Abstract
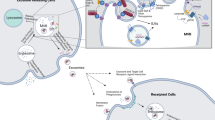
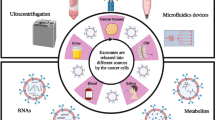
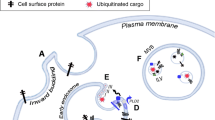
Cancer is a complex disease attributed to genetic distortions and cellular and non-cellular host responses. Tumors contain a variety of cell types that interact in a dynamic manner to maintain cancer-specific signaling networks. Extrinsic vesicles (EVs) are formed and retrieved as part of cell communication. Even tumor cells release exosomes, which are “30–100 nm” membrane vesicles that come from endosomes. Parental cells proteins and nucleic acids enrich their repertoire, and intercellular signals are thought to be transmitted by them. DNA and RNA are released into all body fluids as well as protein biomarkers that can be used to identify tumors and therapeutic targets. Patients with cancer may be screened for tumors based on the presence of exosomes secreted by tumor cells. Biomarkers for clinical diagnoses, such as exosomal proteins and microRNAs, are attracting considerable interest. The unique biogenesis of exosomes, their pervasive production by all cell types, and their biological features in liquid biopsy samples have all contributed to a growing interest in exosomes as cancer biomarkers. Cancer ‘prognosis’, ‘diagnosis’ and ‘progression’ may be more comprehensively assessed using these biomarkers, which reflect the heterogeneous biological changes associated with tumor growth. This chapter provides a brief overview of exosomal initiation, function, isolation, and the current roles of computation in oncology in the context of multi-omic technologies.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., & Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians, 68(6), 394–424.
Li, W., Li, C., Zhou, T., et al. (2017). Role of exosomal proteins in cancer diagnosis. Molecular Cancer, 16(1), 145.
Bu, H., He, D., He, X., & Wang, K. (2019). Exosomes: Isolation, analysis, and applications in cancer detection and therapy. ChemBioChem, 20(4), 451–461.
Thind, A., & Wilson, C. (2016). ExosomalmiRNAs as cancer biomarkers and therapeutic targets. Journal of Extracellular Vesicles, 5, 31292.
Mathieu, M., Martin-Jaular, L., Lavieu, G., & Thery, C. (2019). Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nature Cell Biology, 21(1), 9.
Dakubo, G. D. (2016). Advanced technologies for body fluid biomarker analyses. In G. D. Dakubo (Ed.), Cancer biomarkers in body fluids: Principles (pp. 55–74). Springer Nature.
Ruivo, C. F., Adem, B., Silva, M., & Melo, S. A. (2017). The biology of cancer exosomes: Insights and new perspectives. Cancer Research, 77, 6480–6488.
SkotlandT, S. K., & Llorente, A. (2017). Lipids in exosomes: Current knowledge and the way forward. Progress in Lipid Research, 66, 30–41.
Denzer, K., Kleijmeer, M. J., Heijnen, H. F. G., Stoorvogel, W., & Geuze, H. J. (2000). Exosome: From internal vesicle of the multivesicular body to intercellular signaling device. Journal of Cell Science, 113(19), 3365–3374.
Clayton, A., TurkesA, N. H., Mason, M. D., & Tabi, Z. (2005). Induction of heat shock proteins in B-cell exosomes. Journal of Cell Science, 118(16), 3631–3638.
LespagnolA, D. D., Beekman, C., et al. (2008). Exosome secretion, including the DNA damage-induced p53-dependent secretory pathway, is severely compromised in TSAP6/Steap3-null mice. Cell Death and Differentiation, 15(11), 1723–1733.
Sun, D., Zhuang, X., Zhang, S., et al. (2013). Exosomes are endogenous nanoparticles that can deliver biological information between cells. Advanced Drug Delivery Reviews, 65(3), 342–347.
Olver, C., & Vidal, M. (2007). Proteomic analysis of secreted exosomes. Sub Cell Biochemistry, 43, 99–131.
Huotari, J., & Helenius, A. (2011). Endosome maturation. EMBO Journal, 30(17), 3481–3500.
Del Conte-Zerial, P., Brusch, L., Rink, J. C., Collinet, C., Kalaidzidis, Y., Zerial, M., et al. (2008). Membrane identity and GTPase cascades regulated by toggle and cut-out switches. Molecular Systems Biology, 4, 206.
Mukherjee, S., & Maxfield, F. R. (2004). Membrane domains. Annual Review Cell Development Biology, 20, 839–866.
Trajkovic, K., Hsu, C., Chiantia, S., Rajendran, L., Wenzel, D., Wieland, F., et al. (2008). Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science, 319(5867), 1244–1247.
Savina, A., Fader, C. M., Damiani, M. T., & Colombo, M. I. (2005). Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic, 6, 131–143.
Ostrowski, M., Carmo, N. B., Krumeich, S., Fanget, I., Raposo, G., Savina, A., et al. (2010). Rab27a and Rab27b control different steps of the exosome secretion pathway. Nature Cell Biology, 12, 19–30.
Schorey, J. S., & Bhatnagar, S. (2008). Exosome function: From tumor immunology to pathogen biology. Traffic, 9(6), 871–881.
Kharaziha, P., Ceder, S., Li, Q., & Panaretakis, T. (2012). Tumor cell-derived exosomes: A message in a bottle. Biochimica et BiophysicaActa: Reviews on Cancer, 1826(1), 103–111.
Pegtel, D. M., van de Garde, M. D. B., & Middeldorp, J. M. (2011). Viral miRNAs exploiting the endosomal-exosomal pathway for intercellular cross-talk and immune evasion. Biochimica et Biophysica Acta, 1809(11–12), 715–721.
Markopoulos, G. S., Roupakia, E., Tokamani, M., et al. (2017). A step-by-step microRNA guide to cancer development and metastasis. Cellular Oncology, 40(4), 303–339.
van Niel, G., D’angelo, G., & Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nature Reviews Molecular Cell Biology, 19, 213.
Zhou, W., Fong, M. Y., Min, Y., et al. (2014). Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell, 25, 501–515.
Wang, N., & Xie, L. (2017). Diagnostic and therapeutic applications of tumor-associated exosomes. Precision Radiation Oncology, 1, 34–39.
RekkerK, S. M., Roost, A. M., et al. (2014). Comparison of serum exosome isolation methods for microRNA profiling. Clinical Biochemistry, 47(1–2), 135–138.
Chen, J. F., Mandel, E. M., Thomson, J. M., et al. (2006). The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nature Genetics, 38(2), 228–233.
Lee, K., Fraser, K., Ghaddar, B., et al. (2018). Multiplexed profiling of single extracellular vesicles. ACS Nano, 12(1), 494–503.
Momen-Heravi, F., Getting, S. J., & Moschos, S. A. (2018). Extracellular vesicles and their nucleic acids for biomarker discovery. Pharmacology and Therapeutics, 192, 170–187.
Martial, S. (2016). Involvement of ion channels and transporters in carcinoma angiogenesis and metastasis. American Journal of Physiology Cell Physiology, 310, C710–C727.
Rahbarghazi, R., Jabbari, N., Sani, N. A., et al. (2019). Tumor-derived extracellular vesicles: Reliable tools for cancer diagnosis and clinical applications. Cell Communication and Signaling: CCS, 17(1), 73.
Roma-Rodrigues, C., Mendes, R., Baptista, P. V., & Fernandes, A. R. (2019). Targeting tumor microenvironment for cancer therapy. International Journal of Molecular Science, 20, 840.
Zhao, H., et al. (2016). Tumor microenvironment derived exosomespleiotropically modulate cancer cell metabolism. eLife, 5, e10250.
Whiteside, T. L. (2016). Tumor-derived exosomes and their role in cancer progression. Advances in Clinical Chemistry, 74, 103–141.
HaoY, B. D., & Ten Dijke, P. (2019). TGF-beta-mediated epithelial-mesenchymal transition and cancer metastasis. International Journal of Molecular Science, 20, 27–67.
Wang, J., Zheng, Y., & Zhao, M. (2016). Exosome-based cancer therapy: Implication for cancer. Stem Cells Front Pharmacology, 7, 533.
Sharma, A. (2018). Role of stem cell derived exosomes in tumor biology. International Journal of Cancer, 142, 1086–1092.
Ti, D., HaoH, Fu. X., & Han, W. (2016). Mesenchymal stem cells-derived exosomal microRNAs contribute to wound inflammation. Science China Life Science, 59, 1305–1312.
Yong, S. B., et al. (2019). Non-viral nano-immunotherapeutics targeting tumor microenvironmental immune cells. Biomaterials, 219, 119401.
Ramos-Zayas, Y., et al. (2019). Immunotherapy for the treatment of canine transmissible venereal tumor based in dendritic cells pulsed with tumoralexosomes. Immunopharmacology and Immunotoxicology, 41, 48–54.
Muller, L., et al. (2017). Human tumor-derived exosomes (TEX) regulate Treg functions via cell surface signaling rather than uptake mechanisms. Oncoimmunology, 6, e1261243.
Sakai, C., & Nishikawa, H. (2018). Immunosuppressive environment in tumors. Gan Kagaku Ryoho, 45, 222–226.
Yoshioka, Y., Konishi, Y., Kosaka, N., Katsuda, T., Kato, T., & Ochiya, T. (2013). Comparative marker analysis of extracellular vesicles in different human cancer types. Journal of Extracellar Vesicles, 2, 14–23.
Welker, M. W., Reichert, D., Susser, S., et al. (2012). Soluble serum CD81 is elevated in patients with chronic hepatitis c and correlates with alanine aminotransferase serum activity. PLoS ONE, 7, e30796.
Peinado, H., Aleckovic, M., Lavotshkin, S., et al. (2012). Melanoma exosomes educate bone marrow progenitor cells toward a prometastatic phenotype through MET. Nature Medicine, 18, 883–891.
Khan, S., Jutzy, J. M. S., Valenzuela, M. M. A., et al. (2012). Plasma-derived exosomal survivin, a plausible biomarker for early detection of prostate cancer. PLoS ONE, 7, e46737.
Skog, J., Wurdinger, T., van Rijn, S., et al. (2008). Glioblastomamicrovesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature Cell Biology, 10, 1470–1476.
Li, J., Sherman-Baust, C. A., Tsai-Turton, M., Bristow, R. E., Roden, R. B., & Morin, P. J. (2009). Claudin-containing exosomes in the peripheral circulation of women with ovarian cancer. BMC Cancer, 9, 244.
Conde-Vancells, J., Rodriguez-Suarez, E., & Gonzalez, E., et al. (2010). Candidate biomarkers in exosome-like vesicles purified from rat and mouse urine samples. PROTEOMICS—ClinAppl, 4, 416–425.
Smalley, D. M., Sheman, N. E., Nelson, K., & Theodorescu, D. (2008). Isolation and identification of potential urinary microparticle biomarkers of bladder cancer. Journal of Proteome Research, 7, 2088–2096.
Nilsson, J., Skog, J., Nordstrand, A., et al. (2009). Prostate cancer-derived urine exosomes: A novel approach to biomarkers for prostate cancer. British Journal of Cancer, 100, 1603–1607.
Zhou, H., Cheruvanky, A., Hu, X., et al. (2008). Urinary exosomal transcription factors, a new class of biomarkers for renal disease. Kidney International, 74, 613–621.
ValadiH, E. K., Bossios, A., Sjostrand, M., Lee, J. J., & Lotvall, J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cell. Nature Cell Biology, 9(6), 654–659.
Hunter, M. P., Ismail, N., Zhang, X., et al. (2008). Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE, 3(11), e3694.
Mitchell, P. S., Parkin, R. K., Kroh, E. M., et al. (2008). Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences of the United States of America, 105, 10513–10518.
Tanaka, Y., Kamohara, H., Kinoshita, K., et al. (2013). Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer, 119, 1159–1167.
Taylor, D. D., & Gercel-Taylor, C. (2008). MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecologic Oncology, 110, 13–21.
Corcoran, C., Friel, A. M., Duffy, M. J., Crown, J., & O’Driscoll, L. (2011). Intracellular and extracellular microRNAs in breast cancer. Clinical Chemistry, 57, 18–32.
Silva, J., Garcıa, V., Zaballos, A., et al. (2011). Vesicle-related microRNAs in plasma of non-small cell lung cancer patients and correlation with survival. European Respiratory Journal, 37, 617–623.
Ohshima, K., Inoue, K., Fujiwara, A., et al. (2010). Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS ONE, 5, e13247.
Hong, B. S., Cho, J. H., Kim, H., Choi, E. J., Rho, S., Kim, J., Kim, J. H., Choi, D. S., Kim, Y. K., Hwang, D., & Gho, Y. S. (2009). Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genomics, 10, 556.
Lv, L. L., Cao, Y. H., Pan, M. M., et al. (2014). CD2AP mRNA in urinary exosome as biomarker of kidney disease. Clinica Chimica Acta, 428, 26–31.
Palanisamy, V., Sharma, S., Deshpande, A., Zhou, H., Gimzewski, J., & Wong, D. T. (2010). Nanostructural and transcriptomic analyses of human saliva derived exosomes. PLoS ONE, 5, e8577.
Lau, C., Kim, Y., Chia, D., et al. (2013). Role of pancreatic cancer-derived exosomes in salivary biomarker development. Journal of Biological Chemistry, 288, 26888–26897.
Davis-Turak, J., Courtney, S. M., Hazard, E. S., Glen, W. B., da Silveira, W. A., Wesselman, T., et al. (2017). Genomics pipelines and data integration: Challenges and opportunities in the research setting. Expert Review of Molecular Diagnostics, 17, 225–237.
Maintainer, B. P. (2019). Arrays: Using bioconductor for microarray analysis.
Roy, S., Coldren, C., Karunamurthy, A., Kip, N. S., Klee, E. W., Lincoln, S. E., et al. (2018). Standards and guidelines for validating next-generation sequencing bioinformatics pipelines. Journal of Molecular Diagnostics, 20, 4–27.
Bernstein, B. E., Meissner, A., & Lander, E. S. (2007). The mammalian epigenome. Cell, 128, 669–681.
Hansen, K. D., Langmead, B., & Irizarry, R. A. (2012). BSmooth: From whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biology, 13, R83.
Buenrostro, J. D., Wu, B., Chang, H. Y., & Greenleaf, W. J. (2015). ATAC-seq: A method for assaying chromatin accessibility genome-wide. Current Protocols in Molecular Biology, 109, 21–29.
Harmston, N., Ing-Simmons, E., Perry, M., Barešic, A., & Lenhard, B. (2015). Genomic interactions: An R/bioconductor package for manipulating and investigating chromatin interaction data. BMC Genomics, 16, 963.
Zhang, H., He, L., & Cai, L. (2018). Transcriptome sequencing: RNA-seq. In T. Huang (Ed.), Computational systems biology (pp. 15–27). Humana Press.
Jeong, E., Moon, S. U., Song, M., & Yoon, S. (2017). Transcriptome modeling and phenotypic assays for cancer precision medicine. Archives of Pharmacal Research, 40, 906–914.
Yang, X., Saito, Y., Rao, A., Kim, H. J., Singh, P., Scott, E., et al. (2019). Alignment free filtering for cfNA fusion fragments. Bioinformatics, 35, i225–i232.
Babarinde, I. A., Li, Y., & Hutchins, A. P. (2019). Computational methods for mapping, assembly and quantification for coding and non-coding transcripts. Computer Structure Biotechnology of Journal, 17, 628–637.
Vazquez, A., Kamphorst, J. J., Markert, E. K., Schug, Z. T., Tardito, S., & Gottlieb, E. (2016). Cancer metabolism at a glance. Journal of Cell Science, 129, 3367–3373.
Yang, K., & Han, X. (2016). Lipidomics: Techniques, applications, and outcomes related to biomedical sciences. Trends in Biochemical Sciences, 41, 954–969.
Mohamed, A., Molendijk, J. (2019). Lipidr: Data mining and analysis of lipidomics datasets. R package version 200.
Yakkioui, Y., Temel, Y., Chevet, E., & Negroni, L. (2017). Integrated and quantitative proteomics of human tumors. Methods in Enzymology, 586, 229–246.
Cho, W. C. (2017). Mass spectrometry-based proteomics in cancer research. Expert Review of Proteomics, 14, 725–727.
Cook-Deegan, R., & McGuire, A. L. (2017). Moving beyond Bermuda: Sharing data to build a medical information commons. Genome Research, 27, 897–901.
Jansen, P., van den Berg, L., van Overveld, P., & Boiten, J. W. (2018). Research data stewardship for healthcare professionals. In P. Kubben, M. Dumontier, & A. Dekker (Eds.), Fundamentals of clinical data science (pp. 37–53). Springer.
Grossman, R. L., Heath, A. P., Ferretti, V., Varmus, H. E., Lowy, D. R., Kibbe, W. A., et al. (2016). Toward a shared vision for cancer genomic data. New England Journal of Medicine, 375, 1109–1112.
Wani, N., & Raza, K. (2018). Multiple kernel learning approach for medical image analysis. In: Dey, N., Ashour, A., Shi, F., Balas, E. (Eds.), Soft computing based medical image analysis, (pp. 31–47). Elsevier. https://doi.org/10.1016/B978-0-12-813087-2.00002-6
Gore, J. C. (2020). Artificial intelligence in medical imaging. Magnetic Resonance Imaging, 68, A1-4.
Rodriguez-Ruiz, A., et al. (2019). Detection of breast cancer with mammography: Effect of an artificial intelligence support system. Radiology, 290, 305–314.
Newswire, P. (2020). QuantX artificial intelligence (AI) breast cancer diagnosis system receives 2020 gold edison award. Available: https://www.prnewswire.com/news-releases/quantx-artificial-intelligenceai-breast-cancer-diagnosis-system-receives-2020-gold-edison-award301027112.html
Bera, K., Schalper, K. A., Rimm, D. L., Velcheti, V., & Madabhushi, A. (2019). Artificial intelligence in digital pathology—New tools for diagnosis and precision oncology. Nature Reviews Clinical Oncology, 16, 703–715.
Beck, A. H., et al. (2011). Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Science Translational of Medicine, 108, ra113
Nagpal, K., et al. (2019). Development and validation of a deep learning algorithm for improving gleason scoring of prostate cancer. NPJ Digital Medicine, 2, 48.
Harbeck, N., et al. (2019). Breast cancer. Nature Reviews Disease Primers, 5, 66.
Pokhriyal, R., Hariprasad, R., Kumar, L., & Hariprasad, H. (2019). Chemotherapy resistance in advanced ovarian cancer patient. Biomark Cancer, 11, 1179299X19860815.
Eswaran, J., et al. (2013). RNA sequencing of cancer reveals novel splicing alterations. Science and Reports, 3, 1689.
Vellido, A., Biganzoli, E., Lisboa, P. J. (23–25 April 2008). Machine learning in cancer research: implications for personalised medicine. At: The 16th European symposium on artificial neural networks ESANN.
Sun, Y., Goodison, S., Li, J., Liu, L., & Farmerie, W. (2007). Improved breast cancer prognosis through the combination of clinical and genetic markers. Bioinformatics, 23, 30–37.
Zhang, X., Wang, B., Zhang, X. S., Li, Z. M., Guan, Z. Z., & Jiang, W. Q. (2007). Serum diagnosis of diffuse large B-cell lymphomas and further identification of response to therapy using SELDITOF-MS and tree analysis patterning. BMC Cancer, 7, 235.
Garcia-Bilbao, A., Armananzas, R., Ispizua, Z., et al. (2012). Identification of a biomarker panel for colorectal cancer diagnosis. BMC Cancer, 12, 43.
Bigbee, W. L., Gopalakrishnan, V., Weissfeld, J. L., et al. (2012). A multiplexed serum biomarker immunoassay panel discriminates clinical lung cancer patients from highrisk individuals found to be cancer-free by CT screening. Journal of Thoracic Oncology, 7, 698–708.
Lanara, Z., Giannopoulou, E., & Fullen, M. et al. (2013). Comparative study and meta-analysis of meta-analysis studies for the correlation of genomic markers with early cancer detection, (pp. 7–14). Hum. Genomics.
Zhao, D., & Weng, C. (201). Combining PubMed knowledge and EHR data to develop a weighted bayesian network for pancreatic cancer prediction. Journal Biomedicine Informatics 44, 859–868.
Thompson, C. A., Purushothaman, A., Ramani, V. C., Vlodavsky, I., & Sanderson, R. D. (2013). Heparanase regulates secretion, composition, and function of tumor cell-derived exosomes. Journal of Biological Chemistry, 288, 10093–10099.
Sento, S., Sasabe, E., & Yamamoto, T. (2016). Application of a persistent heparin treatment inhibits the malignant potential of oral squamous carcinoma cells induced by tumor cell-derived exosomes. PLoS ONE, 11, e0148454.
Nishida-Aoki, N., Tominaga, N., Takeshita, F., Sonoda, H., Yoshioka, Y., & Ochiya, T. (2017). Disruption of circulating extracellular vesicles as a novel therapeutic strategy against cancer metastasis. Molecular Theraphy, 25, 181–191.
de la Fuente, A., Alonso-Alconada, L., Costa, C., Cueva, J., Garcia-Caballero, T., LopezLopez, R., & Abal, M. (2015). M-trap: Exosome-based capture of tumor cells as a new technology in peritoneal metastasis. Journal of National Cancer Institute, 107, djv184.
Zhang, Y., Yang, P., & Wang, X. F. (2014). Microenvironmental regulation of cancer metastasis by miRNAs. Trends in Cell Biology, 24, 153–160.
Clancy, C., Khan, S., Glynn, C. L., Holian, E., Dockery, P., Lalor, P., Brown, J. A., Joyce, M. R., Kerin, M. J., & Dwyer, R. M. (2016). Screening of exosomal microRNAs from colorectal cancer cells. Cancer Biomarkers, 17, 427–435.
Zaharie, F., Muresan, M. S., Petrushev, B., Berce, C., Gafencu, G. A., Selicean, S., Jurj, A., Cojocneanu-Petric, R., Lisencu, C. I., Pop, L. A., et al. (2015). Exosome-carried microRNA375 inhibits cell progression and dissemination via Bcl-2 blocking in colon cancer. Journal of Gastrointestinal and Liver Diseases, 24, 435–443.
Dos Anjos, P. B., da Luz Andres Cordero, F., Socorro Faria, S., Peixoto Ferreira de Souza, L., Cristina Brigido Tavares, P., Alonso Goulart, V., Fontes, W., Ricardo Goulart, L., & Jose Barbosa Silva, M. (2017). The multifaceted role of extracellular vesicles in metastasis: Priming the soil for seeding. International Journal of Cancer, 140, 2397–407.
Tian, Y., Li, S., Song, J., Ji, T., Zhu, M., Anderson, G. J., Wei, J., & Nie, G. (2014). A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials, 35, 2383–2390.
Mizrak, A., Bolukbasi, M. F., Ozdener, G. B., Brenner, G. J., Madlener, S., Erkan, E. P., Strobel, T., Breakefield, X. O., & Saydam, O. (2013). Genetically engineered microvesicles carrying suicide mRNA/protein inhibit schwannoma tumor growth. Molecular Theraphy, 21, 101–108.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Kaur, M., Sodhi, H.S. (2022). Exosomes: Supramolecular Biomarker Conduit in Cancer. In: Raza, K. (eds) Computational Intelligence in Oncology. Studies in Computational Intelligence, vol 1016. Springer, Singapore. https://doi.org/10.1007/978-981-16-9221-5_18
Download citation
DOI: https://doi.org/10.1007/978-981-16-9221-5_18
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-9220-8
Online ISBN: 978-981-16-9221-5
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)