. The use of bodily fluid-derived exosomes as biomarkers could address the problems of tumor heterogeneity and invasive procedures. In both cancer patients and healthy individuals, cell-free circulating RNAs, such as messenger RNAs and non-coding RNAs (microRNA, short nuclear RNA, long non-coding RNA, circular RNA, and piwi-interacting RNA), have been identified in reasonably high amounts in plasma, serum, urine, and other bodily fluids [7].
Membrane vesicles with varying contents (nucleic acids, lipids, and proteins) can be released from specific cell populations into the microenvironment. According to the definition, by the International Association of Extracellular Vesicles (ISEV), vesicles isolated from biological fluids and cellular environments are known as extracellular vesicles (EVs). Cell-derived EVs are divided into three distinct categories in terms of size and origin, including apoptotic bodies, microvesicles, and exosomes [8]. Exosomes can function as carriers of circulating nucleic acids and are involved in almost all pathological and physiological processes of any organism. Exosomes are one of the most important sources of microRNAs (miRNA) in human serum or plasma [6]. Many miRNAs are significantly dysregulated in different types of cancers (over-expressed or under-expressed) and have therefore been investigated as diagnostic biomarkers and therapeutic targets in many studies over the last three decades [7]. However, there is still a lack of accurate biomarkers for the diagnosis and prognosis of several cancers. For this reason, many scientists have turned their attention to another type of ncRNAs, called long non-coding RNAs (lncRNA), which in turn regulate the expression of specific miRNAs. Circulating lncRNAs could be new biomarkers for the diagnosis and prognosis of different types of cancers. Disruption of lncRNA expression levels is associated with the progression and metastasis of cancers, according to recent studies. LncRNAs are involved in regulating the expression of oncogenes, tumor suppressor genes, and cellular signaling pathways [9,10,11,12]. Given the presence of ncRNAs, especially lncRNAs, contained within exosomes as circulating ncRNAs, the simultaneous detection of both species could provide biomarkers for the diagnosis and prognosis of various cancers, as well as possible therapeutic targets.
Biogenesis of exosomes in cancer
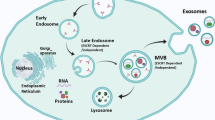
Exosomes are small membrane vesicles secreted by all types of cells and can be found in all bodily fluids [13]. Exosomes are smaller (30–150 nm) compared to microvesicles (100–1000 nm) and apoptotic bodies (800–5000 nm) and have a similar size to viruses. In addition to the size, exosomes differ from other EVs in terms of their biogenesis mechanism. Endosomal components are involved in the biogenesis of exosomes, which play a potential role in maintaining cellular homeostasis [14]. This process begins with an extrusion of the cell plasma membrane and endosome formation. The early endosomes then mature and form late endosomes. Next, the late endosome membrane is invaginated in an inwards direction, which leads to the generation of multiple intraluminal vesicles. The endosome is then transformed into a multivesicular body (MVB), which contains several vesicles, each encapsulating a tiny portion of the cytoplasm, including nucleic acids and proteins [15]. MVBs are next fused to the plasma membrane, and then the exosomes are released out of the cell. However, not all the MVBs combine with the plasma membrane to release exosomes, because some are destroyed during fusion with lysosomes. Because of the high percentage of the membrane lipid ceramide, the exosomes are protected from lysosomal degradation. Through this cellular process, millions of exosomes are released by each tumor cell, containing several cellular components, including nucleic acids, lipids, enzymes, proteins, and cytokines [13]. The exosomal contents do not exactly match that of the parent cells, but there are many similarities in the molecular and genetic profiles. This partial resemblance has led to the concept of exosomes as potential cancer biomarkers [16]. According to recent reports, the rate of production of exosomes is higher in cancer cells, so that the number of exosomes released from cancer cells is many times that of normal cells. Indeed, exosome levels in bodily fluids of cancer patients were found to be significantly higher. Cellular stress, including the hypoxic tumor microenvironment, is the leading cause of increased exosome secretion by tumor cells. Many tumors show an increased expression of p53 and heparanase enzyme, as well as increased activity of Rab GTPase proteins, which could explain the higher secretion of exosomes from tumor cells [17]. The biogenesis of exosomes is illustrated in Fig. 1.
Exosome biogenesis. The first step involves endocytosis producing early endosomes. Endosomes and specific cargos such as nucleic acids, proteins, and lipids are then encased in multivesicular bodies (MVBs). Finally, these multivesicular bodies merge with the plasma membrane and the exosomes are released into the extracellular space. Exosomal contents (proteins, lipids, DNAs, RNAs and non-coding RNAs) are delivered to recipient cells by direct cell membrane fusion, receptor contact, and endocytosis
The importance of tumor-derived exosomes as biomarkers
The absence of severe symptoms before metastasis occurs, as well as the high heterogeneity of tumors, has significant challenges in cancer diagnosis. Although cancer treatment methods have made tremendous progress over the past few decades, none of them have been able to reduce cancer-related mortality entirely. More precisely, cancer mortality reduction is directly related to its detection in the early stages, and definitely before metastasis. Current diagnostic methods mainly include general tests and biopsies, lacking sufficient sensitivity and specificity to identify cancer in the early stages of its formation. In addition, some of these diagnostic methods, including biopsy, are invasive and pose a threat to patient health [18]. For example, the risk of bleeding, infection, and trauma in surgical biopsies is high. On the other hand, sampling and biopsy of some tumors is impossible or difficult due to the inaccessibility of many organs. Urogenital malignancies, for example, are challenging to treat due to their position deep within the pelvic area [19].
Tumor-derived exosomes (TDEs) can be found in the blood and in other bodily fluids, including urine, semen, and breast milk. The molecular and genetic cargos, at least in part, mirror the contents of the parental tumor cells. Therefore, these exosomes may be used as a “liquid biopsy,” allowing for noninvasive tumor detection in real time. Examining the contents of these exosomes could allow scientists to discover more accurate diagnostic biomarkers, especially ncRNAs [20]. TDEs have properties that make them particularly suitable surrogates for tumor-derived proteins and RNAs. Given the importance of ncRNAs (especially miRNAs and lncRNAs) as cancer biomarkers, tumor-derived exosomes containing RNA have attracted much attention as discussed in this review. Exosomal miRNAs could be highly valuable biomarkers for early stage diagnosis of cancer. Exosomes are capable of transferring miRNAs to other cells in order to regulate specific gene expression by suppressing or degrading the relevant mRNAs. Horizontal transfer of miRNAs from tumor cells to normal recipient cells causes transcriptional reprogramming, which can affect tumor progression and metastasis [21]. Although both free circulating ncRNAs and tumor-derived exosomal ncRNAs have the same origin, the higher stability of exosomal ncRNAs has made them more powerful biomarkers. Exosomal ncRNAs are encapsulated within the vesicular (phospholipid) membrane and are therefore protected from the degrading enzymes such as RNAses [22]. On the other hand, exosomes can also contain proteins attached to DNA, RNA or lipoprotein complexes. Therefore, the exosomes are more stable under adverse physical conditions, such as extreme pH, repeated freeze-thawing, and prolonged storage time [23].
On the other hand, due to the absence of contaminating matrix proteins and other biomacromolecules, exosomes show reduced assay noise and therefore have higher sensitivity and specificity compared to other biomarkers. Several studies have shown the superiority of exosome-based biomarkers compared to conventional serum and urine biomarkers [24, 25]. For example, exosomes containing miRNAs isolated from sera showed 90% sensitivity in colorectal cancer, while the sensitivity of carcinoembryonic antigen (CEA) and carbohydrate antigen 19–9 (CA19-9) both typical biomarkers in CRC diagnosis was 30.7% and 16%, respectively [26]. Furthermore, in transcriptome analysis of RNA, it is difficult to differentiate the noise signals from normal cells from the signals of malignant cells. Furthermore, because of the strong RNA background from megakaryocytes created by platelet-derived RNA, RNA obtained directly from plasma is much more difficult to evaluate. Because exosomes carry tumor-specific surface proteins, they are capable of eliminating normal cell noise [27].
Another advantage of using exosomes as biomarkers is the possibility of monitoring the patient’s condition and response to treatment. The abundance and easy access to exosomes in bodily fluids make it possible to monitor and track the patient’s response to anticancer treatment in real-time. If the cancer marker directly corresponds to the clinical status, the amount of the biomarker should alter if the patient responds to treatment [28]. Despite the advantages of exosomal RNAs as cancer diagnostic biomarkers, their effective use in clinical applications requires rapid and effective isolation methods and extraction from patient samples, which remains challenging. We discuss exosome isolation methods and the advantages and disadvantages of each below. The selection of exosome-based biomarkers and their clinical applications are illustrated in Fig. 2, adopted and modified from [29].
Validation steps for exosomal biomarkers for clinical applications. A possible flowchart for the validation and clinical use of extracellular vesicle-based biomarkers. The flowchart depicts a step-by-step procedure for analyzing and discovering exosomal cargo molecules, which might eventually be turned into a therapeutically useful biomarker signature. To advance to the next level, stated goals and criteria must be accomplished at each phase. The figure is adopted and modified from [29]
Isolation of exosomes, methods, and challenges
Various methods are used to separate exosomes based on their morphology, size, protein markers, and flotation density. Conventional methods for isolating exosomes from cell culture medium or bodily fluids include ultracentrifugation, ultrafiltration, immunoaffinity, size exclusion chromatography (SEC), commercial kits, and microfluidic-based techniques. The progress in this field, especially in the last decade, has been significant; however, the isolation of exosomes with high speed and purity is challenging for the following reasons: first and most importantly, the high complexity of biological samples; secondly the heterogeneity of exosomes; and thirdly the similarity of the biochemical and physiochemical properties of exosomes, which makes it challenging to work with them compared to other EVs [30].
Currently, ultracentrifugation is the gold standard method for exosome separation, which separates similar exosomes from other components based on the difference in density and size. However, the disadvantages of this method are time-consuming, poor reproducibility, low stability, and lack of accurate separation of exosomes (based on size and not contents). In addition, ultracentrifugation has a high possibility of impurities, with protein and lipoprotein contamination in the final product [31].
In the ultrafiltration method (the simplest separation method), exosomes are passed through a membrane and separated based on size and molecular weight. However, despite its simplicity, ultrafiltration is very time-consuming and reduces the life and stability of the exosome membrane. On the other hand, in this method, the possibility of binding exosomes to the membrane is high, which leads to insufficient amounts of exosomes for subsequent analyses. In other words, the final product can be insufficient to produce good results [32].
In the SEC technique, gravity flow is used, and as a result, the structure and integrity of exosomes remain intact, and their biological activity is maintained. In addition, SEC is highly reproducible, but its major limitation is the requirement for complex equipment. Moreover, the SEC process is time-consuming and this restricts its application on a large scale for clinical purposes [33]. In the immunoaffinity method, a recognition between antibody and antigen is used. In general, the surface of exosomes is rich in various receptors and proteins, and these molecules have the ability to bind to their specific antibodies. Unlike the previous methods, this method has high specificity. However, the final product, although pure, is confined to the isolation of a particular subset of exosomes [34]. Furthermore, tumor heterogeneity in antigen expression and antigen modification as the tumor progresses may result in underestimation and false negative results. The antigenic epitope may also be inhibited or disguised. Although the initial steps are simple in exosome-antibody precipitation methods, the sample preparation and purification steps are tedious and time-consuming [34]. Finally, if the purity is poor, it can cause errors in the data analysis steps. For example, exosome precipitation in serum and plasma samples leads to simultaneous precipitation of other cellular particles such as protein aggregates and other EVs. Furthermore, the varying viscosity of the sample matrix leads to distinct exosome precipitation stringency requirements, compromising the standardization of precipitation techniques.
Microfluidics is a revolutionary technique for exosome isolation that has advantages of rapidity, high purity, high recovery rates, and has a wide range of applications [35]. Despite significant advances in microfluidics techniques, these devices are not yet suitable for clinical applications. Significant obstacles such as lack of validation, scalability, and standardization have limited their use. In addition, some devices require pretreatment of the samples, which takes more time. Some types of microfluidics devices suitable for intact samples have low separation efficiency. All microfluidics devices are designed to isolate a specific class of exosomes; therefore, the volume of their products is small and specific. In addition, their processing capacity is low, and the lack of sufficient DNA, RNA or protein inside the extracted exosomes limits the subsequent analysis steps [36].
Exosomes have been widely studied in the laboratory using mass spectrometry, fluorescent cell activation sorting (FACS), Western blotting, flow cytometry, electron microscopy, and liquid chromatography. Although nanoparticle tracking analysis and dynamic light scattering approaches are rapid, dependable, and semi-quantitative, they are not be able to distinguish between different exosome types. Microarray analysis, surface plasmon resonance, and using specific antibodies against proteins in the extracellular domain of exosomes have been applied for detection and characterization of tumor-derived exosomes quantitatively without any purification and/or enrichment, resulting in a simple and a more effective novel technique for exosome detection and their characterization [37]. The next-generation sequencing (NGS) method and high-throughput RNA sequencing (RNA-seq) have made it possible to quickly and accurately scan the nucleic acid contents of exosomes. Using RNA-seq, the isolated exosome contents can be accurately determined, and the cancer diagnostic markers monitored and analyzed on a large scale [38].
Breast cancer exosomal miRNAs (Exo-miRNAs)
There have been many reports of cell-derived exosomes occurring in breast cancer. Based on these results, exosomes containing cancer-derived miRNAs could be involved in tumor initiation, angiogenesis, epithelial-mesenchymal transition (EMT), tumor-stromal communication, and treatment resistance [39]. In addition, these exosomes are highly heterogeneous, and in a single cancer sample, there may be several types of exosomes with different miRNA profiles. MiRNAs are clustered in a non-random manner within exosomes [40]. The cargo carried by each exosome is governed by the type of cell from which it was produced. Exosomal information is transferred in various ways to distinct types of cells in the tumor microenvironment [41]. NGS techniques and microarrays can make it easier to study exosomes, and determine the role of their contents in breast cancer. These findings will be useful for early diagnosis, prognosis, and prediction of treatment response in breast cancer patients.
Tumor-derived miR-9 was detected inside exosomes secreted from breast cancer by Baroni et al. [42]. miR-9 is able to transform normal fibroblasts into cancer-associated fibroblasts (CAFs), which promote breast tumor growth and metastasis. In addition, the role of other miRNA-containing exosomes in the cellular processes involved in tumorigenesis and progression has been elucidated. For example, miR-143, miR-378, and miR-21 contained in CAF-derived exosomes can induce the EMT in breast cancer cells [43]. CAFs can also secrete exosomes containing miR-181d-5p, which promotes EMT, cell division, cell migration, invasion, and suppresses apoptosis by inhibiting HoxA5 and CDX2 in breast cancer [44, 45].
In another study, exosomal-miR-222 was associated with increased metastasis in breast cancer cells. This was explained by the fact that miR-222 directly targets the PDLIM2-tumor suppressor gene, and consequently activates the NF-κB signaling pathway [46]. One study by Camacho et al. also showed that exosomes derived from MDA-MB-231 breast cancer cells contained high levels of miR-210, which promoted angiogenesis and brain metastasis in breast cancer. Survival rates are usually poor in patients with breast cancer brain metastases with high levels of miR-210 [47]. Surprisingly, Le et al. showed that the metastatic potential could be transferred from metastatic cancer cells to non-metastatic cancer cells through exosomal miRNAs [48]. In their study, exosomes containing miR-200 secreted by breast cancer cells transferred metastatic properties to non-metastatic cells as demonstrated in human xenograft mouse models [48, 49]. The transfer of miR-770 and miR-105 via exosomes can regulate the migration and metastasis of breast cancer cells [50, 51]. These findings illustrate the role of tumor-derived exosomes in cancer progression and metastasis and highlight the importance of studying exosomal miRNAs as diagnostic and predictive biomarkers. Higher levels of these exosomes could act as a warning sign that the cancer is progressing towards metastasis.
Elevated serum exosomal miR-373 levels have been found in receptor-negative breast tumors, and could be used as a diagnostic biomarker [52]. An increase in exosomes containing miR-21 and miR-1246 was found in the plasma of breast cancer patients compared with healthy individuals in a study by Hannafon et al. [53]. A recent study by Shen et al. also showed that exo-miR-7641 could promote the progression and metastasis of breast cancer [54]. MiR-7641 plasma levels in patients with distant metastases were much higher than in healthy individuals. Increased exo-miR-7641 could explain the connection between tumor growth and metastasis. In addition, Shen et al. suggested that exo-miR-7641 could be a non-invasive diagnostic biomarker, and could also be a target for breast cancer treatment [54].
Some exosomal miRNAs have been associated with survival and response to treatment and could therefore be used as predictive markers for patient response to treatment. For example, increased levels of exo-miR-455-5p and exo-miR-255 were associated with low patient survival rates, and could be considered to be therapeutic targets [55]. In a study by Jaiswal and colleagues it was indicated that exosomes derived from doxorubicin-resistant breast cancer cells contained high levels of miR-362, which was involved in the drug resistant phenotype [56,57,58]. According to other studies, some exosomal RNAs, such as miR-100, miR-30a, miR-222, and miR-17 derived from breast cancer cells, have been implicated in resistance to paclitaxel and doxorubicin [59]. A study by Shah and coworkers showed that exosomes derived from breast cancer associated fibroblasts (CAFs) suppressed estrogen receptor expression in ER-positive breast cancer cells, which may explain the effects of CAFs on tumorigenesis and treatment resistance to treatment in breast cancer [60]. Exosome-mediated cell reprogramming could alter the metabolic profiles of cancer cells and the tumor microenvironment, leading to the acquisition of properties in cancer cells that make them more resistant to therapy. Cancer cells can transmit these properties to each other with the help of exosomal cargos. According to Fong et al. breast, cancer-derived exosomes containing miR-122 could alter glucose metabolism in favor of cancer progression, and inhibition of miR-122 could reduce lung and brain metastasis in breast cancer [61]. MiR-155 is also involved in altering energy metabolism in cancer cells, and acts as an oncogenic signal transmitted through secreted exosomes in breast cancer. Exo-miR-155 led to cell reprogramming and cancer-associated cachexia by targeting PPARγ [62]. In addition to reprogramming of cellular metabolism, cancer-derived exosomal miRNAs can also induce the cancer stem cell (CSC) phenotype, thus enhancing tumor progression. According to a study by Shen et al., exo-miRNAs (miR-203a-3, miR-195-5p, and miR-9-5p) were released in response to neoadjuvant chemotherapy. These exo-miRNAs reduced the expression of the One Cut Homeobox 2 (ONECUT2) transcription factor to induce the CSC phenotype in adjacent breast cancer cells [63] (Fig. 3)
Role of exosomal ncRNAs and their related pathways in breast cancer progression. miR-455 activates the TGF-beta receptor, ultimately leading to activation of its canonical pathway and increases the division, migration, and EMT of breast cancer cells. miR-222, miR-9, and miR-155, by inhibiting PTEN lead to the activation of Bcl2 (apoptosis inhibitor), which ultimately promotes cell division and survival. On the other hand, LncRNA SNHGH14 enhances apoptosis by inhibiting Bcl2, and was found to be reduced as a tumor suppressor in breast cancer. lncRNA XIST leads to increased cell division and tumor progression by regulating the miR-362/UBAP1 axis. CircWHSC1 increases EMT and invasiveness of cancer cells by inhibiting FASN/miR-455-5P axis. miR-373 acts as an estrogen receptor inhibitor and is overexpressed in breast cancer. EMT epithelial–mesenchymal transition, UBAP1 ubiquitin associated protein 1, PETN Phosphatase and tensin homolog, FASN fatty acid synthase
Exosomal lncRNAs in breast cancer
Dysregulation of lncRNA expression in breast cancer patients has been the subject of extensive research [64]. For example, increased exosomal lncRNA-UCA1 in breast cancer cells has been associated with increased resistance to tamoxifen. This resistant phenotype can be transferred to other cells via exosomal-UCA1 [65]. Similar results were obtained for SNHG14 in a study by Dong et al. [66]. In trastuzumab-resistant cells, the expression of SNHG14 was higher, suggesting that exosomal SNHG14 could be a suitable biomarker for monitoring treatment response [66]. As mentioned above, cancer cells and adjacent tumor cells can exchange their phenotype through the effects of exosomes. Transfer of the endogenous signal recognition particle RNA, RN7SL1 contained in exosomes between breast cancer cells, immune cells, and stromal fibroblasts resulted in increased tumor growth, metastasis, inflammation, and drug resistance [67]. Previous studies have shown high levels of lncRNA MALAT1 expression in breast cancer and confirmed its association with drug resistance. Recently, increased levels of exosomes containing MALAT1 have been shown in breast cancer, and have been proposed to be responsible for disease progression [68]. HOTAIR is a lncRNA found in circulating exosomes, and was found to be positively linked with ErbB2 (HER2/neu) expression, implying that it could be used as a liquid biomarker [69]. Exo-AFAP-AS1 is another known exosomal-lncRNAs in drug resistant breast cancer, which binds to the AU-rich element RNA-binding protein 1 (AUF-1) and promotes trastuzumab resistance by activating ERBB2 translation [69]. Some fibroblast-derived exosomes contain SNHG3, which are involved in cell reprogramming thereby altering cell metabolism and increasing the proliferation of breast cancer cells [70]. Doxorubicin resistance can also be transmitted to breast cancer cells via lncRNA-H19 in exosomes derived from cancer cells [71]. The exosomal-ncRNAs involved in breast cancer progression and their mechanism of action are summarized in Fig. 1.
Exosomal ncRNAs involved in lung cancer
About 20% of cancer deaths are due to lung cancer, which is the most common malignancy worldwide [72]. Lung cancer includes two main histological categories: non-small cell lung cancer (NSCLC) and small cell lung cancer. Eighty-five percent of all lung cancers are NSCLC, underlining the need for further study and improved treatment for this type of cancer [73]. In recent years, the expression profiles of several ncRNAs in lung cancer cells have been investigated. Increased expression of many of these circulating ncRNAs in lung cancer cells is found in EVs, including exosomes. Exosomal lncRNAs are responsible for the increased serum levels of MALAT1 in NSCLC [68]. Similar to breast cancer, increased MALAT1 expression promotes tumor metastasis and invasion. MALAT1 inhibits apoptosis and promotes cell division. Therefore, exosomes containing MALAT1 could serve as biomarkers for the diagnosis and prognosis of NSCLC [68]. Exosomal lncRNAs in lung cancer cells can also be altered following interaction with mesenchymal stem cells (MSCs), according to one report [74, 75]. MSCs are thought to act as tumor-promoting cells by encouraging metastasis and invasion, and also by inhibiting the immune response. One study identified the top ten altered exosomal lncRNAs in the NSCLC cell line A549, following treatment with MSCs. This study suggests that cancer-derived exosomes can be affected by MSCs via aberrant lncRNA expression [74] (Table 1).
Deng and colleagues found the role of exosomes containing lncRNA MSTRG.292666.16 in osimertinib-resistant NSCLC [76]. In recent years, many studies have focused on targeting epidermal growth factor receptors (EGFR), and EGFR tyrosine kinase inhibitors (TKIs) have shown promise in the treatment of lung cancer. However, in many cases, mutations occurring in EGFR alleles can cause resistance to TKIs, such as osimertinib [77]. Osimertinib is a third-generation anti-EGFR inhibitor that can inhibit a variety of mutated alleles. Understanding TKI resistance mechanisms could pave the way to find better treatments to increase patient survival [78]. In a study by Deng et al., exosomes derived from patients with NSCLC were isolated before and after treatment with osimertinib, and their lncRNA profiles were determined. Differentially expressed lncRNAs were found in osimertinib-resistant and osimertinib-sensitive exosomes, and the activity of one key lncRNA in promoting osimertinib resistance was investigated in vivo [76]. This study showed that exosomes containing MSTRG.292666.16 in plasma of osimertinib-resistant patients were significantly increased compared to drug-sensitive patients. Based on these results, exosomal-MSTRG.292666.16 was implicated in the development of a drug-resistant phenotype. Therefore, it could be used as a biomarker to predict treatment response, and to identify TKI-resistant patients, as well as a possible therapeutic target [12]. In another study, Zhou et al. [79] examined the molecular mechanism of EGFR TKI resistance in the NSCLC cell line H1975. The study investigated the relationship between macrophages and cancer cells in the tumor microenvironment mediated by exosomes. Macrophages have two different phenotypes, M1 and M2 macrophages. M1 macrophages are responsible for producing proinflammatory cytokines, interleukin-6 (IL-6), IL-12, and TNF-α. On the other hand, M2 macrophages produce anti-inflammatory cytokines, including IL-10 and TGF-β [80]. In the tumor microenvironment, macrophages show a M2 phenotype and are known as tumor-associated macrophages (TAMs), which can suppress the immune response and promote angiogenesis. According to studies, a high ratio of M2/M1 is associated with a high rate of cell division, invasion, migration, as well as drug resistance in lung cancer cells [80, 81]. It has recently been shown that the lncRNA SOX2 overlapping transcript (SOX2-OT) is present at a high concentration in serum-derived exosomes from cancer patients [82]. A study by Zhou et al. showed that NSCLC cells can increase the M2 polarization of macrophages by the transfer of SOX2-OT exosomes, thereby increasing resistance to EGFR-TKIs. The mechanism of action of SOX2-OT is to regulate the expression of genes involved in M2 polarization. This study showed that SOX2-OT could target miR-627-3p, thereby increasing the expression of genes, such as Smad 2, Smad 3, and Smad 4. These studies underline the importance of investigating tumor-derived exosomes in treatment-resistant tumors [79]. On the other hand, the treatment response could be predicted by determining the concentration of specific ncRNAs in exosomes as biomarkers. As a result, more effective decisions could be made to select the best treatment. Due to the limited time available to treat cancer and prevent its metastasis, the identification of such biomarkers could play a vital role in time management during cancer treatment.
Distal‑less homeobox 6 antisense RNA 1 (lncRNA DLX6‑AS1) expression was found to be increased in various solid tumors. Zheng et al. demonstrated its role in the diagnosis of NSCLC by measuring its expression in tumor-derived exosomes [83]. The level of DLX6‑AS1 expression in lung cancer tissue samples was significantly higher compared to healthy controls. In addition, it was shown that the knockdown of DLX6‑AS1 in the NSCLC cell line could reduce proliferation and migration. Interestingly, the results showed that the sensitivity and specificity of DLX6‑AS1 were higher compared to CYFRA21‑1 when used as a diagnostic biomarker. CYFRA21‑1 is considered to one of the most reliable biomarkers for the diagnosis of NSCLC. Therefore, the higher DLX6‑AS1 levels in circulating tumor-derived exosomes from patients with NSCLC could be an acceptable diagnostic biomarker for early detection [83]. Current studies have shown that the expression of lncRNA-GAS5 (a tumor suppressor lncRNA) is lower in most cancers. In a study by Li et al., exo-GAS5 was studied in NSCLC patients and showed a significant reduction compared to exosomes derived from healthy individuals. Based on these findings, the expression of exo-GAS5 was related to the cancer stage, so that in more advanced cancer the expression level of exo-GAS5 was sharply lower. Exo-GAS5 might therefore be used to differentiate stage I NSCLC patients from other stages. Finally, exo-GAS5 might be a non-invasive serum-based biomarker for detecting patients with early NSCLC [84].
One non-invasive approach to diagnose cancer, especially in the early stages of the disease, is to measure the concentration of biomarkers in bodily fluids, such as plasma, serum, sputum, and urine. Examination of exosomes is doubly useful, because ribonucleases usually degrade circulating RNAs in the bloodstream, while exosomes can protect them. Although much effort has focused on diagnostic biomarkers in lung cancer, only a handful of these studies have reported specific miRNAs profiles for early-stage NSCLC [85]. Detection of exosomal miRNAs in bodily fluids has potential for clinical diagnosis and prognosis of NSCLC. In a study by Jin et al., tumor-derived exosomes were isolated from patient plasma [86]. Using the miRNA-seq technique, the profile of these exosomes in patients was compared with healthy individuals. In this study, in addition to determining the miRNA expression profile, six specific exosomal-miRNAs for lung adenocarcinoma were identified, which had much higher expression than healthy controls. Jin et al. showed that these tumor-derived exosomes (TMEs) were only elevated in patients with lung cancer and were not detected in healthy individuals. Comparison of miRNAs in TMEs with the whole plasma miRNA profile can eliminate the interference of exosomal miRNAs secreted by normal cells. The levels of circulating plasma miRNAs were not correlated with tumor-derived exosomal miRNAs, indicating that the miRNA content of cell-free plasma samples could be different from that of plasma exosomes. In both circulating RNA and exosomal RNA, the miRNA expression levels employed for cancer diagnosis or prognosis might be different producing unreliable results. The findings of this study might explain some of the differences in expression patterns observed between serum, plasma, and cell-free exosomes, which have been identified in prior investigations. Their results finally identified exosomal miR-21-5p and miR-24-3p as prognostic and diagnostic biomarkers for lung cancer [41]. Analysis of TMEs in NSCLC patients showed a significant increase in the expression of miR-4257 and miR-21 in patients with recurrent disease [87]. In contrast, the expression of miR-51 and miR-373 was lower in TMEs from lung cancer patients and was associated with a poor prognosis [88]. Other exosomal miRNAs have been identified as biomarkers of therapeutic response in lung cancer, including miR1246 and miR-208a, which bind to DR5mRNA and P21, respectively. These miRNAs can lead to faster tumor growth and increased resistance to radiotherapy [89, 90]. Moreover, in another study, exo-miR-23b, exo-miR-21-5p, exo-miR-146a-5p, and exo-miR-10b-5p were investigated as prognostic biomarkers in NSCLC patients [91]. Zhang et al. studied tumor-derived exosomes containing miR-5684 and miR-125b-5p as biomarkers for NSCLC using a microarray method. They found a lower expression of exo-miR-125b-5p, which was also associated with a poor response to chemotherapy, suggesting that this exosomal miRNA could be a prognostic marker in NSCLC [92].
Tang et al. investigated whether miR-620 could be a diagnostic biomarker in NSCLC [93]. Recent studies have shown that miR-620 is involved in various biological processes, including cell division, and its expression was lower in various cancers [94]. Exo-miR-620 in lung cancer patients showed a pronounced difference compared to healthy individuals, and was significantly lower in patients with metastatic NSCLC. In addition, Tang et al. found that exo-miR-620 expression was associated with a good response to chemotherapy. Based on this finding, this research group suggested that exo-miR-620 could be a suitable diagnostic biomarker for lung cancer [93]. Kim et al. showed the role of miR-619-5p in angiogenesis and metastasis in NSCLC. They found that NSCLC-derived-miR-619-5p targeted the RCAN 1.4 gene [95]. The endogenous protein human regulator of calcineurin 1 (RCAN1) binds to calcineurin, and subsequently suppresses its activity by interfering with the calcineurin–nuclear factor of activated T-cells (NFAT) pathway [96]. RCAN1.4 has a variety of biological functions, such as VEGF-mediated angiogenesis, cardiac hypertrophy, and defending against calcium-mediated oxidative stress [97, 98]. In NSCLC, RCAN1.4 is targeted by miR-619-5p resulting in its lower expression. Based on the results of a study by Kim et al. using exosomes extracted from the plasma of NSCLC patients, miR-619-5p expression was higher than expected. Finally, hypoxic conditions caused more miR-619-5p to be incorporated into exosomes from NSCLC cells. Exosomal miR-619-5p promotes the development and metastasis of NSCLC through regulating RCAN1.4, suggesting that it might be a diagnostic marker for lung cancer [95]. miR-3157-3P is another tumor-derived exosomal miRNA associated with lung cancer. According to recent studies, exo-miR-3157-3P promotes angiogenesis, vascular permeability, and metastasis. In patients with metastatic lung cancer, the expression level of miR-3157-3P in circulating exosomes was much higher than in patients with non-metastatic lung cancer [99]. Ma and colleagues found that miR-3157-3P could be transported in exosomes from lung cancer cells to vascular epithelial cells, and contributed to formation of a pre-metastatic niche. MiR-3157-3P targeted the TIMP/KLF2 pathway, and thus led to increased expression of matrix metalloproteinases (MMP) 2, MMP9, and VEGF in endothelial cells. Ma et al. suggested that tumor-derived exo-miR-3157-3P could be used as a blood-based biomarker to diagnose metastatic lung cancer [100].
Colorectal cancer (CRC)
Colorectal cancer (CRC) is the third most common cancer worldwide and ranks second in mortality. Early diagnosis of CRC is vital for more effective treatment and improved patient survival [101]. Several studies have shown that dysregulation of exosomal-ncRNA expression is involved in various tumorigenic processes in CRC [3, 102]. Therefore, exosomal-ncRNAs have the potential to be used as diagnostic and prognostic biomarkers. The studies on exosomal-ncRNAs associated with CRC have been relatively extensive, compared to other cancers.
Exosomal miRNAs in CRC
According to a study by Li et al., decreased expression of miR-149 and miR-96 was observed in both CRC tissues, and exosomes derived from patients. This was associated with decreased apoptosis, increased cell division, and increased GPC1 expression [103, 104]. The exo-miR-92 expression was also significantly lower in CRC patients compared to healthy individuals. Decreased expression of exo-miR-92 was observed primarily in patients with more advanced CRC stages, and not in non-cancerous lesions [105]. In contrast, tumor-derived exosomes containing miR-320, miR-181, miR-17, miR-125a, miR-18a, and miR-18b were significantly higher in CRC patients [106]. According to recent findings, exo-miR-125a and exo-miR-320 have a higher expression in the early stages of CRC and could therefore be considered as diagnostic biomarkers [107]. Typically, two blood-based biomarkers are screened for colorectal cancer diagnosis, carbohydrate antigen 19–9 (CA19-9) and carcinoembryonic antigen (CEA) [108]. Studies have shown that the efficiency and sensitivity of these diagnostic biomarkers, were significantly higher when combined with some exosomal miRNAs [109]. For example in patients with metastatic CRC, miR-320d has been reported as a biomarker with 64.7% specificity and 62% sensitivity. The combination of exo-miR-320d and CEA increased the specificity up to 91.3% [110]. Due to the differences in the expression profile of these exosomal miRNAs between the early or advanced stages of the disease, they can be used for early detection or diagnosis of metastatic CRC. Some other exo-miRNAs have been studied as prognostic biomarkers in CRC. For example, the expression level of exo-miR-193a was higher in CRC metastatic cell lines, and in plasma from patients with CRC lung metastasis. Increased expression of exo-miR-193a has been associated with decreased survival of CRC-bearing mice [103]. Another study by Liu et al. showed an association between lower expression of exo-miR-4772-3p and recurrence in CRC patients [111]. Decreased expression of exo-miR-4772-3p was associated with a higher risk of cancer recurrence and death in patients. A connection between decreased expression of exo-miR-6869 and toll-like receptor 4 (TLR4) expression has been observed in serum exosomes from CRC patients. Decreased expression of exo-miR-6869 is related to increased cell division, inflammatory cytokines, activity of the NF-κB pathway, and decreased apoptosis. Therefore, a decrease in serum exo-miR-6869 levels was associated with poor three-year survival rates in CRC patients [112]. In contrast, higher expression of some exo-miRs, such as exo-miR-1229, has been observed in CRC patients compared with healthy individuals, and was associated with lymph node metastasis and poor survival, High levels of exo-miR-19a were associated with CRC progression, and increased liver and lymph node metastasis [113]. In addition, other exo-miRNAs can predict the response to treatment. The applications of CRC-derived exosomes as biomarkers are listed in Table 2.
Exosomal lncRNAs in CRC
Recent studies have shown the role of various lncRNAs in cellular processes, including tumorigenesis and tumor growth, by regulating miRNA expression. For example, urothelial carcinoma-associated 1 (UCA1) was higher in CRC tissue samples, but lower in the serum exosomes of CRC patients compared to healthy individuals [141]. The sensitivity of using the UCA1 level to distinguish CRC patients from healthy individuals was 100%, while the specificity was more than 40%. In addition, UCA1 was found to reduce the expression level of miR-135a, miR-143, miR-214, and miR-1271 [141]. In a study by Liu et al., the level of the lncRNA called colorectal neoplasia differentially expressed–h (CRNDE-h) was higher in CRC cell lines and exosomes derived from patients, compared to healthy individuals. Moreover the high level of CRNDE-h was associated with regional lymph node metastasis, distant metastasis, and poor patient survival. Exosomal CRNDE-h levels were found to be an independent predictive factor for five-year overall survival rates [117]. They also looked at the effect of combining serum exosomal CRNDE-h and CEA levels to improve the prognostic prediction. It was discovered that patients with low levels of both exosomal CRNDE-h and CEA, followed by low CRNDE-h and high CEA, then high exosomal CRNDE-h alone, and finally patients with high levels of both biomarkers had progressively worse overall survival rates [117]. Like lung cancer, exo-GAS5 levels in CRC were associated with disease progression. GAS5 inhibited the expression of miR-221, which is involved in cell division, migration, and invasion of CRC cells [116]. A study by Oehme et al. demonstrated a decrease in lncRNA HOTTIP expression in CRC patients. Therefore, HOTTIP could be a prognostic biomarker for a poor prognosis in CRC patients [119].
Circular RNAs (circRNA) are a class of ncRNAs with a closed loop-like structure and are highly resistant to exonucleases. Some recent studies have examined the level of exosomal circRNAs in CRC [142]. For example, exosomal-circFMN2 and exosomal-circIFT-80 were both higher in CRC, and were correlated with tumor size, tumor stage, and distant metastasis [140, 143]. However, a human colonic epithelial cell line did not respond to circFMN2 knock-down. Cell proliferation was not reduced by circFMN2 knock-down or enhanced by circFMN2 over-expression. Because normal cells were not affected, it was suggested that circFMN2 could be a possible therapeutic target. Furthermore, knock-down of circIFT80 increased CRC cell apoptosis, but knock-down of circFMN2 had no effect on CRC cell apoptosis. Furthermore, circIFT80 was shown to promote the EMT. CRC cell migration and invasion were increased by exogenous over-expression of circIFT80. CircFMN2 was found to affect the miR-1182/hTERT axis, while circIFT80 affected the miR-1236-3p/HOXB7 axis [140, 143]. The expression of hTERT and HOXB7 was both higher in CRC tissues than in normal tissue. Furthermore, in both cell lines and serum exosomes from CRC patients, there was an inverse association between the expression levels of these circRNAs and their corresponding miR targets. Furthermore, knock-down of circFMN2 and circIFT80 reduced the expression of hTERT and HOXB7, respectively. This effect was counteracted by using a miRNA inhibitor to target their specific miRNA targets. These findings suggest that the exosomal circRNAs circFMN2 and circIFT80 could be used as prognostic biomarkers [64, 143].
Hon et al. studied the role of hsa-circ-000338 in predicting the response to the treatment in CRC patients [138]. The level of exosomes containing hsa-circ-000338 was two times higher in CRC patients showing resistance to 5-FU and FOLFOX (oxaliplatin). According to this study, exosomes containing hsa-circ-0000338 could transmit it to non-resistant cells, thus imparting 5-FU resistance to the CRC cell line. Therefore, exo-hsa-circ-0000338 could be a predictive biomarker for response to treatment in CRC patients [138]. According to a study by Pan et al., exo-hsa-circ-0004771 levels were > 14-fold higher in CRC patients compared with healthy individuals. Exo-hsa-circ-0004771 was associated with disease severity and metastasis and had more than 80% specificity and sensitivity to differentiate CRC patients from healthy controls. Therefore, exo-hsa-circ-0004771 could be a diagnostic biomarker for CRC [139].
Exosomal lncRNAs in bladder cancer
Bladder cancer is the sixth most common cancer in men worldwide. The diagnosis of bladder cancer relies on the histopathological evaluation of cystoscopic biopsy specimens. The high percentage of false positives or false negatives, high cost, and patient discomfort are the main disadvantages of this approach [101]. Non-invasive biomarkers that can be detected in urine samples, such as bladder tumor antigen (BTA) and nucleus matrix protein 22 (NMP 22) have been used to detect early-stage bladder cancer [144]. However, scientists are still seeking to identify more efficient and less expensive biomarkers for the diagnosis of early-stage bladder cancer. Similar to studies in other cancers, exosomal-ncRNAs are being investigated in bladder cancer as promising diagnostic biomarkers in urine. Higher expression of several lncRNAs, such as PCAT-1, PVT-1, and ANRIL, has been reported in bladder cancer specimens [145]. Abbastabar et al. examined the expression level of these exosomal lncRNAs in patients with bladder cancer. The level of urinary exosomes containing PCAT-1 and ANRIL was significantly higher in patients with bladder cancer compared to healthy individuals. PVT-1 can play an oncogenic role in cancer by interacting with MYC, regulating miRNAs, and participating in DNA rearrangement. However, exosomal PVT-1 in bladder cancer patients was not significantly different from exosomes in healthy controls. However, increased ANIRL and PCAT-1 in exosomes from bladder cancer patients could be valid diagnostic biomarkers [145]. Zheng et al. evaluated exosomal PTENP1 in bladder cancer patients as a diagnostic biomarker [146]. According to this study, exo-PTENP1 could be transmitted to bladder cancer cells, thereby increasing apoptosis, and preventing invasion and migration. Exo-PTENP1 controlled tumor growth in vivo, by regulating the expression of the tumor suppressor PTEN (phosphatase and tensin homolog) by competitively binding to miR-17. Therefore, Zheng et al. suggested that lower exosomal PTENP1 levels could be a biomarker for detecting bladder cancer progression [146]. Berrondo et al. [147] measured the expression of lncRNA HOTAIR in urinary exosomes of patients with bladder cancer. HOTAIR can regulate several genes involved in the EMT, including SNAIL, LAMB3, and LAMC2. HOTAIR has also been associated with cell migration and invasion, and the recurrence of high-grade non-muscle invasive UBC (NMIBC) could be predicted by its high expression. This study showed that urinary exosomes containing HOTAIR could be a diagnostic biomarker in bladder cancer patients. They also identified four additional lncRNAs in their exosomes, including OTX2-AS1, HYMAI, LINC00477, and Loc100506688 [147].
BCYRNI is a lncRNA involved in lymphangiogenesis in bladder cancer [148]. BCYRNI promotes the increased activity of VEGF/VEGFR3 via the hnRNPA1/WNT5A/VEGFR3 axis. Zheng et al. [148] showed that exosomes containing BCYRNI were associated with lymph node metastasis and poor prognosis in bladder cancer patients. They suggested that exo-BCYRNI, in addition to its role as a diagnostic biomarker, could also be an effective therapeutic target. They demonstrated that silencing of exo-BCYRNI suppressed VEGF/VEGFR3 signaling, and prevented lymphangiogenesis and lymph node metastasis in bladder cancer, both in vivo and in vitro [148]. A panel of bladder cancer-derived exosomes was recently studied to identify biomarkers for diagnosis and prediction of relapse. Altogether 11 candidate lncRNAs were screened inside the isolated urinary exosomes in this study, the only highly accurate biomarkers were SNHG16, UBC1, and PCAT-1. The findings of this study also suggested exo-lncRNAs could be accurate biomarkers with greater availability and faster processing for detecting bladder cancer and predicting its recurrence [149].
Exosomal miRNAs in bladder cancer
Recent studies have compared miRNAs in tumor tissue, urinary exosomes, plasma, and white blood cells taken from bladder cancer patients. The results identified 7 hallmark miRNAs (21-5p, 205-5p, 29b-3p, 200c-3p, 4454, and 200b-3p) with higher expression levels in bladder cancer-derived exosomes [150]. These exosomal miRNAs reflected the molecular profile from the source tumor cells, and could be biomarkers for tumor detection. Studies in recent years have examined the expression level of exosomal miRNAs isolated from bladder cancer patients. For example, one study found that 22 separate exo-miRNAs were increased, while 23 exo-miRNAs were decreased in bladder cancer patient urine samples [151]. In a microarray technique-based study, miR-375 and miR-146a were tested to identify high and low stages of bladder cancer compared to healthy individuals [152]. Significant increases in the expression of five miRNAs (miR-31-5p, miR-155-5p, miR-21-5p, miR-132-3p, and miR-15a-5p) in exosomes from bladder cancer patients were reported compared to healthy controls. Of these, exo-miR-21-5p has shown potential as a diagnostic biomarker in bladder cancer [153]. Some exosomal miRNAs have also been studied as prognostic and predictive biomarkers in bladder cancer. For example, low expression of exo-miR-103a-3p and exo-miR-486-3p were associated with worse patient survival [154]. According to current studies, some exosomal miRNAs can induce resistance to cytotoxic drugs. For example, exo-miR-155 promoted resistance to paclitaxel and doxorubicin in a variety of cancers, including bladder cancer [155]. One study by Fanous et al. [156] demonstrated the role of exosomal miRNAs in causing chemotherapy drug resistance in bladder cancer, including gemcitabine and cisplatin. In this study, a total of 759 exosomal miRNAs were screened, of which 16 showed significant changes in expression, which were directly related to drug resistance [156]. While there have been few studies of exosomal miRNAs as treatment response prediction markers, they can provide information about their originating cells, such as the molecular target, degree of malignancy, and potential therapy resistance. As a result, exosomal miRNA biomarkers may be used to monitor and even to reduce tumor resistance, to achieve a personalized therapy. Some studies on the role of bladder cancer-derived exosomal miRNAs as biomarkers are discussed in ref [157].
Gastric cancer
Gastric cancer is the fourth most common malignancy globally and the second leading cause of cancer death in the world, and is still considered a severe challenge to oncologists. Early detection of gastric cancer significantly increases the chance of effective treatment and consequently prolongs the survival rate of patients [158]. Conventional invasive biopsy procedures for diagnosis are often painful and have low sensitivity. The likelihood of precursor or premalignant lesions occurring in gastric cancer makes it challenging to diagnose gastric cancer using non-invasive biomarkers. These predisposing lesions include Helicobacter pylori infection, chronic atrophic gastritis, and intestinal metaplasia [159]. Therefore, investigating gastric tumor-derived exosomes could provide a non-invasive diagnostic method with a low error rate. Exosomal RNAs, in particular, are promising candidates for this purpose [160, 161]. In recent studies, exosomal lncRNAs, such as HOTTIP [162], ZFAS1 [163], and LINC00152 [164], were found to be markedly increased in patients with gastric cancer compared to healthy individuals. A comprehensive study by Lin et al. [165] used RNA-Seq techniques to screen novel potential biomarkers. Due to the importance of early stage diagnosis, the analysis was performed on early gastric cancer (EGC) specimens. Exosomal RNA profiles were determined in EGC samples, and 79 separate exo-RNAs showed a pattern of increased expression in patient samples compared to controls. In addition, this study performed RNA-seq analysis on four gastric cancer cell lines and found an increased expression of 289 separate exo-RNAs. Due to the commonality of lncRNA-UEGC1 between patient samples and cell lines, the follow on study focused on UEGC1. The results showed that almost all of the plasma UEGC1 was carried inside exosomes, and thus avoided degradation by RNase. Because UEGC1 was more accurate than CEA (a non-invasion diagnostic biomarker for gastric cancer), Lin et al. suggested that exo-UEGC1 could be a non-invasive, stable, and highly sensitive diagnostic biomarker for ECG [165].
Some recent studies have focused on the role of exosomal lncRNAs in the progression, angiogenesis, metastasis, and drug resistance of gastric cancer. For example, exosomes containing lncRNA HEIH secreted from gastric cancer cells could transform normal gastric cells into a malignant phenotype [166]. Exosomal miR-221 [167] and exo-miR-15b-3p [168] could both promote the migration and invasion of gastric cancer cells by targeting caspase-3 and -9, and inhibiting apoptosis. Exosomes could transport miR-130a from gastric cancer, to target c-MYB and increase angiogenesis in gastric tumors [169]. In general, tumor-derived exosomes can transfer ncRNAs and appear to play a key role in tumorigenesis and metastasis in gastric cancer, suggesting they could be biomarkers and practical therapeutic targets.
Recently, exosomal lncRNAs have been considered as clinical diagnostic and prognostic biomarkers, and therapeutic targets to reverse drug resistance. In a multi-phase study in over 800 gastric cancer patients, lncRNA GC1 was identified as a valid predictive biomarker [170]. In addition, the serum exosomes of gastric cancer patients showed significantly higher expression of miR-106a-5p and miR-19b-3p. These exo-miRNAs showed better sensitivity and specificity compared to CEA and AFP (accepted diagnostic biomarkers for gastric cancer), and could be non-invasive biomarkers for EGC [171]. Exo-LncRNA GNAQ-6 [172] has also recently been suggested as a new diagnostic biomarker. In addition to diagnostic biomarkers, exo-miRNAs could be used to predict metastasis and recurrence of gastric cancer. For example, Kumata et al. [173] found that the expression level of exo-miR-23b was directly related to tumor size, liver metastasis, depth of invasion, and TNM stage. They suggested that exo-miR-23b levels in gastric cancer could predict patient overall survival [173].
Peritoneal metastasis is the most common site of metastasis in gastric cancer, and is usually associated with short survival rates. In patients with peritoneal metastasis, the expression levels of exo-miR 1225-e5p, exo-miR-21, and exo-miR-29 were all significantly altered compared with healthy individuals, or those with localized EGC [174]. According to a study by Yang et al. [175], high levels of exosomal miR-423-5p was associated with a poor prognosis in patients with gastric cancer. Exosome-derived PIWI (P-element induced wimpy testis)-interacting RNAs (piRNAs) were found to be more frequent than cell miRNAs by Ge and colleagues [176], and could be employed as non-invasive diagnostic biomarkers with significant specificity and sensitivity. These exosomal ncRNAs can be combined with other biomarkers for detecting gastric cancer more specifically. Recent studies on the role of exosomal ncRNAs in gastric cancer are exhibited in Table 3.
Liver cancer
Hepatocellular carcinoma (HCC) is one of the most deadly malignancies, and its high mortality rate and resistance to treatment represents a severe challenge [195]. Recent studies have shown that cancer cells acquire a more malignant phenotype as infiltration progresses, and exosomes appear to contribute to the progression and metastasis of HCC by transferring their contents from premalignant cells to cancer cells [196]. According to a study by Liu et al. [197], exosomes derived from liver cancer increase the invasion and metastasis of cancer cells by transferring miR-25-5p. High expression of miR-25-5p reduces the expression of leucine-rich repeat-containing 7 (LRRC7). LRRC7 is involved in intercellular attachment and therefore reduced expression leads to increased cell motility. Moreover, increased exosomal miR-25-5p levels in HCC were associated with cancer progression and could be a prognostic biomarker [197].
Highly-metastatic HCC cells have a stronger ability to convert normal fibroblasts into cancer-associated fibroblasts (CAFs) in the lung metastatic niche, according to Fang et al. [198]. They found that highly-metastatic HCC cells could release exosomal miR-1247-3p, which specifically targeted B4GALT3 in fibroblasts, and activated β1 integrin–NF-κB signaling. Activated CAFs secrete pro-inflammatory cytokines, including IL-6 and IL-8, which promote cancer growth. Exosomal miR-1247-3p levels in serum were linked to lung metastasis in HCC patients, according to clinical studies. Therefore, tumor-derived exosomes can mediate intercellular interactions between tumor cells and fibroblasts to regulate HCC lung metastasis, and could be prospective targets for prevention of metastasis and improved therapy [198]. Tumor-derived exosomes affect cell behavior by carrying a variety of ncRNAs as epigenetic factors. NcRNAs can alter the tumor phenotype, and exosomes act as their transporters. Conversely, ncRNAs also appear to affect the secretion and regulation of exosomes. For example, Cao et al. [199] found that the lncRNA HULC was not only contained in exosomes to modulate metastasis and TNM in HCC, but it also increased exosome secretion in HCC cells.
Ovarian cancer
Ovarian cancer often has a poor prognosis and a high mortality rate [200]. Therefore, the identification of diagnostic biomarkers for ovarian cancer is important to save patients' lives and increase survival rates. The recent adoption of exosomes as biomarkers has led researchers to study ovarian cancer-derived exosomal ncRNAs, particularly exo-miRNAs. For example, increased expression levels of exo-miR-200c and exo-miR-214 in ovarian cancer were found compared to control samples, suggesting they could be used as diagnostic biomarkers [200]. A study by Vaksman et al. [201] also examined the expression of exosomal miRNAs in ovarian cancer patients. They found that the expression levels of exo-miR-23b, exo-miR-21b, and exo-miR-29a were associated with lower survival rates. Interestingly, ovarian cancer exosomes could transfer miR-21 from stromal cells to cancer cells, leading to the induction of therapeutic resistance. Therefore, exo-miR-21 has been suggested as a predictive biomarker for treatment response. Furthermore, when the miRNA profiles of exosomes acquired from patients were compared to tumor tissue profiles from the same patient, similar miRNAs were found, indicating that the exosomal miRNA profile might be used as a biomarker. In addition, Ma and colleagues [202] showed that tumor-derived exo-circRNA051239 promoted the division and migration of epithelial ovarian cancer cells. They found that exo-circRNA051239 expression levels were significantly higher in patients with metastatic ovarian cancer. Therefore, increasing levels during the course of the disease could be a warning signal for metastasis [202]. In previous studies, the expression profile of exosomal miRNAs in ovarian cancer has been investigated. Some exosomal miRNAs were found to elevated in ovarian cancer versus healthy controls, were associated with invasion or metastasis, and had the potential to be used as diagnostic biomarkers. These exosomal miRNAs are summarized in Table 4.
Other cancers
The role of exosomal ncRNAs in kidney and prostate cancer has also been investigated [224]. Some exosomal miRNAs, including exo-miR-151, exo-miR-650, and exo-miR-29a, were found to be involved in metastasis and invasion [225], while others have been reported as diagnostic biomarkers (Fig. 4) [36]. Studies on the role of exo-lncRNAs as biomarkers in kidney and prostate cancer are limited. According to Qu et al. [226] exo-lncARSR increased the expression of AXL and c-MET in RCC cells by competitively binding to miR-34/miR-449, resulting in sunitinib resistance. Exosomal lncRNAs have a crucial role in tumor biology, and further research is needed to fully understand their function in renal cancer. Moreover, Xiao et al. [227]. discovered that circ400068 was elevated in exosomes secreted from RCC cells using circRNA expression array data. CircRNAs in exosomes produced from RCC cells have only been reported in a few studies so far, so further research is needed.
Exosomal ncRNAs in kidney cancer. miR-19-3b, by activating the TGF/SMAD3,2/KLF10 pathway, leads to the inhibition of cell division and drug resistance (sunitinib resistance). In addition, miR-15a enhances cell division in cancer cells by activating the non-canonical TGF-beta/PI3K/AKT pathway. LncRNA ARSR activates the PI3K/AKT pathway by activating the c-MET receptor, leading to drug resistance. While miR-29a inhibits angiogenesis, miR-549 leads to increased angiogenesis. KLF10 Kruppel Like Factor 10, Vasohibin 1, VEGF vascular endothelial growth factors
Guo et al. [228] examined exosomes derived from pancreatic tumors. These exosomes had high levels of lncRNA-UCA1, which was associated with poor patient survival. In addition, UCA1 promoted tumor growth and angiogenesis in pancreatic cancer cell lines and in a rat model. UCA1 acts as a sponge for miR-96-5p and inhibits its expression. miR-96-5p targets AMOTL2, a protein involved in the formation of new blood vessels, and suppresses its expression. Overall, exosomal UCA1 promoted angiogenesis and tumor growth by regulating miR-96-5p expression, and could be a predictive biomarker for metastatic pancreatic cancer.
Challenges and advantages of tumor derived exosomes
Many studies based on tumor-derived ncRNAs, including solid and hematological tumors, have shown their role as factors involved in tumor progression. The role of exosomes in tumors can be described as a double-edged sword. Sometimes exosomes can prevent the division and invasion of tumor cells by transferring tumor suppressor genes to tumor cells, which shows the potential of using them for cancer treatment and therapeutic vectors. Moreover, tumor-derived exosomes can accelerate the process of tumorigenesis and invasion and drug resistance by transferring oncogenes to non-cancerous or pre-cancerous cells. From this aspect, tumor-derived exosomes can be considered promising therapeutic targets. However, most recent studies have focused on their potential as diagnostic biomarkers. So far, many studies have been conducted on the contents of exosomes derived from tumors and have shown that countless exosomal ncRNAs are dysregulated in all types of cancers. For several reasons, tumor-derived ncRNA exosomes could be ideal diagnostic and prognostic biomarkers for cancer studies. Firstly, the specificity of the exosomal contents reflects the contents of the source cells. The second reason is the high sensitivity of these EVs. Finally, according to the results of studies, cancer patients have higher levels of circulating exosomes than healthy people. Furthermore, functional ncRNAs are always found in exosomes. When compared to free ncRNAs in bodily fluids, exosomes will give greater sensitivity for ncRNA identification in clinical samples [229]. The next factor is the stability of the exosomal contents. As mentioned earlier, the exosome membrane can protect ncRNAs from RNase activity. In addition, the level of ncRNAs in exosomes remains constant despite exposure to extreme stress conditions. Another advantage mentioned earlier is the easy availability of exosomes in fluids throughout the body. Therefore, it seems that continued efforts in identifying biomarkers based on exosomes derived from tumors will be valuable and promising. However, many challenges still limit the use of exosomal ncRNAs in clinical applications. So far there is no Food and Drug Administration (FDA)-approved exosomal ncRNA biomarker for cancers. Nevertheless, some possible exosomal ncRNAs which have been investigated in human studies or clinical trials are listed in Table 5.
From an investigative standpoint, the activities of ncRNAs reported in current investigations are often based on experiments involving exogenous ncRNA overexpression in vitro (such as transfecting miRNA mimics) or ncRNA knockdown in overexpressing cell lines. Therefore, it is still unclear if the body generates a comparable number of ncRNAs which promote the same activities. In addition, many studies that have reported ncRNAs function and mechanism of action have only been conducted in vitro and on cultured cells. In these methods, pure and prepared exosomes are incubated with recipient cells. It is unclear whether the applied exosomal ncRNAs correspond to the amounts found in physiological conditions. Exogenous exosomes may exaggerate the function of native exosomal ncRNAs, although the role of exosomal ncRNAs in tumor progression and invasion has been proven. To use exosomal ncRNAs as key biomarkers in clinical applications, ncRNAs that are consistently dysregulated in exosomes should be identified [230]. Next-generation sequencing (NGS) and RNA-seq techniques are powerful tools for profiling and analyzing these biomolecules. In any case, the varying origin of circulating exosomes limits the accurate detection of patient-specific exosomes in peripheral blood samples. Other exosomes released from normal cells could reduce the concentration of tumor-derived exosomes, leading to a decrease in the actual contribution of tumor-associated ncRNAs in sequencing libraries [231]. Most importantly, there is still no standardized and fully validated method for the extraction and isolation of exosomes, which was discussed in the previous sections. Exosome separation procedures differ between studies, and purity is difficult to maintain within a particular range; these issues may impact the precision and reproducibility of NGS or other exosome study outcomes.
The small amounts of RNA extracted from exosomes compared to tissues and cells are among the limitations of the isolation methods. Therefore, ultra-sensitive commercial kits are needed to achieve accurate and sufficient measurements. Apart from the high price of these kits, each kit with a different brand produces a different quantity and quality of the product, which can ultimately affect the test results. That is why, to date, finding techniques for pure exosome isolation and in sufficient quantities is the most crucial step before tumor-derived exosomes can translate into clinical applications.
Another obstacle is the contamination of exosomes collected from blood and plasma with extracellular vesicles derived from non-tumor cells. How to identify and enrich tumor-derived exosomes from the complex microenvironment is another problem preventing their clinical use. The expression of specific proteins on the cell surface provides a suitable method for their isolation with the help of specific antibodies [232]. It has been shown that the epithelial cell adhesion molecule (EpCAM) may be employed as a marker to identify epithelium-derived extracellular vesicles from CRC patient blood samples via immunoaffinity capture. Rupp et al. [233], on the other hand, found that EpCAM may be cleaved from exosomes by serum metalloproteinases, which could impede tumor exosome enrichment by immune-affinity isolation in breast cancer. More research is needed to identify and develop specific and stable indicators for exosome enrichment. Furthermore, while the stability of the exosome contents allows them to be used in gene therapy, further study is needed to confirm that exosomes harboring therapeutic ncRNAs can safely evade the immune system and precisely home to malignant sites.
Conclusion
In both normal and pathological conditions, exosomes can mediate communication between the same or different cell types, and with their surroundings. Tumor-derived exosomes are gaining traction as a likely mode of communication between malignant cells and the tumor microenvironment, with implications for tumor growth and metastasis. The data gathered thus far employing analysis of tumor-derived exosomes as possible biomarkers for cancer diagnosis and prognosis has provided much information, but further study is still needed. In the near future, the clinical use of tumor-derived exosomes is expected to create a new window for cancer diagnosis, management and therapy.
Data availability
Enquiries about data availability should be directed to the authors.
References
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F et al (2016) Cancer statistics in China, 2015. CA Cancer J Clin 66(2):115–132
Miller KD, Goding Sauer A, Ortiz AP, Fedewa SA, Pinheiro PS, Tortolero-Luna G et al (2018) Cancer statistics for hispanics/latinos, 2018. CA Cancer J Clin 68(6):425–445
Ebrahimi N, Akbari M, Ghanaatian M, Roozbahani Moghaddam P, Adelian S, Borjian Boroujeni M et al (2022) Development of neoantigens: from identification in cancer cells to application in cancer vaccines. Expert Rev Vac 21(7):941–955
Zhu Q, Li N, Zeng X, Han Q, Li F, Yang C et al (2015) Hepatocellular carcinoma in a large medical center of China over a 10-year period: evolving therapeutic option and improving survival. Oncotarget 6(6):4440
Pasut A, Matsumoto A, Clohessy JG, Pandolfi PP (2016) The pleiotropic role of non-coding genes in development and cancer. Curr Opin Cell Biol 43:104–113
Ono S, Lam S, Nagahara M, Hoon DS (2015) Circulating microRNA biomarkers as liquid biopsy for cancer patients: pros and cons of current assays. J Clin Med 4(10):1890–1907
Rapisuwon S, Vietsch EE, Wellstein A (2016) Circulating biomarkers to monitor cancer progression and treatment. Comput Struct Biotechnol J 14:211–222
Babaei M, Rezaie J (2021) Application of stem cell-derived exosomes in ischemic diseases: opportunity and limitations. J Transl Med 19(1):1–11
Huarte M, Rinn JL (2010) Large non-coding RNAs: missing links in cancer? Hum Mol Genet 19(R2):R152–R161
Shen X, Qi P, Du X (2015) Long non-coding RNAs in cancer invasion and metastasis. Mod Pathol 28(1):4–13
Chang Y-N, Zhang K, Hu Z-M, Qi H-X, Shi Z-M, Han X-H et al (2016) Hypoxia-regulated lncRNAs in cancer. Gene 575(1):1–8
Qi P, Zhou X-y, Du X (2016) Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol Cancer 15(1):1–11
Brinton LT, Sloane HS, Kester M, Kelly KA (2015) Formation and role of exosomes in cancer. Cell Mol Life Sci 72(4):659–671
Van der Pol E, Böing AN, Harrison P, Sturk A, Nieuwland R (2012) Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev 64(3):676–705
Cocucci E, Meldolesi J (2015) Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol 25(6):364–372
Cocucci E, Racchetti G, Meldolesi J (2009) Shedding microvesicles: artefacts no more. Trends Cell Biol 19(2):43–51
Marks MS, Heijnen HF, Raposo G (2013) Lysosome-related organelles: unusual compartments become mainstream. Curr Opin Cell Biol 25(4):495–505
Verma S, Bhavsar AS, Donovan J (2014) MR imaging-guided prostate biopsy techniques. Magn Reson Imaging Clin 22(2):135–144
Vaidyanathan R, Soon RH, Zhang P, Jiang K, Lim CT (2019) Cancer diagnosis: from tumor to liquid biopsy and beyond. Lab Chip 19(1):11–34
Rolfo C, Castiglia M, Hong D, Alessandro R, Mertens I, Baggerman G et al (2014) Liquid biopsies in lung cancer: the new ambrosia of researchers. Biochimica et Biophysica Acta (BBA) Rev Cancer 1846(2):539–546
Whiteside TL (2016) Tumor-derived exosomes and their role in cancer progression. Adv Clin Chem 74:103–141
Momen-Heravi F, Getting SJ, Moschos SA (2018) Extracellular vesicles and their nucleic acids for biomarker discovery. Pharmacol Ther 192:170–187
Zhou B, Xu K, Zheng X, Chen T, Wang J, Song Y et al (2020) Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct Target Ther 5(1):1–14
Pham X-H, Hahm E, Huynh K-H, Son BS, Kim H-M, Jun B-H (2020) Sensitive colorimetric detection of prostate specific antigen using a peroxidase-mimicking anti-PSA antibody coated Au nanoparticle. BioChip J 14(2):158–168
Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K et al (2014) Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 9(4):e92921
Yu W, Hurley J, Roberts D, Chakrabortty S, Enderle D, Noerholm M et al (2021) Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol 32(4):466–477
Brinkman K, Meyer L, Bickel A, Enderle D, Berking C, Skog J et al (2020) Extracellular vesicles from plasma have higher tumour RNA fraction than platelets. J Extracell Vesicles 9(1):1741176
Sonoda H, Yokota-Ikeda N, Oshikawa S, Kanno Y, Yoshinaga K, Uchida K et al (2009) Decreased abundance of urinary exosomal aquaporin-1 in renal ischemia-reperfusion injury. Am J Physiol Renal Physiol 297(4):F1006–F1016
Nawaz M, Camussi G, Valadi H, Nazarenko I, Ekström K, Wang X et al (2014) The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nat Rev Urol 11(12):688–701 (PubMed PMID: 25403245. Epub 2014/11/19. eng)
Zhang S, Yang Y, Jia S, Chen H, Duan Y, Li X et al (2020) Exosome-like vesicles derived from Hertwig’s epithelial root sheath cells promote the regeneration of dentin-pulp tissue. Theranostics 10(13):5914
Liu J, Chen Y, Pei F, Zeng C, Yao Y, Liao W et al (2021) Extracellular vesicles in liquid biopsies: potential for disease diagnosis. Biomed Res Int 2021:1–17
Soares Martins T, Catita J, Martins Rosa I, AB da Cruz e Silva O, Henriques AG (2018) Exosome isolation from distinct biofluids using precipitation and column-based approaches. PLoS ONE 13(6):e0198820
Mateescu B, Kowal EJ, van Balkom BW, Bartel S, Bhattacharyya SN, Buzás EI et al (2017) Obstacles and opportunities in the functional analysis of extracellular vesicle RNA–an ISEV position paper. J Extracell Vesicles 6(1):1286095
Mathivanan S, Lim JW, Tauro BJ, Ji H, Moritz RL, Simpson RJ (2010) Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol Cell Proteomics 9(2):197–208
Ghossoub R, Chéry M, Audebert S, Leblanc R, Egea-Jimenez AL, Lembo F et al (2020) Tetraspanin-6 negatively regulates exosome production. Proc Natl Acad Sci 117(11):5913–5922
Mao W, Wang K, Wu Z, Xu B, Chen M (2021) Current status of research on exosomes in general, and for the diagnosis and treatment of kidney cancer in particular. J Exp Clin Cancer Res 40(1):1–13
Mao Y, Wang Y, Dong L, Zhang Y, Zhang Y, Wang C et al (2019) Hypoxic exosomes facilitate angiogenesis and metastasis in esophageal squamous cell carcinoma through altering the phenotype and transcriptome of endothelial cells. J Exp Clin Cancer Res 38(1):1–14
Hosseini K, Ranjbar M, Pirpour Tazehkand A, Asgharian P, Montazersaheb S, Tarhriz V et al (2022) Evaluation of exosomal non-coding RNAs in cancer using high-throughput sequencing. J Transl Med 20(1):1–15
Miraghel SA, Ebrahimi N, Khani L, Mansouri A, Jafarzadeh A, Ahmadi A et al (2022) Crosstalk between non-coding RNAs expression profile, drug resistance and immune response in breast cancer. Pharmacol Res 176:106041
Ahmadi M, Rezaie J (2021) Ageing and mesenchymal stem cells derived exosomes: molecular insight and challenges. Cell Biochem Funct 39(1):60–66 (PubMed PMID: 33164248. Epub 2020/11/10. eng)
Lakshmi S, Hughes TA, Priya S (2021) Exosomes and exosomal RNAs in breast cancer: a status update. Eur J Cancer 144:252–268
Baroni S, Romero-Cordoba S, Plantamura I, Dugo M, D’ippolito E, Cataldo A et al (2016) Exosome-mediated delivery of miR-9 induces cancer-associated fibroblast-like properties in human breast fibroblasts. Cell Death Dis 7(7):e2312-e
Luga V, Zhang L, Viloria-Petit AM, Ogunjimi AA, Inanlou MR, Chiu E et al (2012) Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 151(7):1542–1556
Wang H, Wei H, Wang J, Li L, Chen A, Li Z (2020) MicroRNA-181d-5p-containing exosomes derived from CAFs promote EMT by regulating CDX2/HOXA5 in breast cancer. Mol Therapy-Nucleic Acids 19:654–667
Ebrahimi N, Adelian S, Shakerian S, Afshinpour M, Chaleshtori SR, Rostami N et al (2022) Crosstalk between ferroptosis and the epithelial-mesenchymal transition: Implications for inflammation and cancer therapy. Cytokine Growth Factor Rev 64:33–45
Ding J, Xu Z, Zhang Y, Tan C, Hu W, Wang M et al (2018) Exosome-mediated miR-222 transferring: an insight into NF-κB-mediated breast cancer metastasis. Exp Cell Res 369(1):129–138
Camacho L, Guerrero P, Marchetti D (2013) MicroRNA and protein profiling of brain metastasis competent cell-derived exosomes. PLoS ONE 8(9):e73790
Le MT, Hamar P, Guo C, Basar E, Perdigão-Henriques R, Balaj L et al (2014) miR-200–containing extracellular vesicles promote breast cancer cell metastasis. J Clin Investig 124(12):5109–5128
Ebrahimi N, Nasr Esfahani A, Samizade S, Mansouri A, Ghanaatian M, Adelian S et al (2022) The potential application of organoids in breast cancer research and treatment. Hum Genet 141(2):193–208
Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR et al (2014) Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 25(4):501–515
Li Y, Liang Y, Sang Y, Song X, Zhang H, Liu Y et al (2018) MiR-770 suppresses the chemo-resistance and metastasis of triple negative breast cancer via direct targeting of STMN1. Cell Death Dis 9(1):1–12
Eichelser C, Stückrath I, Müller V, Milde-Langosch K, Wikman H, Pantel K et al (2014) Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget 5(20):9650
Hannafon BN, Trigoso YD, Calloway CL, Zhao YD, Lum DH, Welm AL et al (2016) Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res 18(1):1–14
Shen S, Song Y, Zhao B, Xu Y, Ren X, Zhou Y et al (2021) Cancer-derived exosomal miR-7641 promotes breast cancer progression and metastasis. Cell Commun Signal 19(1):1–13
Xin Y, Wang X, Meng K, Ni C, Lv Z, Guan D (2020) Identification of exosomal miR-455-5p and miR-1255a as therapeutic targets for breast cancer. Biosci Rep 40(1):BSR20190303
Jaiswal R, Gong J, Sambasivam S, Combes V, Mathys JM, Davey R et al (2012) Microparticle-associated nucleic acids mediate trait dominance in cancer. FASEB J 26(1):420–429
Jafari R, Rahbarghazi R, Ahmadi M, Hassanpour M, Rezaie J (2020) Hypoxic exosomes orchestrate tumorigenesis: molecular mechanisms and therapeutic implications. J Transl Med 18(1):474 (PubMed PMID: 33302971. Pubmed Central PMCID: PMC7731629. Epub 2020/12/12. eng)
Soraya H, Sani NA, Jabbari N, Rezaie J (2021) Metformin increases exosome biogenesis and secretion in U87 MG human glioblastoma cells: a possible mechanism of therapeutic resistance. Arch Med Res 52(2):151–162 (PubMed PMID: 33059952. Epub 2020/10/17. eng)
Chen W, Liu X, Lv M, Chen L, Zhao J, Zhong S et al (2014) Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PLoS ONE 9(4):e95240
Shah SH, Miller P, Garcia-Contreras M, Ao Z, Machlin L, Issa E et al (2015) Hierarchical paracrine interaction of breast cancer associated fibroblasts with cancer cells via hMAPK-microRNAs to drive ER-negative breast cancer phenotype. Cancer Biol Ther 16(11):1671–1681
Fong MY, Zhou W, Liu L, Alontaga AY, Chandra M, Ashby J et al (2015) Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol 17(2):183–194
Santos JC, da Silva LN, Sarian LO, Matheu A, Ribeiro ML, Derchain SFM (2018) Exosome-mediated breast cancer chemoresistance via miR-155 transfer. Sci Rep 8(1):1–11
Shen M, Dong C, Ruan X, Yan W, Cao M, Pizzo D et al (2019) Chemotherapy-induced extracellular vesicle miRNAs promote breast cancer stemness by targeting ONECUT2. Can Res 79(14):3608–3621
Miraghel SA, Ebrahimi N, Khani L, Mansouri A, Jafarzadeh A, Ahmadi A et al (2021) Crosstalk between non-coding RNAs expression profile, drug resistance and immune response in breast cancer. Pharmacol Res 176:106041
Xu C, Yang M, Ren Y, Wu C, Wang L (2016) Exosomes mediated transfer of lncRNA UCA1 results in increased tamoxifen resistance in breast cancer cells. Eur Rev Med Pharmacol Sci 20(20):4362–4368
Dong H, Wang W, Chen R, Zhang Y, Zou K, Ye M et al (2018) Exosome-mediated transfer of lncRNA-SNHG14 promotes trastuzumab chemoresistance in breast cancer. Int J Oncol 53(3):1013–1026
Nabet BY, Qiu Y, Shabason JE, Wu TJ, Yoon T, Kim BC et al (2017) Exosome RNA unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell 170(2):352–66. e13
Zhang R, Xia Y, Wang Z, Zheng J, Chen Y, Li X et al (2017) Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun 490(2):406–414
Wang Y-L, Liu L-C, Hung Y, Chen C-J, Lin Y-Z, Wu W-R et al (2019) Long non-coding RNA HOTAIR in circulatory exosomes is correlated with ErbB2/HER2 positivity in breast cancer. Breast 46:64–69
Li Y, Zhao Z, Liu W, Li X (2020) SNHG3 functions as miRNA sponge to promote breast cancer cells growth through the metabolic reprogramming. Appl Biochem Biotechnol 191(3):1084–1099 (PubMed PMID: 31956955. Pubmed Central PMCID: PMC7320061. Epub 2020/01/21. eng)
Wang X, Pei X, Guo G, Qian X, Dou D, Zhang Z et al (2020) Exosome-mediated transfer of long noncoding RNA H19 induces doxorubicin resistance in breast cancer. J Cell Physiol 235(10):6896–6904 (PubMed PMID: 31994191. Epub 2020/01/30. eng.)
Bray F, Ferlay J, Soerjomataram I, Siegel R, Torre L, Jemal A (2020) Erratum: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 70(4):313
Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM et al (2019) Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin 69(5):363–385
Wang S, Li X, Zhu R, Han Q, Zhao RC (2016) Lung cancer exosomes initiate global long non-coding RNA changes in mesenchymal stem cells. Int J Oncol 48(2):681–689
Ebrahimi N, Parkhideh S, Samizade S, Esfahani AN, Samsami S, Yazdani E et al (2022) Crosstalk between lncRNAs in the apoptotic pathway and therapeutic targets in cancer. Cytokine Growth Factor Rev 65:61–74
Deng Q, Fang Q, Xie B, Sun H, Bao Y, Zhou S (2020) Exosomal long non-coding RNA MSTRG. 292666.16 is associated with osimertinib (AZD9291) resistance in non-small cell lung cancer. Aging (Albany NY) 12(9):8001
Lee DH (2017) Treatments for EGFR-mutant non-small cell lung cancer (NSCLC): the road to a success, paved with failures. Pharmacol Ther 174:1–21
Zheng L-P, Chen L-Y, Liao X-Y, Xu Z-H, Chen Z-T, Sun J-G (2018) Case report: primary resistance to osimertinib in erlotinib-pretreated lung adenocarcinoma with EGFR T790 M mutation. BMC Cancer 18(1):1–4
Zhou D, Xia Z, Xie M, Gao Y, Yu Q, He B (2021) Exosomal long non-coding RNA SOX2 overlapping transcript enhances the resistance to EGFR-TKIs in non-small cell lung cancer cell line H1975. Hum Cell 34(5):1478–1489
Isidro RA, Appleyard CB (2016) Colonic macrophage polarization in homeostasis, inflammation, and cancer. Am J Physiol Gastrointest Liver Physiol 311(1):G59–G73
Ruffell B, Coussens LM (2015) Macrophages and therapeutic resistance in cancer. Cancer Cell 27(4):462–472
Teng Y, Kang H, Chu Y (2019) Identification of an exosomal long noncoding RNA SOX2-OT in plasma as a promising biomarker for lung squamous cell carcinoma. Genet Test Mol Biomarkers 23(4):235–240
Zhang X, Guo H, Bao Y, Yu H, Xie D, Wang X (2019) Exosomal long non-coding RNA DLX6-AS1 as a potential diagnostic biomarker for non-small cell lung cancer. Oncol Lett 18(5):5197–5204
Li C, Lv Y, Shao C, Chen C, Zhang T, Wei Y et al (2019) Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J Cell Physiol 234(11):20721–20727
Blenkiron C, Miska EA (2007) miRNAs in cancer: approaches, aetiology, diagnostics and therapy. Hum Mol Genet 16(R1):R106–R113
Jin X, Chen Y, Chen H, Fei S, Chen D, Cai X et al (2017) Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non–small cell lung cancer using next-generation sequencing. Clin Cancer Res 23(17):5311–5319
Dejima H, Iinuma H, Kanaoka R, Matsutani N, Kawamura M (2017) Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncol Lett 13(3):1256–1263
Alipoor SD, Adcock IM, Garssen J, Mortaz E, Varahram M, Mirsaeidi M et al (2016) The roles of miRNAs as potential biomarkers in lung diseases. Eur J Pharmacol 791:395–404
Cazzoli R, Buttitta F, Di Nicola M, Malatesta S, Marchetti A, Rom WN et al (2013) microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol 8(9):1156–1162
Munagala R, Aqil F, Gupta RC (2016) Exosomal miRNAs as biomarkers of recurrent lung cancer. Tumor Biol 37(8):10703–10714
Liu Q, Yu Z, Yuan S, Xie W, Li C, Hu Z et al (2017) Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget 8(8):13048
Zhang Z, Tang Y, Song X, Xie L, Zhao S, Song X (2020) Tumor-derived exosomal miRNAs as diagnostic biomarkers in non-small cell lung cancer. Front Oncol 10:2236
Tang Y, Zhang Z, Song X, Yu M, Niu L, Zhao Y et al (2020) Tumor-derived exosomal miR-620 as a diagnostic biomarker in non-small-cell lung cancer. J Oncol 2020:1–9
Berindan-Neagoe I, Monroig PdC, Pasculli B, Calin GA (2014) MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J Clin 64(5):311–336
Kim DH, Park S, Kim H, Choi YJ, Kim SY, Sung KJ et al (2020) Tumor-derived exosomal miR-619–5p promotes tumor angiogenesis and metastasis through the inhibition of RCAN1. 4. Cancer Lett 475:2–13
Chan B, Greenan G, McKeon F, Ellenberger T (2005) Identification of a peptide fragment of DSCR1 that competitively inhibits calcineurin activity in vitro and in vivo. Proc Natl Acad Sci 102(37):13075–13080
van Rooij E, Doevendans PA, Crijns HJ, Heeneman S, Lips DJ, van Bilsen M et al (2004) MCIP1 overexpression suppresses left ventricular remodeling and sustains cardiac function after myocardial infarction. Circ Res 94(3):e18–e26
Feghhi M, Rezaie J, Akbari A, Jabbari N, Jafari H, Seidi F et al (2021) Effect of multi-functional polyhydroxylated polyhedral oligomeric silsesquioxane (POSS) nanoparticles on the angiogenesis and exosome biogenesis in human umbilical vein endothelial cells (HUVECs). Mater Des 197:109227
Sánchez CA, Andahur EI, Valenzuela R, Castellón EA, Fullá JA, Ramos CG et al (2016) Exosomes from bulk and stem cells from human prostate cancer have a differential microRNA content that contributes cooperatively over local and pre-metastatic niche. Oncotarget 7(4):3993
Ma Z, Wei K, Yang F, Guo Z, Pan C, He Y et al (2021) Tumor-derived exosomal miR-3157-3p promotes angiogenesis, vascular permeability and metastasis by targeting TIMP/KLF2 in non-small cell lung cancer. Cell Death Dis 12(9):1–13
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424
Toiyama Y, Okugawa Y, Fleshman J, Boland CR (1870) Goel A (2018) MicroRNAs as potential liquid biopsy biomarkers in colorectal cancer: A systematic review. Biochimica et Biophysica Acta (BBA) Rev Cancer 2:274–282
Teng Y, Ren Y, Hu X, Mu J, Samykutty A, Zhuang X et al (2017) MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun 8(1):1–16
Hassanpour M, Rezabakhsh A, Rezaie J, Nouri M, Rahbarghazi R (2020) Exosomal cargos modulate autophagy in recipient cells via different signaling pathways. Cell Biosci 10:92 (PubMed PMID: 32765827. Pubmed Central PMCID: PMC7395405. Epub 2020/08/09. eng)
Min L, Chen L, Liu S, Yu Y, Guo Q, Li P et al (2019) Loss of circulating exosomal miR-92b is a novel biomarker of colorectal cancer at early stage. Int J Med Sci 16(9):1231
Zhang H, Zhu M, Shan X, Zhou X, Wang T, Zhang J et al (2019) A panel of seven-miRNA signature in plasma as potential biomarker for colorectal cancer diagnosis. Gene 687:246–254
Wang J, Yan F, Zhao Q, Zhan F, Wang R, Wang L et al (2017) Circulating exosomal miR-125a-3p as a novel biomarker for early-stage colon cancer. Sci Rep 7(1):1–9
Hon KW, Abu N, Ab Mutalib N-S, Jamal R (2017) Exosomes as potential biomarkers and targeted therapy in colorectal cancer: a mini-review. Front Pharmacol 8:583
Rapado-González Ó, Álvarez-Castro A, López-López R, Iglesias-Canle J, Suárez-Cunqueiro MM, Muinelo-Romay L (2019) Circulating microRNAs as promising biomarkers in colorectal cancer. Cancers 11(7):898
Tang Y, Zhao Y, Song X, Song X, Niu L, Xie L (2019) Tumor-derived exosomal miRNA-320d as a biomarker for metastatic colorectal cancer. J Clin Lab Anal 33(9):e23004
Liu C, Eng C, Shen J, Lu Y, Takata Y, Mehdizadeh A et al (2016) Serum exosomal miR-4772-3p is a predictor of tumor recurrence in stage II and III colon cancer. Oncotarget 7(46):76250
Yan S, Liu G, Jin C, Wang Z, Duan Q, Xu J et al (2018) MicroRNA-6869-5p acts as a tumor suppressor via targeting TLR4/NF-κB signaling pathway in colorectal cancer. J Cell Physiol 233(9):6660–6668
Liang Z, Liu H, Wang F, Xiong L, Zhou C, Hu T et al (2019) LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis 10(11):1–17
Deng X, Ruan H, Zhang X, Xu X, Zhu Y, Peng H et al (2020) Long noncoding RNA CCAL transferred from fibroblasts by exosomes promotes chemoresistance of colorectal cancer cells. Int J Cancer 146(6):1700–1716
Huang Y, Luo Y, Ou W, Wang Y, Dong D, Peng X et al (2021) Exosomal lncRNA SNHG10 derived from colorectal cancer cells suppresses natural killer cell cytotoxicity by upregulating INHBC. Cancer Cell Int 21(1):528
Liu L, Meng T, Yang X-H, Sayim P, Lei C, Jin B et al (2018) Prognostic and predictive value of long non-coding RNA GAS5 and mircoRNA-221 in colorectal cancer and their effects on colorectal cancer cell proliferation, migration and invasion. Cancer Biomark 22(2):283–299
Liu T, Zhang X, Gao S, Jing F, Yang Y, Du L et al (2016) Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget 7(51):85551
Hu D, Zhan Y, Zhu K, Bai M, Han J, Si Y et al (2018) Plasma exosomal long non-coding RNAs serve as biomarkers for early detection of colorectal cancer. Cell Physiol Biochem 51(6):2704–2715
Oehme F, Krahl S, Gyorffy B, Muessle B, Rao V, Greif H et al (2019) Low level of exosomal long non-coding RNA HOTTIP is a prognostic biomarker in colorectal cancer. RNA Biol 16(10):1339–1345
Chen X, Liu Y, Zhang Q, Liu B, Cheng Y, Zhang Y et al (2021) Exosomal long non-coding RNA HOTTIP increases resistance of colorectal cancer cells to mitomycin via impairing MiR-214-mediated degradation of KPNA3. Front Cell Dev Biol 8:582723
Barbagallo C, Brex D, Caponnetto A, Cirnigliaro M, Scalia M, Magnano A et al (2018) LncRNA UCA1, upregulated in CRC biopsies and downregulated in serum exosomes, controls mRNA expression by RNA-RNA interactions. Mol Therapy Nucleic Acids 12:229–241
Ostenfeld MS, Jensen SG, Jeppesen DK, Christensen L-L, Thorsen SB, Stenvang J et al (2016) miRNA profiling of circulating EpCAM+ extracellular vesicles: promising biomarkers of colorectal cancer. J Extracell Vesicles 5(1):31488
Li J, Chen Y, Guo X, Zhou L, Jia Z, Peng Z et al (2017) GPC 1 exosome and its regulatory mi RNA s are specific markers for the detection and target therapy of colorectal cancer. J Cell Mol Med 21(5):838–847
Karimi N, Feizi MAH, Safaralizadeh R, Hashemzadeh S, Baradaran B, Shokouhi B et al (2019) Serum overexpression of miR-301a and miR-23a in patients with colorectal cancer. J Chin Med Assoc 82(3):215–220
Matsumura T, Sugimachi K, Iinuma H, Takahashi Y, Kurashige J, Sawada G et al (2015) Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer 113(2):275–281
Hu H-Y, Yu C-H, Zhang H-H, Zhang S-Z, Yu W-Y, Yang Y et al (2019) Exosomal miR-1229 derived from colorectal cancer cells promotes angiogenesis by targeting HIPK2. Int J Biol Macromol 132:470–477
Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J et al (2018) Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun 9(1):1–14
Tsukamoto M, Iinuma H, Yagi T, Matsuda K, Hashiguchi Y (2017) Circulating exosomal microRNA-21 as a biomarker in each tumor stage of colorectal cancer. Oncology 92(6):360–370
Cheng WC, Liao TT, Lin CC, Yuan LTE, Lan HY, Lin HH et al (2019) RAB27B-activated secretion of stem-like tumor exosomes delivers the biomarker microRNA-146a-5p, which promotes tumorigenesis and associates with an immunosuppressive tumor microenvironment in colorectal cancer. Int J Cancer 145(8):2209–2224
Kral J, Korenkova V, Novosadova V, Langerova L, Schneiderova M, Liska V et al (2018) Expression profile of miR-17/92 cluster is predictive of treatment response in rectal cancer. Carcinogenesis 39(11):1359–1367
Fu F, Jiang W, Zhou L, Chen Z (2018) Circulating exosomal miR-17-5p and miR-92a-3p predict pathologic stage and grade of colorectal cancer. Transl Oncol 11(2):221–232
Jin G, Liu Y, Zhang J, Bian Z, Yao S, Fei B et al (2019) A panel of serum exosomal microRNAs as predictive markers for chemoresistance in advanced colorectal cancer. Cancer Chemother Pharmacol 84(2):315–325
Peng ZY, Gu RH, Yan B (2019) Downregulation of exosome-encapsulated miR-548c-5p is associated with poor prognosis in colorectal cancer. J Cell Biochem 120(2):1457–1463
Yan S, Jiang Y, Liang C, Cheng M, Jin C, Duan Q et al (2018) Exosomal miR-6803-5p as potential diagnostic and prognostic marker in colorectal cancer. J Cell Biochem 119(5):4113–4119
Zou S-L, Chen Y-L, Ge Z-Z, Qu Y-Y, Cao Y, Kang Z-X (2019) Downregulation of serum exosomal miR-150-5p is associated with poor prognosis in patients with colorectal cancer. Cancer Biomark 26(1):69–77
Yagi T, Iinuma H, Hayama T, Matsuda K, Nozawa K, Tsukamoto M et al (2019) Plasma exosomal microRNA-125b as a monitoring biomarker of resistance to mFOLFOX6-based chemotherapy in advanced and recurrent colorectal cancer patients. Mol Clin Oncol 11(4):416–424
Ren D, Lin B, Zhang X, Peng Y, Ye Z, Ma Y et al (2017) Maintenance of cancer stemness by miR-196b-5p contributes to chemoresistance of colorectal cancer cells via activating STAT3 signaling pathway. Oncotarget 8(30):49807
Hon KW, Ab-Mutalib NS, Abdullah NMA, Jamal R, Abu N (2019) Extracellular Vesicle-derived circular RNAs confers chemoresistance in Colorectal cancer. Sci Rep 9(1):1–13
Pan B, Qin J, Liu X, He B, Wang X, Pan Y et al (2019) Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet 10:1096
Li Y, Li C, Xu R, Wang Y, Li D, Zhang B (2019) A novel circFMN2 promotes tumor proliferation in CRC by regulating the miR-1182/hTERT signaling pathways. Clin Sci 133(24):2463–2479
Barbagallo C, Brex D, Caponnetto A, Cirnigliaro M, Scalia M, Magnano A et al (2018) LncRNA UCA1, upregulated in CRC biopsies and downregulated in serum exosomes, controls mRNA expression by RNA-RNA interactions. Mol Therapy Nucl Acids 12:229–241
Salzman J (2016) Circular RNA expression: its potential regulation and function. Trends Genet 32(5):309–316
Feng W, Gong H, Wang Y, Zhu G, Xue T, Wang Y et al (2019) circIFT80 functions as a ceRNA of miR-1236-3p to promote colorectal cancer progression. Mol Therapy Nucl Acids 18:375–387
Bhat A, Ritch CR (2019) Urinary biomarkers in bladder cancer: where do we stand? Curr Opin Urol 29(3):203–209
Abbastabar M, Sarfi M, Golestani A, Karimi A, Pourmand G, Khalili E (2020) Tumor-derived urinary exosomal long non-coding RNAs as diagnostic biomarkers for bladder cancer. EXCLI J 19:301
Zheng R, Du M, Wang X, Xu W, Liang J, Wang W et al (2018) Exosome–transmitted long non-coding RNA PTENP1 suppresses bladder cancer progression. Mol Cancer 17(1):1–13
Berrondo C, Flax J, Kucherov V, Siebert A, Osinski T, Rosenberg A et al (2016) Expression of the long non-coding RNA HOTAIR correlates with disease progression in bladder cancer and is contained in bladder cancer patient urinary exosomes. PLoS ONE 11(1):e0147236
Zheng H, Chen C, Luo Y, Yu M, He W, An M et al (2021) Tumor-derived exosomal BCYRN1 activates WNT5A/VEGF-C/VEGFR3 feedforward loop to drive lymphatic metastasis of bladder cancer. Clin Transl Med 11(7):e497
Zhang S, Du L, Wang L, Jiang X, Zhan Y, Li J et al (2019) Evaluation of serum exosomal Lnc RNA-based biomarker panel for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med 23(2):1396–1405
Armstrong DA, Green BB, Seigne JD, Schned AR, Marsit CJ (2015) MicroRNA molecular profiling from matched tumor and bio-fluids in bladder cancer. Mol Cancer 14(1):1–9
Yasui T, Yanagida T, Ito S, Konakade Y, Takeshita D, Naganawa T et al (2017) Unveiling massive numbers of cancer-related urinary-microRNA candidates via nanowires. Sci Adv 3(12):e1701133
Andreu Z, Oshiro RO, Redruello A, López-Martín S, Gutiérrez-Vázquez C, Morato E et al (2017) Extracellular vesicles as a source for non-invasive biomarkers in bladder cancer progression. Eur J Pharm Sci 98:70–79
Matsuzaki K, Fujita K, Jingushi K, Kawashima A, Ujike T, Nagahara A et al (2017) MiR-21-5p in urinary extracellular vesicles is a novel biomarker of urothelial carcinoma. Oncotarget 8(15):24668
Jiang X, Du L, Duan W, Wang R, Yan K, Wang L et al (2016) Serum microRNA expression signatures as novel noninvasive biomarkers for prediction and prognosis of muscle-invasive bladder cancer. Oncotarget 7(24):36733
Bayraktar R, Van Roosbroeck K (2018) miR-155 in cancer drug resistance and as target for miRNA-based therapeutics. Cancer Metastasis Rev 37(1):33–44
Fanous H, Sullivan T, Rieger-Christ K (2017) MP88-15 distinct exosomal mirna profiles in chemoresistant bladder carcinoma cell lines. J Urol 197(4S):e1179–e1180
Piao X-M, Cha E-J, Yun SJ, Kim W-J (2021) Role of exosomal miRNA in bladder cancer: a promising liquid biopsy biomarker. Int J Mol Sci 22(4):1713
Rahib L, Wehner MR, Matrisian LM, Nead KT (2021) Estimated projection of US cancer incidence and death to 2040. JAMA Netw Open 4(4):e214708-e
Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F (2014) Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Prevent Biomarkers 23(5):700–713
Tkach M, Théry C (2016) Communication by extracellular vesicles: where we are and where we need to go. Cell 164(6):1226–1232
Huang Y-K, Yu J-C (2015) Circulating microRNAs and long non-coding RNAs in gastric cancer diagnosis: an update and review. World J Gastroenterol WJG 21(34):9863
Zhao R, Zhang Y, Zhang X, Yang Y, Zheng X, Li X et al (2018) Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer 17(1):1–5
Pan L, Liang W, Fu M, Huang Z, Li X, Zhang W et al (2017) Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J Cancer Res Clin Oncol 143(6):991–1004
Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L et al (2015) Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumor Biol 36(3):2007–2012
Lin L-Y, Yang L, Zeng Q, Wang L, Chen M-L, Zhao Z-H et al (2018) Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol Cancer 17(1):84
Lu Y, Hou K, Li M, Wu X, Yuan S (2020) Exosome-Delivered LncHEIH Promotes Gastric Cancer Progression by Upregulating EZH2 and Stimulating Methylation of the GSDME Promoter. Front Cell Dev Biol 8:571297 (PubMed PMID: 33163491. eng)
Wang M, Zhao C, Shi H, Zhang B, Zhang L, Zhang X et al (2014) Deregulated microRNAs in gastric cancer tissue-derived mesenchymal stem cells: novel biomarkers and a mechanism for gastric cancer. Br J Cancer 110(5):1199–1210 (PubMed PMID: 24473397. Pubmed Central PMCID: PMC3950864. Epub 2014/01/30. eng.)
Wei S, Peng L, Yang J, Sang H, Jin D, Li X et al (2020) Exosomal transfer of miR-15b-3p enhances tumorigenesis and malignant transformation through the DYNLT1/Caspase-3/Caspase-9 signaling pathway in gastric cancer. J Exp Clin Cancer Res CR. 39(1):32 (PubMed PMID: 32039741. Pubmed Central PMCID: PMC7011526. Epub 2020/02/11. eng)
Deng Z, Wu J, Xu S, Chen F, Zhang Z, Jin A et al (2020) Exosomes-microRNAs interacted with gastric cancer and its microenvironment: a mini literature review. Biomark Med 14(2):141–150 (PubMed PMID: 32064893. Epub 2020/02/18. eng)
Guo X, Lv X, Ru Y, Zhou F, Wang N, Xi H et al (2020) Circulating exosomal gastric cancer-associated long noncoding RNA1 as a biomarker for early detection and monitoring progression of gastric cancer: a multiphase study. JAMA Surg 155(7):572–579 (PubMed PMID: 32520332. eng)
Wang N, Wang L, Yang Y, Gong L, Xiao B, Liu X (2017) A serum exosomal microRNA panel as a potential biomarker test for gastric cancer. Biochem Biophys Res Commun 493(3):1322–1328 (PubMed PMID: 28986250. Epub 2017/10/08. eng.)
Li S, Zhang M, Zhang H, Hu K, Cai C, Wang J et al (2020) Exosomal long noncoding RNA lnc-GNAQ-6:1 may serve as a diagnostic marker for gastric cancer. Clinica chimica acta: Int J Clin Chem 501:252–257 (PubMed PMID: 31730812. Epub 2019/11/16. eng)
Kumata Y, Iinuma H, Suzuki Y, Tsukahara D, Midorikawa H, Igarashi Y et al (2018) Exosome-encapsulated microRNA-23b as a minimally invasive liquid biomarker for the prediction of recurrence and prognosis of gastric cancer patients in each tumor stage. Oncol Rep 40(1):319–330 (PubMed PMID: 29749537. Epub 2018/05/12. eng)
Tokuhisa M, Ichikawa Y, Kosaka N, Ochiya T, Yashiro M, Hirakawa K et al (2015) Exosomal miRNAs from peritoneum lavage fluid as potential prognostic biomarkers of peritoneal metastasis in gastric cancer. PLoS ONE 10(7):e0130472 (PubMed PMID: 26208314. Pubmed Central PMCID: PMC4514651. Epub 2015/07/25. eng)
Yang H, Fu H, Wang B, Zhang X, Mao J, Li X et al (2018) Exosomal miR-423–5p targets SUFU to promote cancer growth and metastasis and serves as a novel marker for gastric cancer. Mol Carcinog 57(9):1223–1236 (PubMed PMID: 29749061. Epub 2018/05/12. eng)
Ge L, Zhang N, Li D, Wu Y, Wang H, Wang J (2020) Circulating exosomal small RNAs are promising non-invasive diagnostic biomarkers for gastric cancer. J Cell Mol Med 24(24):14502–14513 (PubMed PMID: 33169519. Pubmed Central PMCID: PMC7753781. Epub 2020/11/11. eng)
Ohshima K, Inoue K, Fujiwara A, Hatakeyama K, Kanto K, Watanabe Y et al (2010) Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS ONE 5(10):e13247
Tsai CC, Chen TY, Tsai KJ, Lin MW, Hsu CY, Wu DC et al (2020) NF-κB/miR-18a-3p and miR-4286/BZRAP1 axis may mediate carcinogenesis in Helicobacter pylori-Associated gastric cancer. Biomedicine pharmacotherapy= Biomedecine pharmacotherapie. 132:110869 (PubMed PMID: 33113427. Epub 2020/10/29. eng)
Feng C, She J, Chen X, Zhang Q, Zhang X, Wang Y et al (2019) Exosomal miR-196a-1 promotes gastric cancer cell invasion and metastasis by targeting SFRP1. Nanomedicine (Lond) 14(19):2579–2593 (PubMed PMID: 31609675. Epub 2019/10/15. eng)
Zhu M, Zhang N, He S, Lu X (2020) Exosomal miR-106a derived from gastric cancer promotes peritoneal metastasis via direct regulation of Smad7. Cell cycle (Georgetown, Tex). 19(10):1200–1221 (PubMed PMID: 32267797. Pubmed Central PMCID: PMC7217357. Epub 2020/04/09. eng)
Xia X, Wang S, Ni B, Xing S, Cao H, Zhang Z et al (2020) Hypoxic gastric cancer-derived exosomes promote progression and metastasis via MiR-301a-3p/PHD3/HIF-1α positive feedback loop. Oncogene 39(39):6231–6244 (PubMed PMID: 32826951. Epub 2020/08/23. eng)
Zhou W, Wang L, Miao Y, Xing R (2018) Novel long noncoding RNA GACAT3 promotes colorectal cancer cell proliferation, invasion, and migration through miR-149. Onco Targets Ther 11:1543–1552 (PubMed PMID: 29593420. Pubmed Central PMCID: PMC5865577. Epub 2018/03/30. eng)
Zhang H, Deng T, Liu R, Ning T, Yang H, Liu D et al (2020) CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol Cancer 19(1):43 (PubMed PMID: 32106859. Pubmed Central PMCID: PMC7045485. Epub 2020/02/29. eng)
Shi L, Wang Z, Geng X, Zhang Y, Xue Z (2020) Exosomal miRNA-34 from cancer-associated fibroblasts inhibits growth and invasion of gastric cancer cells in vitro and in vivo. Aging (Albany NY). 12(9):8549–8564 (PubMed PMID: 32391804. Pubmed Central PMCID: PMC7244055. Epub 2020/05/12. eng)
Ren W, Zhang X, Li W, Feng Q, Feng H, Tong Y et al (2019) Exosomal miRNA-107 induces myeloid-derived suppressor cell expansion in gastric cancer. Cancer Manage Res 11:4023–4040 (PubMed PMID: 31190980. Pubmed Central PMCID: PMC6511657. Epub 2019/06/14. eng)
Shi Y, Wang Z, Zhu X, Chen L, Ma Y, Wang J et al (2020) Exosomal miR-1246 in serum as a potential biomarker for early diagnosis of gastric cancer. Int J Clin Oncol 25(1):89–99 (PubMed PMID: 31506750. Epub 2019/09/12. eng)
Li Q, Li B, Li Q, Wei S, He Z, Huang X et al (2018) Exosomal miR-21–5p derived from gastric cancer promotes peritoneal metastasis via mesothelial-to-mesenchymal transition. Cell Death Dis 9(9):854 (PubMed PMID: 30154401. Pubmed Central PMCID: PMC6113299. Epub 2018/08/30. eng)
Wang M, Qiu R, Yu S, Xu X, Li G, Gu R et al (2019) Paclitaxel-resistant gastric cancer MGC-803 cells promote epithelial-to-mesenchymal transition and chemoresistance in paclitaxel-sensitive cells via exosomal delivery of miR-155-5p. Int J Oncol 54(1):326–338 (PubMed PMID: 30365045. Pubmed Central PMCID: PMC6254863. Epub 2018/10/27. eng)
Wang L, Bo X, Yi X, Xiao X, Zheng Q, Ma L et al (2020) Exosome-transferred LINC01559 promotes the progression of gastric cancer via PI3K/AKT signaling pathway. Cell Death Dis 11(9):723 (PubMed PMID: 32895368. Pubmed Central PMCID: PMC7477231. Epub 2020/09/09. eng)
Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F et al (2020) Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582–3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer 19(1):112 (PubMed PMID: 32600329. Pubmed Central PMCID: PMC7322843. Epub 2020/07/01. eng)
Piao HY, Guo S, Wang Y, Zhang J (2021) Exosome-transmitted lncRNA PCGEM1 promotes invasive and metastasis in gastric cancer by maintaining the stability of SNAI1. Clin Transl Oncol 23(2):246–256 (PubMed PMID: 32519176. Epub 2020/06/11. eng)
Wang S, Ping M, Song B, Guo Y, Li Y, Jia J (2020) Exosomal CircPRRX1 enhances doxorubicin resistance in gastric cancer by regulating MiR-3064–5p/PTPN14 signaling. Yonsei Med J 61(9):750–761 (PubMed PMID: 32882759. Pubmed Central PMCID: PMC7471080. Epub 2020/09/04. eng)
Wang J, Lv B, Su Y, Wang X, Bu J, Yao L (2019) Exosome-mediated transfer of lncRNA HOTTIP promotes cisplatin resistance in gastric cancer cells by regulating HMGA1/miR-218 axis. Onco Targets Ther 12:11325–11338 (PubMed PMID: 31908497. Pubmed Central PMCID: PMC6930390. Epub 2020/01/08. eng)
Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L et al (2015) Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol 36(3):2007–2012 (PubMed PMID: 25391424. Epub 2014/11/14. eng)
Rutherford MJ, Arnold M, Bardot A, Ferlay J, De P, Tervonen H et al (2021) Comparison of liver cancer incidence and survival by subtypes across seven high-income countries. Int J Cancer 149(12):2020–2031
Hoshino A, Costa-Silva B, Shen T-L, Rodrigues G, Hashimoto A, Mark MT et al (2015) Tumour exosome integrins determine organotropic metastasis. Nature 527(7578):329–335
Liu H, Chen W, Zhi X, Chen E-J, Wei T, Zhang J et al (2018) Tumor-derived exosomes promote tumor self-seeding in hepatocellular carcinoma by transferring miRNA-25-5p to enhance cell motility. Oncogene 37(36):4964–4978
Fang T, Lv H, Lv G, Li T, Wang C, Han Q et al (2018) Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun 9(1):1–13
Cao S-Q, Zheng H, Sun B-C, Wang Z-L, Liu T, Guo D-H et al (2019) Long non-coding RNA highly up-regulated in liver cancer promotes exosome secretion. World J Gastroenterol 25(35):5283
Ferlay J, Colombet M, Soerjomataram I, Parkin DM, Piñeros M, Znaor A et al (2021) Cancer statistics for the year 2020: an overview. Int J Cancer. https://doi.org/10.1002/ijc.33588
Vaksman O, Tropé C, Davidson B, Reich R (2014) Exosome-derived miRNAs and ovarian carcinoma progression. Carcinogenesis 35(9):2113–2120
Ma R, Ye X, Cheng H, Cui H, Chang X (2021) Tumor-derived exosomal circRNA051239 promotes proliferation and migration of epithelial ovarian cancer. Am J Transl Res 13(3):1125
Yang C, Kim HS, Park SJ, Lee EJ, Kim SI, Song G et al (2019) Inhibition of miR-214-3p aids in preventing epithelial ovarian cancer malignancy by increasing the expression of LHX6. Cancers 11(12):1917
Taylor DD, Gercel-Taylor C (2008) MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 110(1):13–21 (PubMed PMID: 18589210. Epub 2008/07/01. eng)
He L, Zhu W, Chen Q, Yuan Y, Wang Y, Wang J et al (2019) Ovarian cancer cell-secreted exosomal miR-205 promotes metastasis by inducing angiogenesis. Theranostics. 9(26):8206–8220 (PubMed PMID: 31754391. Pubmed Central PMCID: PMC6857047. Epub 2019/11/23. eng)
Sun Z, Liu B, Liu Z-H, Song W, Wang D, Chen B-Y et al (2020) Notochordal-cell-derived exosomes induced by compressive load inhibit angiogenesis via the miR-140-5p/Wnt/β-catenin axis. Mol Therapy Nucl Acids 22:1092–1106
Ebrahimi N, Kharazmi K, Ghanaatian M, Miraghel SA, Amiri Y, Seyedebrahimi SS et al (2022) Role of the Wnt and GTPase pathways in breast cancer tumorigenesis and treatment. Cytokine Growth Factor Rev
Ying X, Wu Q, Wu X, Zhu Q, Wang X, Jiang L et al (2016) Epithelial ovarian cancer-secreted exosomal miR-222–3p induces polarization of tumor-associated macrophages. Oncotarget 7(28):43076–43087 (PubMed PMID: 27172798. Pubmed Central PMCID: PMC5190009. Epub 2016/05/14. eng)
Alharbi M, Zuñiga F, Elfeky O, Guanzon D, Lai A, Rice GE et al (2018) The potential role of miRNAs and exosomes in chemotherapy in ovarian cancer. Endocr Relat Cancer 25(12):R663–R685
Chen J, Wu S, Wang J, Sha Y, Ji Y (2022) Hsa_circ_0074269-mediated Upregulation of TUFT1 Through miR-485-5p Increases Cisplatin Resistance in Cervical Cancer. Reprod Sci. https://doi.org/10.1007/s43032-022-00855-9
Liu O, Wang C, Wang S, Hu Y, Gou R, Dong H et al (2021) Keratin 80 regulated by miR-206/ETS1 promotes tumor progression via the MEK/ERK pathway in ovarian cancer. J Cancer 12(22):6835–6850 (PubMed PMID: 34659572. Pubmed Central PMCID: PMC8517993. Epub 2021/10/19. eng)
Wang J, Liu L (2021) MiR-149-3p promotes the cisplatin resistance and EMT in ovarian cancer through downregulating TIMP2 and CDKN1A. J Ovarian Res 14(1):1–12
Yu H, Pan S (2020) MiR-202-5p suppressed cell proliferation, migration and invasion in ovarian cancer via regulating HOXB2. Eur Rev Med Pharmacol Sci 24:2256–2263
Meng X, Müller V, Milde-Langosch K, Trillsch F, Pantel K, Schwarzenbach H (2016) Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget 7(13):16923–16935 (PubMed PMID: 26943577. Pubmed Central PMCID: PMC4941360. Epub 2016/03/05. eng)
Li F, Liang Z, Jia Y, Zhang P, Ling K, Wang Y et al (2022) microRNA-324-3p suppresses the aggressive ovarian cancer by targeting WNK2/RAS pathway. Bioengineered 13(5):12030–12044
Kobayashi M, Salomon C, Tapia J, Illanes SE, Mitchell MD, Rice GE (2014) Ovarian cancer cell invasiveness is associated with discordant exosomal sequestration of Let-7 miRNA and miR-200. J Transl Med 12:4 (PubMed PMID: 24393345. Pubmed Central PMCID: PMC3896684. Epub 2014/01/08. eng)
Cao J, Zhang Y, Mu J, Yang D, Gu X, Zhang J (2021) Exosomal miR-21–5p contributes to ovarian cancer progression by regulating CDK6. Hum Cell 34(4):1185–1196
Mateescu B, Batista L, Cardon M, Gruosso T, De Feraudy Y, Mariani O et al (2011) miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat Med 17(12):1627–1635
Samuel P, Carter DRF (2017) The diagnostic and prognostic potential of microRNAs in epithelial ovarian carcinoma. Mol Diagn Ther 21(1):59–73
Wang X, Jiang L, Liu Q (2022) miR-18a-5p derived from mesenchymal stem cells-extracellular vesicles inhibits ovarian cancer cell proliferation, migration, invasion, and chemotherapy resistance. J Transl Med 20(1):1–19
Yin J, Huang H-Y, Long Y, Ma Y, Kamalibaike M, Dawuti R et al (2021) circ_C20orf11 enhances DDP resistance by inhibiting miR-527/YWHAZ through the promotion of extracellular vesicle-mediated macrophage M2 polarization in ovarian cancer. Cancer Biol Ther 22(7–9):440–454
Liu J, Yoo J, Ho JY, Jung Y, Lee S, Hur SY et al (2021) Plasma-derived exosomal miR-4732–5p is a promising noninvasive diagnostic biomarker for epithelial ovarian cancer. J Ovarian Res 14(1):59
Taheri M, Shoorei H, Anamag FT, Ghafouri-Fard S, Dinger ME (2021) LncRNAs and miRNAs participate in determination of sensitivity of cancer cells to cisplatin. Exp Mol Pathol 123:104602
Amirmahani F, Ebrahimi N, Askandar RH, Rasouli Eshkaftaki M, Fazeli K, Hamblin MR (2021) Long noncoding RNAs CAT2064 and CAT2042 may function as diagnostic biomarkers for prostate cancer by affecting target MicrorRNAs. Indian J Clin Biochem. https://doi.org/10.1007/s12291-021-00999-6
Grange C, Tapparo M, Collino F, Vitillo L, Damasco C, Deregibus MC et al (2011) Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Can Res 71(15):5346–5356
Qu L, Ding J, Chen C, Wu Z-J, Liu B, Gao Y et al (2016) Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 29(5):653–668
Xiao H, Shi J (2020) Exosomal circular RNA_400068 promotes the development of renal cell carcinoma via the miR-210-5p/SOCS1 axis. Mol Med Rep 22(6):4810–4820
Guo Z, Wang X, Yang Y, Chen W, Zhang K, Teng B et al (2020) Hypoxic tumor-derived exosomal long noncoding RNA UCA1 promotes angiogenesis via miR-96-5p/AMOTL2 in pancreatic cancer. Mol Therapy Nucl Acids 22:179–195
Zhou R, Chen KK, Zhang J, Xiao B, Huang Z, Ju C et al (2018) The decade of exosomal long RNA species: an emerging cancer antagonist. Mol Cancer 17(1):1–14
Buschmann D, Kirchner B, Hermann S, Märte M, Wurmser C, Brandes F et al (2019) Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J Extracell Vesicles 8(1):1581487
Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M et al (2013) Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics 14(1):1–14
Sempere LF, Keto J, Fabbri M (2017) Exosomal MicroRNAs in breast cancer towards diagnostic and therapeutic applications. Cancers (Basel). 9(7) PubMed PMID: 28672799. Pubmed Central PMCID: PMC5532607. Epub 2017/07/05. eng
Rupp AK, Rupp C, Keller S, Brase JC, Ehehalt R, Fogel M et al (2011) Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol Oncol 122(2):437–446 (PubMed PMID: 21601258. Epub 2011/05/24. eng)
Işın M, Uysaler E, Özgür E, Köseoğlu H, Şanlı Ö, Yücel ÖB et al (2015) Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Front Genet 6:168
Dijkstra S, Birker IL, Smit FP, Leyten GH, de Reijke TM, van Oort IM et al (2014) Prostate cancer biomarker profiles in urinary sediments and exosomes. J Urol 191(4):1132–1138
Wang J, Zhou Y, Lu J, Sun Y, Xiao H, Liu M et al (2014) Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med Oncol 31(9):1–8
Dong L, Lin W, Qi P, Xu M-d, Wu X, Ni S et al (2016) Circulating long RNAs in serum extracellular vesicles: their characterization and potential application as biomarkers for diagnosis of colorectal cancer. Cancer Epidemiol Biomark Prev 25(7):1158–1166
Acknowledgements
The authors would like to thank all of those whose fruitful research has contributed in any way to the elucidation of tumor-derived exosomes roles and their ncRNA expression profiles as diagnostic biomarkers in oncology. The author(s) received no financial support for the research, authorship, and/or publication of this article.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
NE, FF, and SSF contributed to conceptualization, writing, and original draft preparation; PRM, EY, ZK, FR-T, and SA performed data curation and writing; MR. H and ARA performed supervision and writing—reviewing and editing.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ebrahimi, N., Faghihkhorasani, F., Fakhr, S.S. et al. Tumor-derived exosomal non-coding RNAs as diagnostic biomarkers in cancer. Cell. Mol. Life Sci. 79, 572 (2022). https://doi.org/10.1007/s00018-022-04552-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04552-3