Abstract
Prognosis of lung cancer still remains grim largely due to recurrence and aggressive metastasis of the disease. In this study, we examined the potential of exosomal miRNAs as biomarkers of recurrent lung cancer. Initially, in vitro miRNA profiles of normal lung (Beas-2b) and lung cancer (H1299) cells and of exosomes isolated from conditioned media were determined. In vivo study involved establishing subcutaneous primary and recurrent lung cancer xenografts in nude mouse model and examining tumor and serum exosomal miRNA alteration in secondary/recurrent lung tumors. A total of 77 miRNAs were observed to be significantly modulated in the H1299 cells (47 miRNA upregulated and 30 downregulated) compared to the Beas-2b cells. The exosomes isolated from conditioned media indicated several miRNAs which were in agreement with cells of origin. A similarity was also observed between miRNAs from serum exosomes and tumors, indicating their origin from the lung tumors. Two miRNAs, miR-21 and miR-155, were found to be significantly upregulated in recurrent tumors compared to primary tumors. These miRNAs were also upregulated in serum exosomes of recurrent tumor-bearing animals versus non-tumor- or primary tumor-bearing animals. Increased expression of the recurrent disease markers were also observed in recurrent tumors compared with primary tumors. Serum exosomes from recurrent tumor mice mirrored its tumor profile in expressing higher levels of these proteins compared with exosomes from primary tumor mice. Our data suggest that exosomal miRNA signatures may be a true representation of a pathological profile of lung cancer; thus, miRNAs could serve as promising biomarkers for non-invasive diagnosis of the disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
More people in the USA die of lung cancer than of prostate, breast, and colon cancers combined [1]. Non-small cell lung cancer (NSCLC) is the most common type of lung cancer and accounts for 85 to 90 % of all lung cancers. Eighty-five percent of patients eventually develop advanced or recurrent NSCLC. The lung cancer 5-year survival rate (17.4 %) is lower than most other leading cancer sites, such as the colon (64.2 %), breast (89.4 %), and prostate (98.6 %) [2]. Poor outcomes and relapses indicate the urgent need for development of new screening and early biomarkers for rapid, non-invasive, and sensitive detection of recurrent lung tumors leading to prevention strategies.
Early diagnosis of recurrent lung cancer can have tremendous impact in decreasing mortality rates with timely therapeutic interventions and disease management. Exosomes have been identified as novel mechanisms of cell-cell communications and horizontal transfer of information via its bioactive molecular cargo of miRNA, mRNA, DNA, and proteins [3]. Exosomes are small (30–100 nm) membrane vesicles of endocytic origin that are released into the extracellular environment upon fusion of multivesicular bodies (MVB) with the plasma membrane [4]. Recent studies indicate the role of exosomes in crosstalk between tumor and normal cells resulting in cell recruitment facilitating the malignant process [5]. Exosomes from tumors were shown to condition tumor microenvironment and stromal cells with induction of proliferation and tumor metastasis [6, 7]. Due to their bioactive cargo, exosomes are capable of re-programming the recipient cells in the tumor microenvironment as well as distal cells from their release site.
miRNAs are frequently dysregulated in NSCLC and are implicated in lung cancer growth, recurrence, and metastasis [8–10]. For this reason, miRNAs are considered as valuable biomarkers for diagnosis and therapeutic monitoring of disease [11, 12]. miRNAs are small non-coding RNAs that post-transcriptionally regulate mRNAs. Evidence from recent studies indicates promise of miRNAs in risk assessment, prevention, early diagnosis, and prognosis of NSCLC [13–15]. Intracellular miRNAs are released into extracellular space and body fluids either packaged in lipoproteins [16] or secreted in extracellular vesicles such as exosomes [3], to shielding them from RNase activity. Exosomes are released from both normal as well as tumor cells. Studies indicate that tumor cells secrete increased amounts of exosomes compared to normal cells [17]. A significant increase in circulatory levels of exosomes and miRNA concentration has been reported in lung cancer patients [18]. Recent studies indicate a promising relationship between exosomal miRNA profiles in blood with the pathological condition of patient with AML and colon and gastric cancers [19–21].
In this study, miRNA profiles of normal bronchial epithelial cells, lung cancer cells, and exosomes isolated from the conditioned media were determined. Later, we established subcutaneous primary and recurrent lung cancer xenografts and characterized miRNA profiles of serum exosomes and identified miRNAs that are altered in secondary/recurrent lung cancer and determined if miRNAs carried by the tumor-derived exosomes represent tumor profile and be more stringent circulatory biomarkers.
Results
Isolation and characterization of vesicles from conditioned media and mouse serum
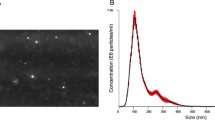
Exosomes were isolated from conditioned media of lung cancer (H1299) and normal bronchial epithelial (Beas-2b) cells after 72 h of culture by ultracentrifugation, and serum exosomes were isolated from nude mice bearing H1299 xenograft by precipitation with ExoQuick reagent (Systems Bioscience, Mountain View, CA). Vesicles isolated from conditioned media and mouse serum were confirmed to be exosomes based on size (30–100 nm) as determined by NanoSight (Fig. 1a) and transmission electron microscopy (TEM) (Fig. 1b). Total RNA isolated from exosomes was analyzed by Bioanalyzer (Agilent Technologies, Santa Clara, CA), and the electropherograms revealed the presence of miRNA species in both cell-derived and serum-derived exosomes, with no detectable 18S and 28S ribosomal RNAs (Fig. 1c). We also observed that the yield of serum exosomal RNA was significantly higher in primary tumor-bearing (25.6 ± 3.7 ng/μl; p = 0.049) versus non-tumorous mice (16.6 ± 4.6 ng/μl); the content was much higher in recurrent tumor-bearing mice (64.9 ± 29.0 ng/μl; p = 0.045) (Fig. 1d).
Exosome characterization. a Size and visualization of serum exosomes by NanoSight. b Visualization of serum exosomes by scanning electron micrography (×53,000). c Digital gel electrophoresis of RNA from Beas-2b and H1299 exosomes and serum exosomes from primary tumor (PT)- and recurrent tumor (RT)-bearing mice; lane1, size markers, and lane 2, synthetic miR-200a. Representative graph showing miRNA peak from cell-derived exosomes (top) and serum exosome-derived (bottom). d The bar graph depicts total serum exosomal RNA yield from non-tumor-, primary tumor-, and recurrent tumor-bearing mice
miRNA expression profile of lung cancer cells and their secreted exosomes
We evaluated miRNA expression profile of lung cancer and normal bronchial epithelial cells as well as in exosomes secreted from these cells in culture media. In order to identify the modulated miRNAs, we analyzed the expression profile of miRNAs in human lung cancer H1299 cells and normal Beas-2b cells using qPCR cancer-pathway finder miRNA array (Qiagen, Valencia, CA) that contained 84 miRNAs. miRNA analysis showed an aberrant miRNA expression in lung cancer cells compared with normal lung cells. After normalizing the values using miR374-5p, values with significant differences (p < 0.05) were analyzed using hierarchical clustering of the log 2 value and displayed in a heat map. These results reflect an inverse miRNA expression with several miRNAs being upregulated and downregulated in cancer cells compared with normal cells (Fig. 2a). Volcano plot shows that nearly 48 miRNAs were upregulated and 30 miRNAs were downregulated in cancer cells compared with normal cells. Significantly upregulated miRNAs included miR-182, miR-185, miR-21, miR-127, miR-142, miR-155, etc., and miRNAs that were downregulated were miR-138, miR-125-5p, let-7e, miR-193b, miR-16, miR-26a, miR-345, and miR-423-5p, among others (Fig. 2b). A complete list of miRNAs significantly dysregulated in H1299 lung cancer cells is shown in Supplementary Table S1. Similar to cells, a dysregulated signature of miRNA expression was observed in H1299-derived exosomes compared with Beas-2b-derived exosomes (Fig. 2c). H1299 exosomes had 14 miRNAs that were significantly upregulated, and 28 miRNAs were under-expressed compared to Beas-2b exosomes (Fig. 2d). Supplementary Table S2 shows the list of significantly modulated miRNAs in H1299 exosomes. Several miRNAs had similar trends of overexpression (e.g., miR-132, miR-155, miR-21, miR-331-5p, and miR-483-5p) or downregulation (e.g., let-7e, miR-193b, miR-16, miR-26a, miR-345, and miR-423-5p) among lung cancer cells and exosomes derived from these cells, suggesting that exosomal miRNAs can reflect the uniqueness of the cells of their origin. Fig. 2e shows the number of shared and specific miRNAs between the cancer and normal cells as well as among cancer and normal cell derived-exosomes. It was observed that 190 miRNAs were detected common in the cancer and normal cells. Meanwhile, 35 and 54 unique miRNAs were detected in the cancer and normal cells, respectively. Remarkably, several high-number miRNAs were selectively identified in the exosomes of normal cells (108 miRNAs) compared with cancer cell exosomes (36 miRNAs). Additionally, 74 miRNAs were shared common between the exosomes from the two cell types.
In vitro miRNA microarray data analysis. a Heat map represents the relative miRNA expression levels in normal lung Beas-2b and lung cancer H1299 cells and (c) exosomes secreted from Beas2b and H1299 cell lines (n = 3 each group). The miRNAs that demonstrated at least 2-fold changes with p < 0.05 were selected for hierarchical cluster analysis to generate the heat map. The color bar depicts the color contrast level of the heat map. Red and green indicate high- and low-expression levels, respectively. b Volcano plots of differentially expressed miRNAs in Beas-2b and H1299 cells and d exosomes from Beas-2b and H1299 cells. Log 2-fold changes in expression are plotted against −log 10-corrected p values of significance. Each point represents an miRNA, with miRNAs shown in red in the upper left and right quadrants differentially expressed (fold change >2 upregulation or downregulation and corrected p < 0.05). The Venn diagram shows the number of miRNAs differentially expressed in normal and lung cancer cells (e) and their exosomes (f) and those common in both
Recurrent lung tumor model in vivo
In order to determine the biomarker significance of serum exosomal miRNAs in vivo, we established for the first time a recurrent lung tumor model using nude mice. Of 25 nude mice bearing subcutaneous H1299 lung xenograft tumors, measuring 350–400 mm3 by volume, tumors from 18 animals were surgically removed while primary tumors in 7 animals continued to grow. These tumors were found to be encapsulated, and we were able to remove them completely surgically. The first recurrent tumor appeared 1 week post-surgery (Fig. 3a), and the incidence rate of recurrent tumors was about 60 % (11/18) with a tumor volume of 431 ± 236 mm3 3 weeks post-surgery at the termination of the study. The recurrent tumors were analyzed for their human cell origin and for the presence of any contaminating mouse cells by qPCR using species-specific GAPDH primers. The H1299 lung cancer cells and mouse liver were used as positive and negative controls, respectively. The data indicated that a human-specific GAPDH primer was amplified in primary and recurrent tumors resulting in a product of 258 bp, while no amplification was observed with mouse-specific primers (Fig. 3b), thus confirming recurrent tumors to be of human origin with essentially no mouse cell contamination.
Recurrent tumor mouse model. a Growth of recurrent tumors in nude mice inoculated with human lung cancer H1299 cells (1.5 × 106 cells), followed by complete removal of the primary tumor (18/25 mice) after 3.5–4 weeks. Number of mice with recurrent tumors = 11/18. Data show mean tumor volume ± SD; n = 7 (primary), n = 11 (recurrent). b RT-PCR analysis of total RNA from H1299 cells, primary tumor xenograft, recurrent tumor, and mouse liver (negative control) with mouse- and human-specific GAPDH primers
Tumor and serum exosomal miRNA profile
Small RNAs enriched from tumor tissues and serum exosomes from animals bearing primary and recurrent tumors were subjected to cancer-pathway finder miRNA array analysis. A heat map of primary and recurrent tumor miRNAs indicates that several miRNAs were common among the two tumor types, indicating that recurrent tumors to a great extent resemble primary tumors with respect to the miRNA profile (Fig. 4a). Further, a Venn diagram shows that at least 221 miRNAs were shared by both primary and recurrent tumors while only 40 and 13 miRNAs were unique to primary and recurrent tumors, respectively (Fig. 4b). We also identified that at least 10 miRNAs were significantly differentially modulated in recurrent tumors compared with primary tumors. Among miRNAs that were upregulated included miR-129-3p, miR-96, miR-21, miR-95, miR-155, and miR-337-5p, while three miRNAs, namely, miR-299-5p, miR-369-3p, and miR-127-5p, were downregulated in recurrent tumors (Fig. 4c, Supplementary Table S3).
miRNA microarray data analysis of primary and recurrent tumors. a Heat map shows the relative miRNA expression levels in primary and recurrent lung tumors. The miRNAs that demonstrated at least 2-fold changes with p < 0.05 were selected for hierarchical cluster analysis to generate the heat map. The color bar depicts the color contrast level of the heat map. Red and green indicate high and low expression levels, respectively. b The Venn diagram depicts the number miRNAs differentially expressed and those common in primary and recurrent lung tumors. c Volcano plots of differentially expressed miRNAs in primary and recurrent versus primary tumors. Log 2-fold changes in expression are plotted against −log 10-corrected p values of significance. Each point represents an miRNA, with miRNAs shown in red in the upper left and right quadrants differentially expressed (fold change >2 upregulation or downregulation and corrected p < 0.05)
After identifying miRNAs that were modulated in recurrent tumors, we examined the serum exosomal miRNA profile in vivo. Serum exosomal RNAs from non-tumor (control) and primary tumor and recurrent tumor-bearing animals were examined. Serum exosomes samples showed low-positive amplification rate, i.e., ct values greater than 35 compared to tumor samples indicating low abundance. The number of miRNAs amplified also varied among replicates and between groups ranging 18–66 % positive amplifications, and 2–11 % of these signals had ct values greater than 35. However, even with low positive signals from serum exosomes, we were able to identify a distinct miRNA modulation between exosomes from primary tumor- and recurrent tumor-bearing animals compared to non-tumor control mice (Fig. 5a). In addition, our findings suggested that at least two miRNAs were significantly upregulated, miR-21 (73.2-fold, p = 0.039) and miR-155 (68.2-fold, p = 0.047), in exosomes from recurrent tumor compared to primary tumor-bearing animals (Fig. 5b). A Venn diagram comparing miRNAs from tumor and its respective serum exosomes shows a majority of miRNAs unique to tumor with nearly 20 miRNAs common to both tumor and serum exosomes (Fig. 5c). This observation could be an artifact due to low positive amplification rate in serum-derived exosomes and should be interpreted with caution.
miRNA microarray data analysis of serum exosomes from primary tumor- and recurrent tumor-bearing mice. a Heat map representing the relative miRNA expression levels in serum exosomes from primary and recurrent lung tumor animals. The miRNAs that demonstrated at least 2-fold changes with p < 0.05 were selected for hierarchical cluster analysis to generate the heat map. The color bar depicts the color contrast level of the heat map. Red and green indicate high and low expression levels, respectively. b The Venn diagram shows the number of miRNAs differentially expressed and those common in serum exosomes from primary and recurrent lung tumor animals. c Volcano plots of differentially expressed miRNAs in serum exosomes from primary and recurrent versus primary tumor-bearing mice. Log 2-fold changes in expression are plotted against −log 10-corrected p values of significance. Each point represents an miRNA, with miRNAs shown in red in the upper left and right quadrants differentially expressed (fold change >2 upregulation or downregulation and corrected p < 0.05)
Expression of recurrent tumor-associated proteins
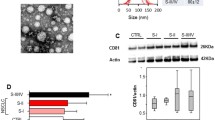
We examined expression of five proteins, namely, Notch1, cyclin D1 (CCND1), vascular endothelial growth factor (VEGF), matrix metalloproteinase-2 (MMP-2), and heat shock protein 90 (Hsp90), associated with aggressiveness, metastasis, recurrence, and poor prognosis of NSCLC [22–24] in primary and recurrent tumors from mice and determined if expression levels of these proteins were reflective of cancer stage. Western blot analysis of primary and recurrent lung tumors (n = 4) indicated an increased expression of the miRNA target proteins, Notch1 (1.4-fold), CCND1 (7.2-fold), VEGF (5.0-fold), MMP-2 (2.9-fold), and Hsp90 (2.1-fold) in recurrent tumor tissue compared to primary tumors (Fig. 6a) which corresponds to an aggressive phenotype. Subsequently, we examined if serum exosomes in circulation would reflect the nature of the tumor from which they are secreted.
Analysis of protein markers in tumors and serum exosomes. a Protein markers associated with recurrence disease were analyzed by western blot in primary H1299 tumor xenografts and recurrent tumors. Data show mean ± SEM. **p < 0.01 and *p < 0.05 compared to primary tumor, t tests. b Serum exosomes from non-tumor (control), primary tumor, and recurrent tumor animals analyzed for select recurrent disease markers. Fold change in various protein markers is expressed after normalization to β-actin control. Data show mean ± SEM. *p < 0.05 compared to primary tumor or no tumor control, t tests
Protein lysates of serum exosomes from non-tumor- and primary tumor- and recurrent tumor-bearing mice were probed for VEGF and MMP-2. The results showed a significant increase in expression of these two proteins in exosomes from recurrent tumor mice compared to primary tumor and non-tumor animals. Serum exosomes exhibited nearly 47- and 213-fold higher expression of VEGF and 21-fold higher levels of MMP-2 in recurrent tumor-bearing animals compared to non-tumor animals (Fig. 6b). Moreover, recurrent serum exosomes were found to have at least 3.8- and 16.2-fold higher levels of VEGF and MMP-2 protein expression compared to exosomes from primary tumor-bearing animals. These findings suggest that circulating serum exosomes could also be reflective of tumor stage.
Discussion
Analysis of exosomal cargo such as miRNAs is being considered as valuable source of markers for diagnosis and therapy monitoring. Since exosomes are actively released from cells, and circulatory exosomes are reported in almost all bodily fluids such as blood, cerebrospinal fluid, saliva, milk, and urine [25], diagnostic tests that involve non-invasive procedures such as simple blood draw are highly desirable and patient friendly. Circulating exosomes in the blood therefore offer a new platform of biomarkers for tumor diagnosis and monitoring of therapy response [26, 27].
The aim of this study was to identify specific serum exosomal miRNAs as biomarkers reflecting the recurrence of lung cancer using mouse model. We hypothesized that lung cancer cells secreted exosomes containing miRNAs into the blood that can be used to differentiate primary from recurrent tumors. Our initial in vitro studies showed several miRNAs to be upregulated in lung cancer cells such as miR-182, miR-185, miR-21, miR-127, miR-142, and miR-155 compared with normal bronchial epithelial cells. Interestingly, several of these miRNAs were also found to be upregulated in exosomes isolated from conditioned media of lung cancer cells suggesting that exosomal miRNA profile can represent the cells of its origin.
This study first reports on the establishment of recurrent lung tumors in athymic nude mice after surgical removal of subcutaneous xenograft tumors. We observed tumor recurrence in 61 % of the mice post-surgery, suggesting an aggressive nature of lung cancer and an occurrence of recurrent disease from microscopic tumor cells at the primary tumor site post-surgery. This phenomenon resembles human scenario where secondary lung tumor recurrence occurs after surgical resection and treatment. miRNA profiles of excised tumors showed that 58 % of miRNAs (221 out of 384 miRNAs) were shared by both primary and recurrent tumors, further suggesting that recurrent tumor resembles the primary tumor to a large extent. Our data further indicated that at least seven miRNAs were significantly upregulated in recurrent tumors compared with primary tumor. Most of these upregulated miRNAs, miR-129-3p, miR-96, miR-21, miR-95, miR-155, and miR-337-5p, have been associated with aggressive disease, metastasis, and recurrence of lung and other cancers [12, 28–30].
Analysis of serum exosomal RNA yield indicated a significantly higher level of RNAs in exosomes from tumor-bearing mice compared with non-tumor control mice. This increase could be attributed to higher secretion of exosomes from tumors compared to normal tissues. A still higher amount of exosomal RNAs was observed in serum from recurrent tumor- than primary tumor-bearing mice; however, it was not statistically significant. In agreement with our observations, a significant difference in total exosomes and the exosomal miRNA levels between patients and healthy controls has been reported [18], indicating that tumor burden increases the serum exosome levels in circulation. miRNA analysis indicated low amplification rates in serum exosome samples. This shortcoming can presumably be addressed by introducing a pre-amplification step prior to PCR to improve the results. However, unavailability of serum from all the groups prevented repeat analysis in our case. Nevertheless, at least two miRNAs (miR-21 and miR-155) were significantly upregulated in exosomes from recurrent tumors compared to primary tumor-bearing animals. It is worth noting that these two miRNAs were also significantly upregulated in recurrent tumors compared with primary tumors, suggesting that serum exosomal miRNA signature is reflective of tumor profile.
Our findings are consistent with recent findings where exosomal miRNA signature was shown to emulate pathological changes in colon [31] and prostate [32] cancer patients with several miRNAs secreted at significantly higher levels compared to normal subjects. Yanaihara et al. revealed an overexpression of 12 specific miRNAs (has-miR-17-3p, hsa-miR-21, hsa-miR-106a, hsa-miR-146, hsa-miR-155, hsa-miR-191, hsa-miR-192, hsa-miR-203, hsa-miR-205, hsa-miR-210, hsa-miR-212, and hsa-miR-214) in lung tumor compared with normal lung tissue [12].
Rabinowits et al. compared 12 specific exosome-derived miRNAs and tumor-derived miRNAs in lung cancer patients and healthy subjects and showed that there was no significant difference between circulatory miRNAs and tumor miRNAs and thus the exosome-derived miRNAs can be used as biomarkers for lung cancer [18]. In another study, overexpression of exosomal miR-19a in serum was identified as a prognostic biomarker for recurrence in colorectal cancer patients [31, 33]. Therefore, the matching miRNA signatures of exosomes with its originating tumor cells, indicating that exosomal miRNA profiling can be performed in the absence of tumor biopsy in patients, can accurately reveal the tumor’s profile.
Advances in molecular biology have led to identification of an extensive arsenal of biomarkers implicated in the literature as having diagnostic and/or prognostic value for NSCLC. We focused on biomarkers primarily involved in growth regulation, cell cycle regulation, apoptosis, and angiogenesis that were associated with predication of early recurrence and poor prognosis in NSCLC [22, 24, 34, 35]. Local tumor expansion requires growth-regulating (EGFR, HSP90) and cell cycle-regulating (CCND1, CCNE, PCNA) proteins. While tumor invasion requires expression of angiogenesis markers such as VEGF and Notch1, the development of distant metastases involves the expression of adhesion proteins (CD-44, e‐cadherin, matrix metalloproteinases (MMPs)) [36].
CCND1 is the one of the most studied cyclin in NSCLC. CCND1 overexpression was noted in approximately 50 % of NSCLC, but its role in the prognosis for lung cancer patients continues to be unclear because of the conflicting results reported in literature. Over the last decade, at least four studies have identified CCND1 overexpression to be a negative prognostic marker [37–40] whereas one study associated it with better prognosis [41] and three reports indicated no association [23, 42, 43]. These marked disparities in prognostic impact of CCND1 can, in part, be attributed to the heterogeneity in patient selection, inclusion of mixed tumor types, or laboratory methodology used in different studies. Heat shock protein 90-beta (Hsp90-β) is associated with cell proliferation, differentiation and apoptosis, and has been investigated as a prognostic factor in many cancers. Recent studies have indicated upregulation of serum Hsp90-β to be associated with pathological grade and clinical stage of lung cancer patients [34, 44].
Overexpression of VEGF has also been associated with tumor progression and poor prognosis in most cancer types, including 60 % of NSCLC. A recent meta-analysis of 74 studies covering 7631 patients reported that overexpression of VEGF resulted in unfavorable prognosis in NSCLC [45]. Another meta-analysis examined 19 studies involving 3663 patients for the role of Notch1 in NSCLC prognosis. Findings of this study indicated that higher expression of Notch1 was not only associated with greater possibility of lymph node metastasis and higher TNM stages but also showed significantly poor overall survival [35]. MMPs have the ability to degrade fibrillar collagens and are made of almost 20 family members. Results from meta-analysis studies have suggested that high expressions of MMP‐2, MMP-7, and MMP‐9 are poor prognostic markers for NSCLC, especially the expression of MMP‐2 [46–48].
Our findings indicated significant higher expression of HSP90, CCND1, VEGF, Notch1, and MMP-2 proteins in recurrent tumors compared to primary tumors, reflecting their ability to indicate recurrent disease or severity of cancer stage. It was interesting to note that serum exosomes mirrored the tumor profile with respect to expression of VEGF and MMP2 proteins. It is presumed that several of the deregulated miRNAs could possibly play a yet unconfirmed mechanistic role in modulating the expression of these tumor biomarkers. These observations also corroborate the assumption that exosomal cargo of macromolecules including miRNAs and proteins can be a true representation of tumor profile and thus have great biomarker potential for screening of asymptomatic individuals and to monitoring disease recurrence. However, validation studies are warranted before bypassing its use with tumor mass biopsies. Reproducibility during validation studies can be challenging due to the possible changes of miRNA expression in the circulation that originate from other tissues and organs. Alternatively, enrichment of serum exosomes using tumor markers, such as EpCAM, can be employed prior to analysis of exosome-associated miRNA profile. This approach could further improve sensitivity and specificity of miRNA-based diagnostic test.
Conclusions
In summary, attempts have recently been made to use exosomal miRNAs as diagnostic and prognostic biomarkers of various cancers, including lung cancer. Exosomes have several unique properties such as biomolecule cargo of miRNAs, stability in circulation, and representation of tumor profile that make them suitable for the development of highly sensitive non-invasive diagnostic strategies for monitoring cancer patients. Our data suggest the important role of exosomal miRNA profiles in distinguishing recurrent from primary lung tumors and identified miR-21 and miR-155 as potential markers of recurrent lung disease. However, further research and validation is required to determine which miRNAs should be selected as markers. Enrichment of serum exosomes for tumor markers is likely to increase the biomarker significance. Exosomal miRNAs can be a true representation of tumor profile and can be applied as functional biomarkers for diagnosis and outcome predictions.
Methods
Development of recurrent lung tumor model
To determine frequency of lung tumor recurrence, female athymic nude mice (4–5 week-old; n = 25) were inoculated with H1299 cells (1.5 × 106 cells) in matrigel. When tumors reached 350–400 mm3, the tumors were excised completely under anesthesia from 18/25 mice; the primary tumors in the remaining animals (n = 7) were allowed to grow. Animals were monitored initially daily to ascertain recovery from the surgery and thereafter biweekly, and volumes of recurrent tumors were measured with a digital caliper. At the end of study, animals were euthanized by CO2 asphyxiation and tumors and serum were collected and stored at −80 °C until use.
Isolation of exosomes
Conditioned media was harvested from lung cancer (H1299) and normal bronchial epithelial (Beas-2b) cells after 72 h of culture and centrifuged using a Sorvall legend RT centrifuge at 300 g at 4 °C for 10 min to remove detached cells. Supernatant was collected and filtered through 0.22-μm filters (Merck Millipore) to remove contaminating apoptotic bodies, microvesicles, and cell debris. Clarified conditioned media was then centrifuged at 100,000 × g in Type 45 Ti fixed angle rotor using Optima LE-80K Ultracentrifuge (Beckman Coulter, Brea, CA) at 4 °C overnight to pellet exosomes. The supernatant was carefully removed, and crude exosome-containing pellets were resuspended in 1 mL of ice-cold PBS and pooled. Exosomes were isolated from 1 ml serum (pool from five mice) using ExoQuick reagent (System Biosciences, Mountain View, CA).
NanoSight
Vesicles isolated from conditioned cell culture media and serum were analyzed by nanoparticle tracking, using the NanoSight LM10 system (NanoSight Ltd, Wiltshire, UK), configured with a 635-nm laser and a high-sensitivity digital camera system (ORCA-Flash2.8, Hamamatsu C11440, NanoSight Ltd). Each sample was diluted appropriately in distilled water to give counts in the linear range of the instrument. The particles in the laser beam undergo Brownian motion, and videos of these particle movements are recorded. Recorded videos were analyzed using the NTA software (version 2.3), with the minimal expected particle size, minimum track length, and blur setting, all set to automatic [49]. All samples were analyzed in triplicates.
Transmission electron microscopy
TEM was performed by the method of Thery et al. [50]. Exosomes isolated by ExoQuick reagent were suspended in PBS. A drop of this solution was allowed to settle on a gold-coated grid, fixed in 1 % glutaraldehyde, washed for 2 min in double-distilled water, and incubated in uranyl oxylate for 5 min, followed by incubation with three separate drops of methyl cellulose with uranyl acetate—5 min with the first two drops and 10 min with the last drop—and finally, methyl cellulose-uranyl acetate was removed by slow-drag on edge on filter paper. Exosomes were visualized by standard TEM with a Philips CM120 microscope.
RNA Isolation
mirVana miRNA Isolation Kit (Applied Biosystems, Foster City, CA) was used to isolate RNA [51]. Small RNA was further enriched from total RNA for qPCR analysis of miRNAs according to the manufacturer’s protocol. The concentration of the total and small RNA was determined with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE), and RNA integrity was verified with a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA).
Bioanalyzer
Total RNA from exosomes and small RNA tissue samples was analyzed on the 2100 Bioanalyzer. Quantification and quality assessment of small RNA including the miRNA fraction were undertaken with the “Small RNA Assay” (Agilent Technologies, Santa Clara, CA). Synthetic miR-200a was used as reference.
RT-PCR
Purified DNA was subjected to PCR amplification using human- and mouse-specific GAPDH primers to amplify 258-bp and 512-bp fragments, respectively. The primers are as follows: human GAPDH forward, 5′-AGA AGG CTG GGG CTC ATT TG-3′; reverse, 5′-AGG GGC CAT CCA CAG TCT TC-3′, and mouse GAPDH forward, 5′-AGG CCG GTG CTG AGT ATG TC-3′; reverse 5′-TGC CTG CTT CAC CAC CTT CT-3′. PCR conditions were as follows: preheating for 2 min at 94 °C was followed by 30 cycles of 30 s at 94 °C, 30 s at 55 °C, and 60 s at 68 °C and a final extension of 10 min at 68 °C. PCR was performed in a final volume of 50 μl, which consisted of 1 μl extracted DNA as template, 5 μl 10× PCR buffer, 1 U Platinum Taq Polymerase High Fidelity (Invitrogen, Carlsbad, CA), 20 pmol of each primer, and 0.2 mM dNTPs. The final concentration of MgCl2 was 2.5 mM. The amplicons were checked on 1.5 % agarose (NuSieve, Cambrex, ME).
miRNA PCR Array
To determine tumor and circulating exosomal miRNA profiles, we analyzed primary and recurrent H1299 tumors and serum exosomes from control (non-tumor) and H1299 lung tumor-bearing nude mice. Small RNAs were enriched from tumors using mirVana Kit (Life technologies, Carlsbad, CA), and total RNA was isolated from exosomes by TRIzol and 500 ng of the total RNA was analyzed by qPCR using cancer-pathway finder miRNA array, containing 84 miRNAs (Qiagen, Valencia, CA).
Western blot analysis
Tumor tissue lysates and serum exosomes from tumor- and non-tumor-bearing animals were prepared in RIPA buffer. Protein concentration was determined using the BCA method. Sample lysates were separated on SDS-PAGE gel, and blots were blocked and then incubated with the primary antibodies overnight at 4 °C. After washing, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody. The transferred proteins were visualized with enhanced chemiluminescence detection kits (Amersham, ECL kits, Sunnyvale, CA).
Statistical analysis
All data are expressed as the mean ± SD or mean ± SEM. Data were compared using Mann-Whitney U test or one-way ANOVA. Values of p < 0.05 were considered to indicate statistical significance. All statistical analyses were performed using GraphPad Prism 5.01 (GraphPad Software, La Jolla, CA).
References
American Cancer Society. Cancer Facts & Figures 2012.
Howlader N, Noone AM, Krapcho M, Garshell J, Miller D, Altekruse SF et al. SEER Cancer Statistics Review, 1975–2012. National Cancer Institute. Bethesda, MD, http://seer.cancer.gov/csr/1975_2012/. based on November 2014 SEER data submission, posted to the SEER web site, April 2015.
Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–U72. doi:10.1038/ncb1596.
Urbanelli L, Magini A, Buratta S, Brozzi A, Sagini K, Polchi A, et al. Signaling pathways in exosomes biogenesis, secretion and fate. Genes. 2013;4(2):152–70. doi:10.3390/genes4020152.
Roma-Rodrigues C, Fernandes AR, Baptista PV. Exosome in Tumour Microenvironment: Overview of the Crosstalk between Normal and Cancer Cells. Biomed Res Int. 2014. doi:Artn 179486 10.1155/2014/179486.
Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883−+. doi:10.1038/nm.2753.
Rana S, Malinowska K, Zoller M. Exosomal tumor microRNA modulates premetastatic organ cells. Neoplasia. 2013;15(3):281–95.
Chen LT, Xu SD, Xu H, Zhang JF, Ning JF, Wang SF. MicroRNA-378 is associated with non-small cell lung cancer brain metastasis by promoting cell migration, invasion and tumor angiogenesis. Med Oncol. 2012;29(3):1673–80. doi:10.1007/s12032-011-0083-x.
Lin PY, Yu SL, Yang PC. MicroRNA in lung cancer. Br J Cancer. 2010;103(8):1144–8. doi:10.1038/sj.bjc.6605901.
Wang X, Ling C, Bai Y, Zhao J. MicroRNA-206 is associated with invasion and metastasis of lung cancer. Anat Rec. 2011;294(1):88–92. doi:10.1002/ar.21287.
Hennessey PT, Sanford T, Choudhary A, Mydlarz WW, Brown D, Adai AT, et al. Serum microRNA biomarkers for detection of non-small cell lung cancer. PLoS One. 2012;7(2), e32307. doi:10.1371/journal.pone.0032307.
Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–98. doi:10.1016/j.ccr.2006.01.025.
Gao W, Xu J, Shu YQ. miRNA expression and its clinical implications for the prevention and diagnosis of non-small-cell lung cancer. Expert Rev Respir Med. 2011;5(5):699–709. doi:10.1586/ers.11.55.
Markou A, Liang Y, Lianidou E. Prognostic, therapeutic and diagnostic potential of microRNAs in non-small cell lung cancer. Clin Chem Lab Med. 2011;49(10):1591–603. doi:10.1515/CCLM.2011.661.
Feng B, Zhang K, Wang R, Chen L. Non-small-cell lung cancer and miRNAs: novel biomarkers and promising tools for treatment. Clin Sci. 2015;128(10):619–34. doi:10.1042/CS20140530.
Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13(4):423–33. doi:10.1038/ncb2210.
Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110(1):13–21. doi:10.1016/j.ygyno.2008.04.033.
Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10(1):42–6. doi:10.3816/CLC.2009.n.006.
Hornick NI, Huan J, Doron B, Goloviznina NA, Lapidus J, Chang BH et al. Serum Exosome MicroRNA as a Minimally-Invasive Early Biomarker of AML. Sci Rep-Uk. 2015;5. doi:Artn 11295 10.1038/Srep11295.
Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K et al. Circulating Exosomal microRNAs as Biomarkers of Colon Cancer. PloS one. 2014;9(4). doi:ARTN e92921 10.1371/journal.pone.0092921.
Tokuhisa M, Ichikawa Y, Kosaka N, Ochiya T, Yashiro M, Hirakawa K et al. Exosomal miRNAs from Peritoneum Lavage Fluid as Potential Prognostic Biomarkers of Peritoneal Metastasis in Gastric Cancer. PloS one. 2015;10(7). doi:ARTN e0130472 10.1371/journal.pone.0130472.
Zhu CQ, Shih W, Ling CH, Tsao MS. Immunohistochemical markers of prognosis in non-small cell lung cancer: a review and proposal for a multiphase approach to marker evaluation. J Clin Pathol. 2006;59(8):790–800. doi:10.1136/jcp.2005.031351.
Zhang LQ, Jiang F, Xu L, Wang J, Bai JL, Yin R, et al. The role of cyclin D1 expression and patient's survival in non-small-cell lung cancer: a systematic review with meta-analysis. Clin Lung Cancer. 2012;13(3):188–95. doi:10.1016/j.cllc.2011.10.003.
Singhal S, Vachani A, Antin-Ozerkis D, Kaiser LR, Albelda SM. Prognostic implications of cell cycle, apoptosis, and angiogenesis biomarkers in non-small cell lung cancer: a review. Clin Cancer Res. 2005;11(11):3974–86. doi:10.1158/1078-0432.CCR-04-2661.
Lasser C. Identification and analysis of circulating exosomal microRNA in human body fluids. Methods Mol Biol. 2013;1024:109–28. doi:10.1007/978-1-62703-453-1_9.
Cazzoli R, Buttitta F, Di Nicola M, Malatesta S, Marchetti A, Rom WN, et al. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol. 2013;8(9):1156–62. doi:10.1097/JTO.0b013e318299ac32.
Lin J, Li J, Huang B, Liu J, Chen X, Chen XM, et al. Exosomes: novel biomarkers for clinical diagnosis. Sci World J. 2015;2015:657086. doi:10.1155/2015/657086.
Lussier YA, Khodarev NN, Regan K, Corbin K, Li H, Ganai S, et al. Oligo- and polymetastatic progression in lung metastasis(es) patients is associated with specific microRNAs. PLoS One. 2012;7(12), e50141. doi:10.1371/journal.pone.0050141.
Wang R, Chen XF, Shu YQ. Prediction of non-small cell lung cancer metastasis-associated microRNAs using bioinformatics. Am J Cancer Res. 2015;5(1):32–51.
Saito M, Schetter AJ, Mollerup S, Kohno T, Skaug V, Bowman ED, et al. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res. 2011;17(7):1875–82. doi:10.1158/1078-0432.CCR-10-2961.
Matsumura T, Sugimachi K, Iinuma H, Takahashi Y, Kurashige J, Sawada G, et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer. 2015;113(2):275–81. doi:10.1038/bjc.2015.201.
Hessvik NP, Sandvig K, Llorente A. Exosomal miRNAs as biomarkers for prostate cancer. Front Genet. 2013;4:36. doi:10.3389/fgene.2013.00036.
Huang LL, Wang XY, Wen CY, Yang XL, Song MM, Chen JX et al. Hsa-miR-19a is associated with lymph metastasis and mediates the TNF-alpha induced epithelial-to-mesenchymal transition in colorectal cancer. Sci Rep-Uk. 2015;5. doi:Artn 13350 10.1038/Srep13350.
Rong BX, Zhao CC, Liu H, Ming ZJ, Cai XG, Gao WL, et al. Identification and verification of Hsp90-beta as a potential serum biomarker for lung cancer. Am J Cancer Res. 2014;4(6):874–85.
Yuan X, Wu H, Xu H, Han N, Chu Q, Yu S, et al. Meta-analysis reveals the correlation of Notch signaling with non-small cell lung cancer progression and prognosis. Sci Rep. 2015;5:10338. doi:10.1038/srep10338.
D'Amico TA, Massey M, Herndon JE, Moore MB, Harpole DH. A biologic risk model for stage I lung cancer: Immunohistochemical analysis of 408 patients with the use of ten molecular markers. J Thorac Cardiov Sur. 1999;117(4):736–42. doi:Doi 10.1016/S0022-5223(99)70294-1.
Sterlacci W, Fiegl M, Hilbe W, Jamnig H, Oberaigner W, Schmid T, et al. Deregulation of p27 and Cyclin D1/D3 control over mitosis is associated with unfavorable prognosis in non-small cell lung cancer, as determined in 405 operated patients. J Thorac Oncol. 2010;5(9):1325–36.
Zhu J, Yu L, Zhan P, Song Y, Wang Q. The relationships between cyclin D1 expression and prognosis of non-small cell lung cancer. Zhongguo fei ai za zhi =Chinese J Lung Cancer. 2010;13(8):803–8. doi:10.3779/j.issn.1009-3419.2010.08.10.
Li R, An SJ, Chen ZH, Zhang GC, Zhu JQ, Nie Q, et al. Expression of cyclin D1 splice variants is differentially associated with outcome in non-small cell lung cancer patients. Hum Pathol. 2008;39(12):1792–801. doi:10.1016/j.humpath.2008.05.008.
Burke L, Flieder DB, Guinee DG, Brambilla E, Freedman AN, Bennett WP, et al. Prognostic implications of molecular and immunohistochemical profiles of the Rb and p53 cell cycle regulatory pathways in primary non-small cell lung carcinoma. Clin Cancer Res. 2005;11(1):232–41.
Khoury T, Alrawi S, Ramnath N, Li Q, Grimm M, Black J, et al. Eukaryotic initiation factor-4E and cyclin D1 expression associated with patient survival in lung cancer. Clin Lung Cancer. 2009;10(1):58–66. doi:10.3816/CLC.2009.n.009.
Dworakowska D, Jassem E, Jassem J, Boltze C, Wiedorn KH, Dworakowski R, et al. Prognostic value of cyclin D1 overexpression in correlation with pRb and p53 status in non-small cell lung cancer (NSCLC). J Cancer Res Clin Oncol. 2005;131(7):479–85. doi:10.1007/s00432-004-0661-9.
Dworakowska D, Jassem E, Jassem J, Karmolinski A, Lapinski M, Tomaszewski D, et al. Prognostic value of the apoptotic index analysed jointly with selected cell cycle regulators and proliferation markers in non-small cell lung cancer. Lung Cancer. 2009;66(1):127–33. doi:10.1016/j.lungcan.2009.01.008.
Wu Y, Huang B, Liu Q, Liu Y. Heat shock protein 90-beta over-expression is associated with poor survival in stage I lung adenocarcinoma patients. Int J Clin Exp Pathol. 2015;8(7):8252–9.
Zheng CL, Qiu C, Shen MX, Qu X, Zhang TH, Zhang JH, et al. Prognostic impact of elevation of vascular endothelial growth factor family expression in patients with non-small cell lung cancer: an updated meta-analysis. Asian Pac J Cancer Prev. 2015;16(5):1881–95.
Passlick B, Sienel W, Seen-Hibler R, Wockel W, Thetter O, Mutschler W, et al. Overexpression of matrix metalloproteinase 2 predicts unfavorable outcome in early-stage non-small cell lung cancer. Clin Cancer Res. 2000;6(10):3944–8.
Qian Q, Wang Q, Zhan P, Peng L, Wei SZ, Shi Y, et al. The role of matrix metalloproteinase 2 on the survival of patients with non-small cell lung cancer: a systematic review with meta-analysis. Cancer Invest. 2010;28(6):661–9. doi:10.3109/07357901003735634.
Liang Y, Guo S, Zhou Q. Prognostic value of matrix metalloproteinase-7 expression in patients with non-small cell lung cancer. Tumour Biol. 2014;35(4):3717–24. doi:10.1007/s13277-013-1491-7.
Dragovic RA, Gardiner C, Brooks AS, Tannetta DS, Ferguson DJP, Hole P, et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomed-Nanotechnol. 2011;7(6):780–8. doi:10.1016/j.nano.2011.04.003.
Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current protocols in cell biology / editorial board, Juan S Bonifacino [et al.]. 2006;Chapter 3:Unit 3 22. doi:10.1002/0471143030.cb0322s30.
Munagala R, Aqil F, Vadhanam MV, Gupta RC. MicroRNA 'signature' during estrogen-mediated mammary carcinogenesis and its reversal by ellagic acid intervention. Cancer Lett. 2013;339(2):175–84. doi:10.1016/j.canlet.2013.06.012.
Acknowledgments
This work was supported from the Agnes Brown Duggan Endowment and Helmsley Trust Funds. Ramesh C. Gupta holds the Agnes Brown Duggan Chair in Oncological Research. The Microarray core is gratefully acknowledged for assisting with sample analysis and Xiaohong Li, Biostatistician, University of Louisville Bioinformatics Core, for her assistance in analysis of the data. We thank Manicka V. Vadhanam for useful discussions during the study.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
Table S1: Significantly modulated miRNAs in cancer (H1299) vs normal (Beas-2b) cell line. Table S2: Significantly modulated miRNAs in serum exosomes from tumor bearing vs normal (non-tumor) mice. Table S3: Significantly modulated miRNAs in recurrent vs primary tumors. (DOCX 32 kb)
Rights and permissions
About this article
Cite this article
Munagala, R., Aqil, F. & Gupta, R.C. Exosomal miRNAs as biomarkers of recurrent lung cancer. Tumor Biol. 37, 10703–10714 (2016). https://doi.org/10.1007/s13277-016-4939-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-016-4939-8