Abstract
CRISPR/Cas technology of genome editing is a powerful tool for making targeted changes in the DNA of various organisms, including plants. The choice of the precise nucleotide sequence (protospacer) in the gene to be edited is important in the design of guide RNA, which can be carried out by specialized software. We review and compare all the known on-line and off-line resources for guide RNA design, with special attention paid to tools capable of searching for off-target edits sites in plant genomes. The use of Cas12a may be preferable to Cas9. Techniques allowing C→T and G→A base editing without DNA cleavage are discussed along with the basic requirements for the design of effective and highly specific guide RNAs. Ways for improving guide RNA design software are presented. We also discuss the lesser risks of off-target editing in plant genomes as opposed to animal genomes. Examples of edited plant genomes including those that do not lead to the creation of transgenic plants are reviewed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
The CRISPR/Cas-genome editing technology is a powerful tool for the introduction of targeted modifications into the DNA of different organisms, including plants. This technology consists of several stages, each of which determines to a certain degree the success of the final result. An important part of plant genome editing is CRISPR/Cas-component delivery to the place of editing, which the genetic (gene modified) status of the plant obtained also depends on [1]. The selection of targets, i.e. specific nucleotide sequences (protospacers) within the genes aimed for editing, is also a key point of this approach. This stage is associated with the design of guide RNAs (gRNAs), using computer programs. At presently, a number of these programs are developed, which are considered in the present review.
Programs for gRNA design are discussed in several reviews [2–9]. We also published a review on this topic, where we considered not only the programs for gRNA design, but also databases for these CRISPR/Cas-editing components, as well as programs aimed at the analyses of the genomes edited [10]. However, because of the fast development of gene engineering and bioinformatics techniques, these reviews already do not fully describe modern approaches to the design of gRNA. Some of the earlier developed programs have been recently updated, and a lot of new resources exist. Therefore, it has become necessary to pay attention again to this drastically important stage of genome editing. In contrast with other reviews, we consider almost all the existing programs that are aimed specifically at editing plant genomes in the design of gRNA. As this article is first of all addressed to the final users of this software, the algorithms used by different designers for the development of such programs are discussed very briefly. Programs for genome editing data analysis will also not be discussed, as this is a topic for a separate article.
As for the programs for the design of gRNA aimed at the editing genomes of organisms other than plants, it should be noted that most of them are suitable for the design of gRNAs of any nucleotide sequences regardless of the forms of life they belong to. However, these programs do not allow assessment of the presence of off-target sites in the genomes of individual plants. For example, the CRISPR-Cas9 guide RNA design checker (https://eu.idtdna.com/site/order/ designtool/index/crispr_sequence), from the well known company “Integrated DNA Technologies”, allows gRNAs to be designed and checked for their efficacy with respect to the absence of off-target sites in the human genome and in the genomes of mice, rats, zebrafish and nematodes. For other species, the “Other species” option may be used by indicating in brackets “no off-target analysis”. In this case, the researcher may either check for both full and non-full matching of the spacer sequences of gRNAs which were chosen with this, or with an alternative program from the GenBank database, using the Nucleotide BLAST service (https://blast.ncbi.nlm.nih.gov/Blast.cgi). It is also noteworthy that to date not many plant genomes have been analyzed, and most of them refer to either model species or industrial agricultural species. When editing a genome, which has not yet been sequenced, any programs may be used to design gRNAs (the list and URLs are given below), including those, which are not aimed at identifying off-target sites in the known genomes of plants.
Before we start discussing the main topic of the article, attention must be paid to designations of the Cas-nucleases used in genome editing, as well as their mutant forms, which were obtained by gene engineering approaches and sometimes designated by 2-3-letter abbreviations that originate from the Latin names of bacterial strains (Table 1). In this article, we designate them in the same manner as the developers of the programs discussed. It should be noted that the designation Cas9 implies a specific enzyme (by default it is the wild type nuclease wtSpCas9 obtained from Streptococcus pyogenes) even if the origination is not specified.
THE CAS9/12A NUCLEASES USED IN GENOME EDITING
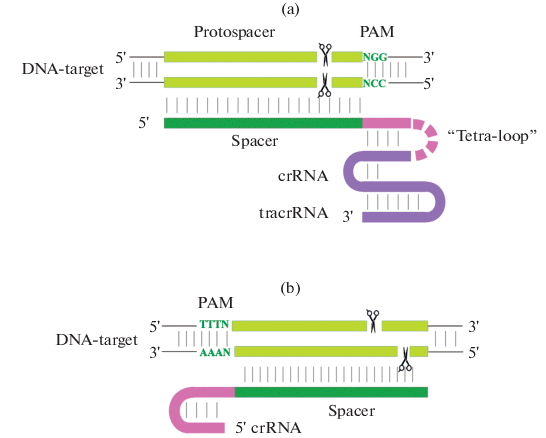
Detailed discussion of the CRISPR/Cas-technology of genome editing, including the detailed mechanisms of the activities of nucleases used, as well as a variety of achievements in the improvement of agriculturally valuable traits of plants, is far beyond the scope of this article, and a number of reviews on these points are available [11–22]. However, we should dwell briefly on the issue of the Cas9 and Cas12a (Cpf1) nucleases. As one may see from Fig. 1, the main differences between them are in the location of the first sequence recognized, which is adjacent to the protospacer (Protospacer Adjacent Motif, PAM) as well as in the sites of DNA cleavage and structural organization of the gRNA basis: CRISPR RNA (crRNA) and trans-activated CRISPR RNA (tracrRNA), which form the corresponding ribonucleoprotein (RNP) with a specific apoenzyme.
The classical variation of the Cas9 nuclease recognizes the NNG-3′ PAM-sequence, whereas the Cpf1 nuclease recognizes the AT-rich 5′‑TTTN PAM-sequence, which is located on the opposite end of the protospacer. Spacer sequences of gRNAs of these nucleases are 20 nt and 23 nt long respectively. The most important for the protospacer recognition part of the spacer sequence, which is adjacent to the PAM-region, is called the “seed sequence”. For nucleases of the Cas9 family, its length is 8–12 nt, whereas in Cas12a nucleases it is shorter—5–6 nt. The Cpf1 nuclease makes a double-strand break with 4–5 nt projected on the 5′‑end at some distance from the PAM-region, in contrast to Cas9, which forms breaks with blunt ends in the seed sequence near the PAM-sequence. The Cpf1 nuclease has a single catalytic domain, RuvC, which introduces breaks into both strands of DNA, whereas the Cas9 nuclease, which cleaves complementary DNA chains, has two catalytic domains, RuvC and HNH, single mutations in which (H840A in the RuvC or D10A in the HNH) transform the Cas9-nuclease into the corresponding nCas9 nickases, as the domain, which remains unchanged, can cleave only one of the DNA chains. If both catalytic domains become modified, the Cas9 nuclease, which, in this case, is called the dCas9 nuclease, loses its ability to cleave DNA. However, if this dCas9 nuclease is linked with the FokI restiction endonuclease monomer, the FokI-dCas9 nuclease is made, which may also be called fCas9. When these chimeric enzymes with different recognition sites are bound to the edited DNA, the FokI dimer is formed, which possesses catalytic activity. FokI-dCas9, alongside with the nCas9-nickases, is also used in genome editing and theoretically decreases the number of off-target editing sites, due to a decrease in the probability of recognition of two long sequences of different protospacers, which are located close to each other.
One more type of chimeric Cas nuclease, which lacks catalytic activity, is dCas or nCas-nickase, which carry linked cytidine- or adenine deaminases, as well as some other enzymes, which allow genome editing of individual nitrogenous bases by the C→T and A→G transitions without formation of double-strand breaks in DNA. It is noteworthy that editing individual nitrogenous bases allows knockout of genes via the transformation of glutamine (CAA and CAG), arginine (CGA) and tryptophan (TGG) codons into terminal codons (TTA, TAG, TGA). Conversely, the recovery of codons encoding these amino acids from termination codons is also possible and is important for genome editing via removal of mutations.
Several bacterial species have been shown to possess the Cas9/12a-nucleases, which recognize different PAM-sequences. A number of such genetically modified Cas-nucleases, which recognize different PAM-motives, have also been obtained. This, on one hand, increases the possibilities to screen genomes for appropriate protospacers, and, on the other hand, decreases the risk of the off-target site occurrence when nucleotides that recognize longer PAM-sequences, the level of which is lower in edited genomes.
A significant difference of the Cpf1 nuclease from the Cas9 is that its function does not require tracrRNA, as the length of crRNA that is sufficient for the formation of the active complex with the Cpf1-apoenzyme, is limited to 42–44 nt, in contrast to the full-size gRNA of about 100 nt, which includes crRNA, tracrRNA and artificial tetranucleotide loop GAAA, and is commonly used for the Cas9-nuclease. Because of the shortened size of the gRNA basis for Cpf1, this RNP component is easier to synthesize chemically [23]. This is however applicable to Cas9 as well [24]. This significantly simplifies the problem of genome editing, taking into account that ready-to-use enzymes are provided by several companies. It was found that chemically synthesized gRNA works better than enzymatically synthesized gRNA, as use of the latter for genome editing with Cpf1-containing RNPs led to unexplainable insertions of DNA fragments [25]. It was also shown that the Cas12a-nuclease demonstrated higher specificity in comparison to Cas9 [26]. Therefore, it is preferable to use Cas12a for genome editing, even though Cas9 has much more widespread use. One may suggest that the high specificity of Cas12a is due to the fact that it cleaves DNA far from the PAM-region. Thus, the process of DNA “melting”, and the progress of a so called R-loop, which are sensitive to mismatch and proceed steadily, may, at a certain point, stuck because of the presence of uncomplementary nucleotides. This results in a break of the RNP-complex with the DNA motive, because of the non-fitting off-target site. The Cas9-nucleases cleave both chains of DNA near the PAM-motive, and, theoretically, this cleavage may take place earlier than the formation of the full-mature R-loop in the off-target site.
THE MAIN REQUIREMENTS FOR THE DESIGN OF GUIDE RNA
For successful genome editing, in order to prevent off-target mutations and effectively produce target mutations, good gRNA must be designed. To achieve this, full-genome analysis of the organism is required (if its sequence is known) in order to reveal the presence of nucleotide sequences similar to the motive to be edited. As mismatch of individual nucleotides and even the presence of small insertions/deletions (indels, which may occur in both DNA and RNA chains) may theoretically not to be obstacles for off-target editing, one should take into account a not only 100% match of gRNA nucleotide sequences to the potential protospacers. Therefore, the gRNAs chosen should not contain motives fully matching the off-target sites in the genome edited, and the presence of partially homologous gRNAs should be minimized. Special attention should be paid to the seed region, in which nucleotide substitutions are most crucial. The location of potential off-target sites in coding regions, introns, promoters and intergenic spacers is also important, because it affects the risk of undesirable editing. To provide more effective knockout of specific genes, it is preferable to choose a region located in the first exon (if there is one) at a 150–500 bp distance from the starting codon. For small proteins this diapazone varies from 100 bp to 300 bp. When striving to exclude the possibility of functional but C-terminal truncated proteins, and to position the edited place closer to the NH2-terminal, one should not forget about possible leader peptides, distortions in which will not affect the protein itself, but will change its compartmentalization.
In 2013, the analysis of more that 700 gRNAs allowed researchers to choose several recommendations for gRNA selection so as to decrease the probability of the off-target editing and to develop the first programs for the design of gRNA [27]. The main requirement was to exclude the location of the PAM-sequence near off-target regions homologous to the targeted region, because the PAM-sequence is where place the nuclease is first guided to. Furthermore, for the PAM-sequence regions, exclusion of areas fully homologous to off-target sites and admission no more than three nucleotide substitutions of which two are located in the area adjacent to the PAM-sequence. It was hoped that such mismatches would be characterized by neighboring nucleotides or separated from each other by no more than 4 positions. Analysis of 1841 variations of gRNA by Doench et al. [28] revealed the best localization of nitrogenous bases in the spacer regions. This allowed the authors to compose a predicative model of their activity and develop rules, which were later called Rule Set 1. For example, it was found that the Cas9-nuclease prefers that the variable nucleotide in the PAM-sequence is cytosine. However, this was shown to be extremely undesirable in the neighboring region of DNA, which interacts with gRNA. It the latter region the preferable variable nucleotide was guanine. Further studies of target and off-target gRNA editing by Doench et al. [29] resulted in the development of the improved Rule Set 2, which became the basis for the development of several programs for the design of gRNA. Further improvements of the programs for the design of gRNA actively used algorithms that were based on deep machine learning, which allowed improved screening of the unique protospacers with a decreased probability of occurrence of off-target sites of editing [30–36].
It will be shown in the article that programs for the design of gRNAs, aimed at plant genome editing, are significantly different from each other, though they share several features. These include the length of gRNA spacer regions, which varies in the design in diapason from 15 nt to 25 nt, but is characterized by lengths of 20 nt and 23 nt pre-installed by default in many programs for the Cas9 and Cas12a (Cpf1) nucleases respectively. It is believed that the use of gRNAs with shortened spacers slightly decreases nonspecific editing, though excessive shortening may lead to total loss of catalytic activity of the Cas-nucleases. The nucleotide composition of protospacers is also important. Choosing protospacers with either high or low levels of GC-pairs is not recommended. It is preferable to choose regions that contain about 40–60% GC-pairs, though 20–80% is considered to be acceptable. Homopolymeric motives that consist of four or more similar nucleotides must be excluded. The presence of four (or even three) thymine residues one after the other should be avoided when obtaining gRNA by transcription, as this sequence terminates the process. One should also take into account expression systems that are used to obtain gRNAs and require the presence of two guanine residues at the 5′-end, when using RNA polymerase under the control of the T7-promoter, or a single guanine if using the U6-promoter, in order to increase the transcription efficiency. The nucleotide sequence of the region of choice should not induce the occurrence of the wrong spatial structure of the whole gRNA molecule. When choosing protospacers, prior to the sequencing of this part of genome, prediction of the presence of recognition sites of restriction endonucleases in the place of the double-stranded break and following repair, which after the cleavage of amplicons obtained by PCR with flanking primers serve as the control for genome editing, is recommended. In knock-in experiments with CRISPR/Cas-editing, the donor DNA, taking into account a small number of substitutions, should be absent from the protospacers chosen.
Recent analysis of a number of ineffective gRNAs showed that the majority of them contain the TT and GCC motives in the four nucleotides adjacent to the PAM-region in the proximal part of spacer sequences [37]. Earlier, the developers of the WU-CRISPR program for the design of gRNA reported that if the four nucleotides of the spacer sequence, adjacent to the PAM-region, are rich with pyrimidines, the efficiency of genome editing with such gRNAs decreases drastically, because these nucleotides may anneal with some nucleotides of the gRNA, increasing the length of one of its loop structures and excluding an important part of the seed sequence from interaction with the site of editing.
SPECIFICITIES OF PLANT GENOME EDITING
When editing plant genomes (in knockout variation), the occurrence of edited sites other than the targeted ones is not as drastic as it is for animals due to the large size of many plant genomes. They contain a considerable level of non-coding regions, and may often demonstrate polyploidy, including so called paleopolyploids (now considered as diploid forms). Many genes in these genomes are represented by many copies, and individual cells and tissues may be characterized by endopolyploidy due to endoreduplication. One more reason for the decreased risk of off-target editing in plant genomes is the possibility of relatively easy segregation of undesirable traits for many species over several generations, and the ability to carry out saturating crossings and backcrossings. Finally, the consequences of spontaneous mutagenesis, which occur under the influence of radiation and chemical agents and leads to a number of mutations in unpredictable places of the genome, are well known for plants. In these cases, some plants not only survive, but may also acquire advantageous traits.
In fact, off-target CRISPR/Cas genome editing, although not very random, may be considered to be similar to chemical or radiation mutagenesis. Theoretically, it may be considered as the introduction of multiple mutations into different positions of the plant genome, using, for example, some degenerated gRNAs (similarly to degenerated primers that anneal at different places [39]), which can edit the same multitude of protospacers in plant DNA. This would allow plants to be obtained with improved agriculturally important traits. “Mild” editing of individual nitrogenous bases by deaminases linked to inactive Cas-nucleases is probably more suitable for this purpose instead of common Cas-nucleases that lead to indel formation. Theoretically, these plants should not be referred to as genetically modified organisms, because of the off-target influence, similarly to mutagenesis induced by chemicals or radiation. However, the laboriousness, duration and cost of this approach for such multiple and almost random editing costs more than treatment of seeds with known mutagens. Therefore, this approach is considered to be uneconomic for obtaining mutant plants with improved characteristics.
The title of a published article “CRISPR/Cas precision: do we need to worry about off-targeting in plants?” is demonstrative. In this article [40] Hahn & Nekrasov asked a direct question: do we need to worry at all about the off-target CRISPR/Cas-editing in plant genomes? They provide references to large-scale studies from other authors, in which no mass mutations were found after genome editing [41]. For example, full-genome sequencing was performed for 69 rice plants, 34 of which were edited with the Cas9, 15 with Cpf1, and 20 were used as the control [41]. It was found out that almost all mutations (from 102 to 148 single nucleotide substitutions and from 32 to 83 indels per plant) occurred naturally, and only one mutation occurred as a result of off-target editing with Cas9, for which 12 gRNAs were used. Moreover, three gRNAs were aimed at Cpf1, which did not cause the off-target editing. This may be considered as evidence of good selection of gRNAs, as well as of the fact that off-target editing of plant genomes occurs less frequently than natural mutations. For example, in Arabidopsis, these are mostly transitions and small indels, which occur with an average frequency of 10–9 per site in one generation [42]. Wolt et al. [43] analyzed data obtained for several plant species and concluded that in most cases editing with the CRISPR/Cas-system, which worked either transiently or after being introduced into the plant genomes, was targeted. Wong et al. [38] also showed that off-target editing occurs more rarely than suggested previously for different organisms.
It is noteworthy that efficacy (completeness) and specificity (influence either preferably or solely on the targeted sites) of genome editing in fact conflict with each other. For example, editing targeted regions of the genome will be more effective the longer the RNP is active in the cell with a specific Cas-nuclease and gRNA, but the probability of off-target editing also increases. The duration of RNP activity depends on the delivery method chosen, which, in plants, may either provide introduction of all or only part of the CRISPR/Cas-components (constant exposure), lead to transient expression (prolonged exposure), or provide only a short-term influence, when a ready-to-use RNP-complex is directly delivered. In the last case, non-plasmid delivery takes place, an additional advantage of which, apart from the speed of the editing procedure, is the prevention of introduction of alien DNA (for knockout editing) into the genome of the plant, which, in this case, cannot become transgenic. These approaches have been developed quickly in recent years [44–47].
Taking into account the increase in the number of copies of individual genes, variations in the nucleotide sequences, and polyploidy in many plant species, it is important to choose several gRNAs for every gene for more effective knockout editing, so that if one gRNA is not involved in editing in different copies of the gene then the other will be.
COMPUTER PROGRAMS AND WEB-RESOURCES FOR THE DESIGN OF GUIDE RNA
For design of gRNA for the CRISPR/Cas genome editing it is important to predict the presence of off-target sites. To meet this requirement, programs aimed at designing gRNAs should have access to the corresponding genome sequences. Table 2 provides brief information on several programs for the design of gRNA, as well as data about the genomes of the plants used for analysis. It should be noted that the developers of individual programs may introduce additional genomes upon the request of users.
Table 3 summarizes the main features (more than 30) of the programs for gRNA design described below. The programs are grouped in blocks (shown in different colors), which refer to the Cas-nucleases (A—F), protospacers (G—K), input variations aimed at editing DNA/RNA sequences (L—N), features of the outcome data (O—W) and producing additional information (X—Z). Apart from this information, Table 3 represents data about the ability of some programs to work off-line and provide validation of previously designed gRNAs, including those designed with other programs. Some programs provide even more variations of analysis of the gRNAs designed than shown in Table 3, and we discuss them in more details.
The algorithm of gRNA design is, on the whole, common for the majority of programs. In some programs, protospacer search allows the choice of certain Cas-nucleases and indication of the features of the spacer region of gRNA (length, mismatch, indels, GC-content, etc.). The potential protospacers should be found in the genome sequence aimed for editing, which is then taken for analysis by different methods: input of the nucleotide sequence through the clipboard in either the FASTA- or text format; file download; indication of genes using an identifier (gene name, accession number); or using the genome coordinates. Several programs allow introduction of multiple sequences for gRNA design, which are suitable for editing similar conservative motives of genomes of different (evolutionarily related) species. When the sequence for editing is input, the Cas-nucleases and other parameters, including optional ones, are chosen, and the search should be run. When the search is finished, the program shows the potential protospacers, both ranked and non-ranked with respect to their suitability, as well as other information.
Some of the columns in the Table 3, which provide similar or interconnected information, repeat each other. For example, the GC-content of protospacers is shown in two columns (K and V), because in some programs it is settled at the stage of the search parameters, while in the others it just follows the selected sequences of protospacers. Therefore, these data refer to different blocks and it would be illogical to combine (even in double-columns) them in the same column. We decided not to overload the table with data on the length of nucleotide sequences that may be searched for protospacers using different programs. However, taking into account the importance of this parameter, we provided these data out of the table. For example, scatter of the lengths of these DNA regions varies widely for different programs and is and is listed here by increasing number of nucleotides: CCTop—500; E‑CRISP—500; CRISPR RGEN Tools—1000; CRISPOR—2000; CRISPR‑P 2.0—5000; CRISPR‑GE—10 000; CRISPRdirect—10 000; DESKGEN—10 000; CHOPCHOP—20 000; Breaking Cas—20 000; CRISPR‑PLANT—30 000; CRISPR MultiTargeter—50 000; PhytoCRISP‑Ex— 2 000 000. Unfortunately, this information is unavailable for some programs (Benchling, CGAT, GT‑Scan, CRISTA, CT‑Finder/CRISPR‑DT/CRISPR‑RT). It should be noted that analysis of 2000 and even 1000 nucleotides may be enough in most cases of targeted knockout editing of a gene in order to choose a specific place (protospacer), whereas analysis of 500 nt may not be enough, because of the possible absence of the most effective gRNA-binding sites in this fragment of the gene. In this case, a second analysis of one more region of the same length is required.
The upper lines of Table 3 are occupied by the most functional programs, which provide users with more information for improved gRNA design. They will be considered in more detail in this sequence.
The CRISPR-P Web-resource was the first specialized program for the design of gRNA for plant genome editing [60]. Later, this program was updated as CRISPR-P 2.0, which retained the former interface [61]. The assessment of targeted and off-target sites is performed on the basis of the latest ideas on their interactions with SpCas9 and other Cas-nucleases, the number of which has increased significantly. The CRISPR-P 2.0 program implies the selection of protospacers for cleavage with the following enzymes (see Table 1 for the full names), apart from SpCas9 (NGG and NRG): StCas9 (NNAGAAW), MmCas9 (NNNNGMTT), and SaCas9 (NNGRRT), as well as with the nucleases Cas12a: AsCpf1 (TTTV), LbCpf1 (TTTV), FnCpf1 (TTN) and others. This widens the possibilities for searching of optimal targets in the plant genome. This program gives the screening data as a linear plot for both strands of DNA, for the places of interaction of spacers with complementary chains of protospacers in three different colors in accordance with their relevance. Besides of the protospacer sequence, the data table includes information about the GC-content, and the position and characteristics of gRNAs selected for the target sites that may be ranked by any of these parameters. For example, it is possible to choose variations by indication of the first nucleotide on the 5′-end (G or A). Pointing the mouse cursor on a protospacer, the operator can obtain information about the number of off-target sites in the genome in the left part of the table. The off-target sites will be ranked in accordance with their specificities (the first 20 are shown), and the mismatched nucleotides will be shown in red. It is possible to get more information about protospacers in the form of data on the microhomology of the regions adjacent to the site of DNA cleavage with the Cas-nuclease. In an individual window, one can see an image of the supposed secondary structure of the gRNA. This allows considering the CRISPR-P 2.0 program, which provides wide possibilities for users, as one of the most promising. One of the improvements of the CRISPR-P 2.0 program is the development of the autonomous version CRISPR-Local [62], in which the assortment of nucleases has been decreased, and, simultaneously, the reference genome database has been increased to 71. All possible gRNAs for editing with Cas9 (NGG) and Cpf1 (TTTV and TTV) nucleases are already selected and available for download as an archive.
The CRISPR-GE (Genome Editing) [57] consists of the following instruments: targetDesign, offTarget, primerDesign-V, primerDesign-A, etc. Design of gRNA with the targetDesign program begins with the choice of nuclease, among which are the SpCas9 (NGG), FnCpf1 (TTN) and AsCpf1 (TTTN), and the corresponding PAM-sequence. Other types of PAM-region maybe chosen. Then, the length of the protospacer should be estimated and a genome chosen from the list. It is also possible not to indicate any genome, choosing the option “None”. The program returns the results of the screening in the form of a data table with the possibility to show restriction sites. However, this information may be hidden as it takes up a lot of space. Boxes in the first column of the table may be marked, and the set of primers for the construction of the required DNA sequence and its cloning in the corresponding vector will be selected for those gRNAs. In the other columns unsuccessful gRNAs are reported, which are indicated with different numbers of exclamation marks. The probability of the off-target editing may be assessed with the offTarget tool. To design primers the primerDesign program, and two of its subprograms, primerDesign-V and primerDesign-A, can be used. The first one responsible for the design of oligonucleotides for cloning, while the other is responsible for amplification of the target region in order to confirm editing. However, this is another step in genome editing, which is beyond the scope of this article. The offTarget program can be used independently from the targetDesign program and can analyze previously input gRNA sequences for suitability for off-target genome editing. To perform such an analysis, the type of nuclease and a genome must be chosen and the spacer sequence of gRNA input, using the “Insert” option for the gRNAs.
A specialized portal, CRISPR RGEN Tools, which contains a set of applied programs, has also been developed. This portal includes several programs, which are used for CRISPR/Cas-editing, including the Cas-OFFinder [65], Microhomology-Predictor [66], Cas-Designer [67] and BE-Designer [69] programs, as well as some other programs that can be used for the post-editing analysis, and, thus, are not considered here. Moreover, this web-portal contains two databases: Cas-Database [67] and Cpf1-Database [70], which contain information on several tens of thousands of gRNAs for five plant species (Arabidopsis thaliana, Musa acuminata, Vitis vinifera, Solanum lycopersicum, Glycine max).
The Cas-Designer program finds all possible protospacers in the sequence analyzed. It proposes a wide spectrum of nucleases, including mutant forms with different PAM-sequences: SpCas9 (NGG for the target sites and NRG for the off-target sites), StCas9 (NNAGAAW), NmCas9 (NNNNGMTT), SaCas9 (NNGRRT), CjCas9 (NNNVRYAC), CjCas9 (NNNNRYAC), wtSpCas9 (NNGTGA), VRER SpCas9 (NGCG), VQR SpCas9 (NGA), AsCpf1 (TTTN), AsCpf1 (TTTV), FnCpf1 (TTN), FnCpf1 (KYTV), BhCas12b (TTN), etc. In the search of protospacers one may indicate if it is necessary to allow the probability of single nucleotide indels. In this case there is a notification about the increase in the search time. However, the search results return quickly if this option is not used. Other parameters are selected by default. To run the search a genome must be chosen from the list, which is divided into groups and includes the group “Plant”. It is possible to change additional options in the Cas-Designer program after installing it on the computer.
The Microhomology-Predictor analyzes microhomology of nucleotide sequences in the sites of double-stand breaks, facilitating the prediction of the type of nonhomologous end-joining (NHEJ), and microhomology-mediated end-joining (MMEJ). The data are returned in the form of a data table, which contains different information, which can be narrowed by changing several parameters.
The Cas-OFFinder program is aimed at finding possible off-target sites in the genomes studied. This program may work independently to screen the off-target sites in the genomes of different organisms for previously selected gRNAs and provide the researcher with more possibilities for this analysis. To perform the analysis, it is necessary to insert the sequences of selected gRNAs, specify the acceptable number of mismatched nucleotides, the number of indels (both in DNA and RNA) and select the genome to be analyzed from the list, similarly Cas-Designer.
The BE-Designer program allows gRNAs for editing individual nitrogenous bases to be designed. This program works similarly to Cas-Designer, though there are some differences. For example, the selection of the PAM-regions and the corresponding nucleases is slightly narrowed, and the “window” size must be selected in order to search the sites of editing and the desired substitution of nucleotides (C→T or A→G). The results of the search are shown in a table, in which the nucleotide substituted is highlighted with a color.
The CRISPOR program [54, 55] is also a user friendly product, which contains a detailed user manual. The updated version CRISPOR V4.7 has been available since January 2019. This version allows the researcher to select optimal protospacers in three steps. In the first step, the sequence to be analyzed is inputted. In the second step, the researcher must choose a genome, which will be screened for off-target sites. Genomes may be omitted in this step by choosing the “No Genome” option. In the third step, the Cas9 nuclease is chosen out of 17 variants that include not only the standard SpCas9 type, but also its mutant forms with modified PAMs, as well as other orthologous nucleases of this type and their mutant forms. In addition, one may choose the Cas12a—AsCpf1 nuclease with two variations of PAM. The results of the search are represented in both graphic and table formats. The sequence analyzed is shown with the potentially edited sites indicated below and highlighted with different colors, corresponding for high, medium and low specificity. The resulting table contains information about the sequences of protospacers, their positions with DNA chains indicated, and reference to the “Cloning/PCR primers”, which allows the researcher to see the information about cloning and expression of a specific gRNA, and primers for detection. The proposed gRNAs are ranked by specificity, and their efficiency is assessed by two different algorithms. Special attention is payed to microhomology. The off-target sites are listed in accordance with the number of mismatches. It is noteworthy that the reference for the search results may be sent to colleagues, and the developers promise to keep the data of all the analyses performed for at least one year.
The Benchling cloud service is provided to academic organizations for free (https://benchling.com/academic), though registration is required. This resource contains a series of bioinformatics programs, one of which allows planning experiments for the CRISPR/Cas-editing of genomes, as well as designing gRNAs for a relatively wide spectrum of Cas9 and Cas12a nucleases, which recognize the following PAM-regions: NGG, NAG, NNNNGATT, NNAGAAW, NAAAAC, NNGRR, NNGRRT and TTTN. The researcher needs to choose the type of editing: simple single Cas-nuclease, nickases or editing individual nitrogenous bases without double-stranded breaks, or using the inactive Cas-nuclease linked to the corresponding deaminase. There are also several additional options, among which there are parameters for demonstration of the target and off-target sites and the nucleotide composition of gRNA. The search results are represented in graphic format, which show the whole nucleotide sequence analyzed with the restriction sites indicated for each endonuclease. Data on each protospacer is also provided in a table to show the localization, DNA chains and assessment of efficacy and specificity of the target and off-target editing. A special feature of the information provided for editing of individual nitrogenous bases is the assessment of efficacy of this process, and indication of nucleotide substitutions in protospacers by color.
The DESKGEN cloud service, from “Desktop Genetics”, is available for free for noncommercial organizations. However, registration is required, after which the use of programs such as Knockout, Knockin, Guide Picker [73] and Genome Editor becomes possible. At present, 26 genomes, including 6 plant genomes, are available for analysis. However, the Guide Picker program works with human and mouse genomes only. Operation is quite simple: first, the operator should choose the reference genome, then choose the gene, type of nuclease and the length of variable region of gRNA. It is not recommended to analyze DNA with a length of more than 10 000 bp, because it slows the program down. It is possible to design gRNAs to form ribonucleoprotein complexes with 10 different nucleases, including three from the Cpf1 group, as well as the catalytically inactive SpdCas9-FokI nuclease, though this program is designed so as to use the classical Cas9-nuclease, and the results provided for other enzymes may be not fully correct. In contrast with some other programs, the DESKGEN service indicates sequences of three (not four) thymines (TTT) as undesirable.
The CCTop (CRISPR/Cas9 Target online predictor) program [49] allows gRNAs to be designed and then screened for target and off-target sites. The program provides a wide spectrum of Cas-9 nucleases from different bacterial strains (Streptococcus pyogenes, Staphylococcus aureus, Neisseria meningitidis, Streptococcus thermophilus, Treponema denticola, Campylobacter jejuni), including mutant forms, as well as Cpf1 from Acadaminoccus/Lachnospiraceae and Francisella novicida. The special feature of this program is the possibility to add nucleotides to the spacer region in the form of one or two guanines at the 5′-end in order to improve in vitro transcription. It is also possible to indicate two desirable nucleotides adjacent to the PAM-region, to avoid them being with the gRNA. The data of gRNA design are represented in both graphic and table format ranked by the CRISPRater prognosis algorithm [32]. gRNAs in exons, introns and intergenic spacers are indicated in different colors.
The CHOPCHOP Web-resource [51, 52] is aimed at the screening of gRNAs, which work in complex with Cas9-nuclease, its nickase variations and with the Cpf1 nuclease. It is possible to run the search either in a specified coding region only or in all exons, splicing sites, 5'- or 3'-non-coding regions and in the promoter sequence, the length of which may be specified individually in a wide range of values. It is also possible to screen the restriction sites in the edited region. To do this, the corresponding enzymes may be found through indication of the manufacturer’s name (several companies are represented). When the search is finished, an adjustable color image and an interactive table with a ranked sequence of gRNA targets, including the GC-composition, DNA chains and some other information, is given.
The Breaking Cas Web-server [48], along with some other programs for gRNA design, allows screening of the off-target sites in all the to date known eukaryotic genomes available in the Ensembl database (http://ensemblgenomes.org), which contained 1335 genomes in January 2019. Design of gRNA with the Breaking Cas web-server begins with the choice of genome from a drop down alphabetic list. Otherwise, one may type the first letters of the organism’s common name or its Latin name in the input line, it is not important whether they are from the genus or species. It is necessary to indicate the type of nuclease of the four variations of the Cas9 and three variations of the Cpf1 with pre-set spacer lengths and the number of mismatches. The choice of a “Custom” nuclease, which is followed by input of the corresponding PAM-sequences, lengths of the spacer sequence of gRNA, and the choice of acceptable number of mismatches, is also possible. This provides wide possibilities for the targeted editing of the specified sites in the chosen genome. It allows the significance of each mismatch to be changed depending on the distance it is located from the PAM-region for the ranking of gRNAs proposed.
The E-CRISP program, which was developed in 2014, has been improved significantly, with an increase in the number of genomes from 12 to 55, more than ten of which belong to plants. In the Design module, the search level must be indicated. Three regimes are available: relaxed, medium and strict. In all three cases, introns, CpG-isles and 5′-non-translated sequences may either be taken into consideration or not. It is necessary to specify whether a single gRNA or its paired variations are required to use nickases or to run knockin-experiments. To make the search more specific, it is possible to choose from a large number of options. The MultiCRISP module suggests alignment of several related sequences. The Evaluation module allows revision of previously selected gRNAs for the presence of their off-target sites in the same genomes. It is also possible to set the acceptable number of mismatches and their localization. The E-CRISP program allows gRNAs for variations of the CRISPR/Cas-technology in the form of repression and activation of the genes—CRISPRi (CRISPR interference) and CRISPRa (CRISPR activation) respectively, to be designed. This possibility is also realized in the CLD [53] and CRISPR-ERA programs (http://crispr-era.stanford.edu) [77], though the latter does not work with plant genomes.
The CT-Finder (CRISPR Target Finder) Web-resource [71] allows screening of gRNAs for genome editing with three enzymes: Cas9-nuclease, nCas9-nickase and the RFN-system (RNA-guided FokI Nuclease) in the form of the fCas9-nuclease. For the Cas9-nuclease, the program finds single gRNAs. When using the nCas9 (D10A) nickase or the fCas9-nuclease, paired gRNAs will be proposed in the search results, and, in the latter case, the distances between them will be given. The number of DNA strains analyzed, one or two, is also shown, and several other options may be chosen.
Zhu and Liang [36] improved the CT-Finder program, which contains descriptions of the gRNA design using the CRISPR-DT (DNA Targeting) program, the first aimed at designing gRNA using the Cpf1-nuclease. For this, the authors first analyzed a large number of gRNAs, classifying the data by the reference vectors in order to provide maximal efficacy and specificity of the design of gRNAs using the CRISPR-DT program.
However, this is only for seven species, of which three are plants: Arabidopsis, rice and soybean. Researchers are provided with two possibilities: to design gRNA de novo or to verify the efficacy of editing of previously selected gRNAs. By default, the TTTV PAM-sequence in used to design gRNA, whereas the TTTN PAM region is used for screening off-target editing sites. The de novo design consists of several steps, the first of which is input of the nucleotide sequence for editing. Then, the organism from the same list as in the CT-Finder program, which shares all other procedures, should be specified. The search sensitivity must be chosen out of four options: low, medium, high and very high. The results of the search are represented in a table. The GC-composition for the protospacers selected is provided for both the whole sequence and individually for the six nucleotides that are adjacent to the PAM-sequence.
The CRISPR-RT (RNA Targeting) program, which was developed by the same group [72] is the first program for the design of gRNA, which uses the C2c2 (Cas13a) nuclease. After inputting the sequence to be analyzed, the operator should choose one out of ten pre-loaded transcriptomes, three of which belong to plants: Oryza sativa, Zea mays and Arabidopsis thaliana. For the last species, two transcriptomes from different databases are available. There are several options that can be used to narrow the dispersion of the data obtained.
The CRISPR MultiTargeter program [58, 59] is also interesting, because apart from gRNA design for single sequences it allows users to perform multiple alignments of homologous sequences for the selection of similar sites as protospacers. The PAM-sequence may be input either by the experimenter or may be NGG, and only in the latter case, will the associated programs perform a search of off-target sites in the selected genome.
Work with the CRISPR Genome Analysis Tool— CGAT program [50] consists of two stages. In the first stage, a gene or a nucleotide sequence should be selected for editing with Cas9. These should be input through the clipboard. Several parameters must be set: gRNA length, the required GC-composition (40–60%) and the acceptable number of repeated nucleotides in homopolymeric regions. Then the search starts and the results are returned in a table with gRNAs ranked by their applicability for effective genome editing. Optionally, one may estimate the potential sites for the off-target editing of genomes selected from the same plant species.
The CRISPRdirect program [56] is a simple and user-friendly web-resource for designing gRNAs for the Cas9 nuclease. To choose the protospacers in the sequence analyzed, it is necessary to indicate the PAM, though the choice is not very large: NGG (by default), NRG or NAG. It contains quite a wide choice of genomes, among which 98 plants, including agricultural species, model objects, and wild plant species. It is possible to input previously selected gRNAs for analysis instead of searching for new ones. The results are returned in both graphic and table formats, which provide the positions of protospacers with indication of DNA chains, the sequence itself with the PAM, the GC-composition and melting temperature (without the PAM), the presence of restriction sites and TTTT-sequences, and the number of off-target sites for the gRNAs selected for the 20-, 12- and 8-mer sequences of the spacers individually. By putting a tick into the corresponding window, one may limit the result to highly specific variations only.
The CRISPR-PLANT Web-resource is aimed at the designing gRNA and finding of the off-target sites in eight plant genomes [35, 63]. A new algorithm for screening of the off-target sites has been developed for the new v2 version of the program. This algorithm significantly increases the confidence of the analysis in order to increase the efficacy of genome editing with the Cas9 nuclease. Only NGG sequences are represented as PAM, because the NAG variations are more predisposed to off-target editing. The results of the protospacer search are returned in table format separately for 0.0 and 1.0 classes of gRNA. 0.0 denotes the absence of off-target sites in the genomes of plant species that undergo editing, and 1.0 denotes the presence of sites those which contain four or more mismatches in the whole target or three mismatches in the region adjacent to the PAM. The first group is represented by good gRNAs with minimal possibility of off-target pairing, whereas the second group is represented by worse gRNAs, which are better not to use. Information about the DNA such as the characteristics of the editing sites are also provided.
The GT-Scan program [75] allows researchers to search for gRNAs for the Cas9 nuclease with two variations of the PAM. The specificity level varies from 0 to 3. The results are returned in a table format, as well as in the form of a list of suitable protospacer sequences. The seed squence and the remaining region are highlighted with in colour. The program provides information about the localization of these sequences, which DNA strand they belong to and the number of potential off-target sites, depending on the specificity of the screening. More detailed information about the off-target site patterns in the genome can be obtained for each sequence. This website contains also other programs, among which the CUNE (Computational Universal Nucleotide Editor) program should be mentioned. Although the CUNE program is for working with the mouse genome only, it is one of the few programs, which allows the planning selective editing of nitrogenous bases [78]. On the whole, the CUNE program is similar to the BE-Designer program from the CRISPR RGEN package. The difference is that only one Cas9-nuclease with canonic the PAM-sequence NGG is used to design gRNA.
The CRISTA (CRISPR Target Assessment) Web-resource [30] provides analysis of previously selected gRNAs, as well as their ranking, in order to edit genomes and identify off-target sites. To design gRNA, the RANK TARGETS IN GENE program must be run, which is searching the protospacers recognizing by Cas9 (NGG) in the sequence inputted through the clipboard. The program does not have any additional parameters and options. When the search is finished, a table with gRNAs ranked by editing efficacy will be formed. For these gRNAs the off-target editing sites in one of the genomes available on the CRISTA Website may be identified using the connected FIND OFF-TARGETS program. It is noteworthy that the FIND OFF-TARGETS program may be used directly for the specificity analysis of previously selected gRNAs. The sgRNA:DNA SCORE program is also capable of analyzing the efficacy of previously selected gRNAs.
The PhytoCRISP-Ex program allows gRNAs for editing the genomes of 13 phytoplankton species using the Cas9 nuclease to be designed [76]. It uses two types of filters, which work in parallel and cut off different variations of gRNA, that carry the potentially off-target sites in the. When the search is finished, the program returns information about the presence of the restriction sites in the protospacers selected in for confirmation of editing by PCR-RFLP (PCR-Restriction Fragment Length Polymorphism) with flanking primers. The capacities of the PhytoCRISP-Ex program are increased by installing it on a personal computer.
The WheatCrispr program is aimed at designing gRNAs, for editing the genome of bread wheat (Triticum aestivum) only. To search the protospacers, a specific gene to edit must be chosen from the IWGSC RefSeq assembly v1.0 (https://wheat-urgi.versailles.inra.fr/ Seq-Repository/Assemblies) database, which was developed by the International Wheat Genome Sequencing Consortium or input the sequence through the clipboard. The length of the sequence is not limited, but we succeeded in designing gRNA of 1 815 nucleotides to find 350 potential protospacers. The results are ranked by efficacy of editing, taking into account possible off-target sites. Bearing in mind that wheat is a hexaploid, one may search for protospacers in the homologous sequences of all three subgenomes (B, A and D), choosing the corresponding option. One more option of this program allows selection of the editing sites, either coding regions or promoters.
Besides the web-resources that allow on-line gRNAs design, there are similar off-line programs, some of which may be used for genome editing in plants. One of them is the above mentioned the CLD (CRISPR Library Designer) program [53], which was developed by the same authors that developed the E-CRISPR program [74]. This explains why some of the options coincide in these programs. To design gRNA with the CLD program, one needs to choose a genome from the Ensembl database in order to screen it for off-target editing sites, a list of genes is formed the parameters are set for the protospacer search for the Cas9 nuclease (length, GC-composition, number of mismatches in the seed sequence and the remained parts). The acceptable gRNAs binding sites are annotated, which is followed by information about the DNA chain, restriction sites and oligonucleotides recommended for cloning. The special feature of the CLD program its ability to design gRNA for variations of the CRISPR/Cas-technology in the form of CRISPRi and CRISPRa.
Another off-line program aimed at editing in plants is CRISPR Primer Designer [64]. Search of the protospacers for the Cas9 is performed in a DNA sequence input via the clipboard (length may exceed 5000 nt) after setting several parameters: seed sequence length, GC-composition, indication of the 5′-nucleotide in the form of “G”, etc. The off-target sites may be traced with the CRISPR Primer Designer program for rice and Arabidopsis only. To do this, the program refers to the BLAST web-resource to generate ranked protospacers for the design of gRNA. The second working regime of this program is aimed at designing gRNAs for marking (visualization) of chromosomal regions in vivo. For this purpose, special search parameters are provided. No other program for the design of gRNA provides this possibility.
Recently, the GRIBCG (Guide RNA Identifier for Balancer Chromosome Generation) program has been developed [79]. This program is aimed at searching for gRNAs that flank a specific chromosomal region within one chromosome, in order to inverse it after the formation of double-strand breaks in DNA by the SpCas9-nuclease. This program works off-line and is available at https://sourceforge.net/projects/gribcg/. Searches, including screening of the off-target editing sites, can be performed on the chromosomes of six species, two of which are plants (Arabidopsis and rice).
In recent years, web-resources have been released, which allows experimenters to not only design gRNA, but also, for a specified price, order either chemical synthesis of the corresponding gRNAs or gene engineering constructions ready for CRISPR/Cas-editing of concrete genes. These programs are discussed below.
The Dharmacon CRISPR Design Tool web-resource, from “Dharmacon, Inc.” allows design of gRNAs, using 10 thousand nucleotide long DNA sequences, which may be input through the clipboard or via the ID of a specific gene. Then, the organism must be chosen from the small list provided, which contains five plants (maize, soybean, rice, apple tree and cotton). The results of the search are represented in both graphic and table formats, which provide different information about the protospacer sequences proposed, including their specificity, localization and the possibility of screening the surrounding regions of DNA. At the next stage, when a specific sequence is chosen, one may order chemical synthesis of the corresponding gRNA also indicating its scale. The CRISPR Specificity Analysis Tool allows identification of off-target sites in the same genomes. To do this, the protospacer sequence must be input and search parameters chosen. The search parameters are: one of the two variations of the PAM (NGG or NAG), seed sequence length and the number of mismatches and indels with indication of the strand type (DNA and/or RNA).
Another similar web-resource is the Synthego CRISPR Design Tool, which was developed by “Syntego” and allows design of gRNA for knockout editing with the SpCas9 nuclease, including those of plants, available in the Ensembl database, taking into account the presence of off-target sites. The name of the organism or the genome ID must be input, next, the name of the gene or its ID is input and the search is run. The result will return the recommended gRNA sequences, the chemical synthesis of which can be ordered for a specified price, for genome editing using ribonucleoprotein compexes.
The CRISPR gRNA Design tool developed by “Atum” provides the possibility of designing gRNA for genome editing using the wild-type Cas9 nuclease or nickases based on it. Choosing the option “I have my own gRNA” one can carry out the analysis of previously designed gRNAs for the presence of the off-target sites in the genomes of five species, one of which is a plant (Arabidopsis thaliana). To do this, the DNA fragment must be input. This may be done by indication of the name of the gene or its coordinates in the genome, or by inputting a nucleotide sequence of no more than 10 thousand nucleotides through the clipboard. The data is represented in both graphic and text format, and the sequences are ranked by suitability for genome editing. At the end of the search procedure a vector for the commercial cloning of gRNAs can be selected.
The wide spectrum of products for the CRISPR/ Cas9-technology can be seen on the “Sigma-Aldrich” Web-site on the MISSION CRISPR/Cas9 Products and Service page (https://www.sigmaaldrich.com/ catalog/product/sigma/crispr). On this site, researchers can fill in a form to order gRNAs to use them with SpCas9-nuclease or with nCas9 (D10A) nickases in order to perform editing in the knockout or other modes of almost any genome, of which the full nucleotide sequence is known.
PROGRAMS, WHICH ARE NOT AIMED AT THE EDITION OF PLANT GENOMES AND WEB-RESOURCES FOR THE DESIGN OF GUIDE RNA
Apart from the programs described above, which are fully or partially aimed at editing plant genomes, there are several programs for the design of gRNA, which can be used to edit plant genomes when there is no possibility to reveal off-target sites or if the presence of these sites is not crucial. Below we provide a list of these programs divided into on-line and off-line ones.
On-line programs for the design of gRNA (listed alphabetically): CasBLASTR (http://www.casblastr.org); CasOT (http://casot.cbi.pku.edu.cn); COSMID (https://crispr.bme.gatech.edu); CREATE Designer (http://www.thebioverse.org); CRISPcut (http://web.iitd.ac.in/crispcut/webserver/index.html); CRISPETa (http://crispeta.crg.eu); CRISPR4P (http://bahlerweb.cs.ucl.ac.uk/cgi-bin/crispr4p/ webapp.py); CRISPR-Cas9 guide RNA design checker (https://eu.idtdna.com/site/order/designtool/ index/crispr_sequence); CRISPR-DO (http://cistrome. org/crispr/); CRISPR Efficiency Predictor (http://www.flyrnai.org/evaluateCrispr/); CRISPR-ERA (http://crispr-era.stanford.edu); CRISPR-FOCUS (http://cistrome.org/crispr-focus/); CrispRGold (http://crisprgold.mdc-berlin.de); CRISPR Mapper (http://crdd.osdd.net/servers/crisprge/mapper.php); CRISPR.ML (https://crispr.ml); CRISPRoff (https://rth.dk/resources/crispr/crisproff/); CRISPR Optimal Target Finder (http://targetfinder.flycrispr. neuro.brown.edu); CRISPR-PN (http://www.crispr-pn.net); CRISPRScan (http://www.crisprscan.org); CRISPR sgRNA Design Tool (http://www.genscript.com/gRNA-design-tool.html); CRISPR-SKIP (http://song.igb.illinois.edu/crispr-skip); CRISPy CHO (http://staff.biosustain.dtu.dk/laeb/crispy); CRISPys (http://multicrispr.tau.ac.il); CRISPy-web (https://crispy.secondarymetabolites.org); CROP-IT (http://www.adlilab.org/CROP-IT/cas9tool.html); DeepCRISPR (http://www.deepcrispr.net); EuPaGDT (http://grna.ctegd.uga.edu); FORECasT (https://partslab. sanger.ac.uk/FORECasT); GB CRISPR Tools (https:// gbcloning.upv.es/tools/crisprs); ge-CRISPR (http:// bioinfo.imtech.res.in/manojk/gecrispr/index.php); GPP Web Portal (https://portals.broadinstitute.org/ gpp/public); Green Listed (http://greenlisted.cmm.ki.se); grID (http://crispr.technology); Guide RNA Generator (http://penchovsky.atwebpages.com/applications. php?page=48); GUIDES (http://guides.sanjanalab. org/#/); GuideScan (http://guidescan.com); inDelphi (https://www.crisprindelphi.design/about); Mojo Hand (http://www.talendesign.org); Off-Spotter (https://cm.jefferson.edu/Off-Spotter/); PAVOOC (https://pavooc.me); sgRNA Designer (CRISPRko) (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design); Stupar Lab’s CRISPR Design (http://stuparcrispr.cfans.umn.edu/CRISPR/); and ZiFiT (http://zifit.partners.org/ZiFiT).
Off-line programs for the design of gRNA (listed alphabetically): Azimuth (https://github.com/MicrosoftResearch/Azimuth); CasFinder (http://arep.med. harvard.edu/CasFinder); CASPER (https://github.com/ trinhlab/casper); Crisflash (https://github.com/crisflash/crisflash); CRISPR-Analyser (https://github.com/ htgt/CRISPR-Analyser); CRISPRer (http://jstacs.de/ index.php/CRISPRer); CRISPRO (https://gitlab.com/ bauerlab/crispro); CRISPR-offinder (https://sourceforge.net/projects/crispr-offinder-v1-2/); CRISSPRpred (https://github.com/khaled-rahman/CRISPRpred); CRISPRseek (http://bioconductor.org/packages/ release/bioc/html/CRISPRseek.html); DeepCas9 (https://github.com/lje00006/DeepCas9); FlashFry (https://github.com/aaronmck/FlashFry); GRIBCG (https://sourceforge.net/projects/gribcg/); MENTHU (http://genesculpt.org/menthu); pgRNAFinder (https://github.com/xiexiaowei/pgRNAFinder); predictSGRNA (http://www.ams.sunysb.edu/~pfkuan/ softwares.html#predictsgrn); sgRNAcas9 (https:// sourceforge.net/projects/sgrnacas9); and WU-CRISPR (http://crispr.wustl.edu).
These lists includes both simple tools with limited capacities and programs with a wide spectrum of capacities, using a number of different nucleases, different options, and providing improved design of gRNAs, predicting the off-target sites in the genomes in these programs that belong to different groups of organisms. Some of these programs were developed rather long ago and are already well acknowledged. Others however, have been recently developed, but still have good potential.
It is noteworthy that sometimes it is reasonable to design gRNA for editing plant genomes, using one of these “non-plant” programs, which allow them to be designed without the indication of a specific genome (options “None”, “No Genome”, etc.). The search of the off-target sites of editing should be performed with several programs, such as CRISPR RGEN Tools, CRISPR-GE, E-CRISP, CRISTA and CRISPR gRNA Design tool, which provide validation of the designed gRNAs via the analysis of specific plant genomes (if they are present in these programs).
Programs aimed at the design of gRNAs not only with the classical variation of the Cas9 nuclease, but also with its orthologues, as well as with the Cas12a (Cpf1) nucleases, which are characterized by AT-rich PAMs, may be of special interest for scientists working on genome editing. These programs also provide far more possibilities in choosing the sites of editing in plant genomes. Moreover, this enzyme is characterized by increased specificity. As an example, the CRISPRScan web-resource [80], apart from the classical variation of the Cas9, allows Cas12a nucleases to be used: LbCpf1 (TTTV), AsCpf1 (TTTV), LbCpf1 (TTTN) and AsCpf1 (TTTN). This resource is aimed at searching for off-target sites in 14 animal and yeast genomes. However, it is possible to choose the “No search” option in order to design gRNAs for other types of organisms, including plants, and obtain protospacer sequences.
CONCLUSION
In the present review, we have discussed about 100 programs, either in detail or briefly, which are aimed at designing gRNA for CRISPR/Cas genome editing. To date, this is the most complete collection of such tools. About a quarter of them provide screening of both target and off-target sites in plant genomes. Although one may not be too afraid of the occurrence of off-target mutations when editing plant genomes, it goes without saying that a researcher should do his best to avoid off-target editing. Therefore, if the full sequences of the genomes are known, increased attention should be paid to the design of gRNAs. Thus, it is not surprising that a number programs for the design of gRNAs for genome editing, including plants, have been developed. However, these programs do not fully follow the development of the genome editing technologies, which use the CRISPR/Cas-system. For example, only a few programs allow gRNAs for Cas12a-nucleases to be designed. The majority of do not have complete ranking and search of off-target sites for this nuclease. Taking into account the availability of plant genome editing with Cas12a (CpfI), it is desirable to develop special algorithms for it to screen editing sites and analyze gRNAs, including off-target sites. This process is already in progress [36]. Nowadays, RNP genome editing using chemically synthesized gRNAs is used more often. Therefore, it also seems important, at the very beginning, when screening potential protospacers, to propose the researcher an alternative choice of variations of genome editing, using either enzymatically or chemically synthesized RNA, and, respectively, to adjust the requirements of the protospacer screening.
It also seems promising to edit individual nitrogenous bases, because the chimeric nucleases used in this methodology, can perform knockout editing without introduction of double-strand breaks and indels. Unfortunately, to date, there are few programs for the design of gRNA that can provide such an opportunity. However, it is expected that the number will increase. This process is also in progress. In fact, a paper, which reports such a program called the beditor (https://github.com/rraadd88/beditor), which allows design of gRNA for editing individual nitrogenous bases and the off-target site screening in 125 genomes, including plants, has been published after almost complete ending of writing this review [81].
There is one more point, which special attention has recently been paid to. This is the localization of the edited sites in nucleosomes in the eu- and heterochromatin. It was shown to affect the efficacy of editing [82–85]. However, there is another information. For example, no differences in the efficacy of editing of genomic DNA located either in euchromatin or heterochromatin, have been observed in maize [86]. At present there is only one the CROP-IT (CRISPR/Cas9 Off-target Prediction and Identification Tool) web resource [87] (http://www.adlilab.org/CROP-IT/cas9tool.html), which is aimed at designing gRNA and identification of the potentially off-target editing sites, taking into account the chromatin state. However, the program operates with the human and mouse genome only. It is stated on the Web site of the program that the number of genomes available will increase, but only for the Cas9 nuclease with two variations of the PAM-sequence. It is expected that the development of new programs for the design of gRNA, as well as the improvements to the existing ones, including those aimed at editing plant genomes, will take into account the chromatin conditions as much as possible. This is expected to increase the efficacy of the CRISPR/Cas-technology. A recently published paper from Zhang S. et al. [88] discusses the synergetic effect of prediction of the editing sites when taking into account the chromatin state.
The thermodynamics of the interaction of the Cas-nucleases with DNA-targets must also be taken into account, in order to predict the efficacy of target site editing and activity with respect to off-target sites. This was shown by Zhang D. et al. [89] for the Cas9-nuclease, to which the “closest neighbors” model was applied, in order to calculate the free energy of intercrossed dinucleotides. The uCRISPR model (http://rna.physics. missouri.edu/uCRISPR/index.html), which was developed by these authors, improved the prediction of sites off-target editing .
We intentionally decided not to show which programs are more suited for beginners, and which for advanced users, because this point is more or less subjective and different goals or different users determine the choice of tools. Therefore, the main goal of this review was to compose a list of programs and briefly describe them. It should be noted that new features and nuances of the Cas-nucleases are continuously found, and the effects of specific nucleotide sequences in target and off-target sites on their interactions are revealed. Program algorithms for the design of gRNA are continuously being improved. Thus, for better results in genome editing, it is necessary to continuously follow their updates. Nevertheless, on the basis of our own experience, we recommend using several programs for the design of gRNA within the scope of one experiment, and to compare the results obtained.
REFERENCES
Kuluev B.R., Gumerova G.R., Mikhaylova E.V., Gerashchenkov G.A., Rozhnova N.A., Vershinina Z.R., Knyazev A.V., Matniyazov R.T., Baymiev An.Kh., Baymiev Al.Kh., Chemeris A.V. 2019. Delivery of CRISPR/Cas components into higher plant cells for genome editing. Russ. J. Plant Physiol.66 (5), 694–706. https://doi.org/10.1134/S0015330319050117
Graham D.B., Root D.E. 2015. Resources for the design of CRISPR gene editing experiments. Genome Biol.16, 260. https://doi.org/10.1186/s13059-015-0823-x
Kanchiswamy C.N., Maffei M., Malnoy M., Velasco R., Kim J.S. 2016. Fine-tuning next-generation genome editing tools. Trends Biotechnol.34, 562‒574. https://doi.org/10.1016/j.tibtech.2016.03.007
Chuai G.H., Wang Q.L., Liu Q. 2017. In silico meets in vivo: Towards computational CRISPR-based sgRNA design. Trends Biotechnol.35, 12‒21. https://doi.org/10.1016/j.tibtech.2016.06.008
Periwal V. 2017. A comprehensive overview of computational resources to aid in precision genome editing with engineered nucleases. Brief Bioinform.18, 698‒711. https://doi.org/10.1093/bib/bbw052
Yennmalli R.M., Kalra S., Srivastava P.A., Garlapati V.K. 2017. Computational tools and resources for CRISPR/Cas 9 genome editing method. MOJ Proteomics Bioinform.5, 00164.
Cui Y., Xu J., Cheng M., Liao X., Peng S. 2018. Review of CRISPR/Cas9 sgRNA design tools. Interdiscip. Sci.10, 455‒465. https://doi.org/10.1007/s12539-018-0298-z
Demirci Y., Zhang B., Unver T. 2018. CRISPR/Cas9: An RNA-guided highly precise synthetic tool for plant genome editing. J. Cell Physiol.233, 1844‒1859. https://doi.org/10.1002/jcp.25970
Yan J., Chuai G., Zhou C., Zhu C., Yang J., Zhang C., Gu F., Xu H., Wei J., Liu Q. 2018. Benchmarking CRISPR on-target sgRNA design. Brief Bioinform.19, 721‒724. https://doi.org/10.1093/bib/bbx001
Chemeris D.A., Kir’yanova O.Yu., Gerashchenkov G.A., Kuluev B.R., Rozhnova N.A., Matniyazov R.T., Baymiev An.Kh., Baymiev Al.Kh., Gubaidullin I.M., Chemeris A.V. 2017. Bioinformatic resources for CRISPR/Cas genome editing. Biomika.9, 203‒208.
Chugunova A.A., Dontsova O.A., Sergiev P.V. 2016. Methods of genome engineering: A new era of molecular biology. Biochemistry (Moscow). 81 (7), 662‒677.
Jiang F., Doudna J.A. 2017. CRISPR-Cas9 structures and mechanisms. Annu. Rev. Biophys.46, 505‒529. https://doi.org/10.1146/annurev-biophys-062215-010822
Bannikov A.V., Lavrov A.V. 2017. CRISPR/CAS9, the king of genome editing tools. Mol. Biol. (Moscow). 51 (4), 514–525. https://doi.org/10.7868/S0026898417040036
Kuluev B.R., Gerashchenkov G.A., Rozhnova N.A., Baymiev An.Kh., Vershinina Z.R., Knyazev A.V., Matniyazov R.T., Gumerova G.R., Mikhailova E.V., Nikonorov Yu.M., Chemeris D.A., Baymiev Al.Kh., Che-meris A.V. 2017. CRISPR/Cas editing of plant genomes. Biomika.9, 155‒182.
Karagyaur M.N., Rubtsov Yu.P., Vasiliev P.A., Tkachuk V.A. 2018. Practical recommendations for improving efficiency and accuracy of the CRISPR/Cas9 genome editing system. Biochemistry (Moscow). 83 (6), 629‒642.
Ahmad H.I., Ahmad M.J., Asif A.R., Adnan M., Iqbal M.K., Mehmood K., Muhammad S.A., Bhuiyan A.A., Elokil A., Du X., Zhao C., Liu X., Xie S. 2018. A review of CRISPR-based genome editing: Survival, evolution and challenges. Curr. Issues Mol. Biol.28, 47‒68. https://doi.org/10.21775/cimb.028.047
Makarova S.S., Khromov A.V., Spechenkova N.A., Taliansky M.E., Kalinina N.O. 2018. Application of the CRISPR/Cas system for generation of pathogen-resistant plants. Biochemistry (Moscow). 83, 1552‒1562. https://doi.org/10.1134/S0006297918120131
Redaktirovanie genov i genomov (Gene and Genome Editing), 2nd ed. Zakiyan S.M., Medvedev S.P., Dement’ev E.V., Pokushalov E.A., Vlasov V.V., Eds. Novosibirsk: Ross. Akad. Nauk, 2018.
Zlobin N.E., Lebedeva M.V., Taranov V.V., Kharchenko P.N., Babakov A.V. 2018. Plant genome editing by targeted nitrogenous base replacement. Biotekhnologiya.34, 59–68. https://doi.org/10.21519/0234-2758-2018-34-6-59-68
Korotkova A.M., Gerasimova S.V., Khlestkina E.K. 2019. Current achievements in modifying crop genes using CRISPR/Cas system. Vavilov. Zh. Genet. Selekts.23, 29‒37. https://doi.org/10.18699/VJ19.458
Molla K.A., Yang Y. 2019. CRISPR/Cas-mediated base editing: Technical considerations and practical applications. Trends Biotechnol. pii: S0167-7799(19)30053-8. https://doi.org/10.1016/j.tibtech.2019.03.008
Wolter F., Schindele P., Puchta H. 2019. Plant breeding at the speed of light: the power of CRISPR/Cas to generate directed genetic diversity at multiple sites. BMC Plant Biol.19, 176. https://doi.org/10.1186/s12870-019-1775-1
Swarts D.C., Jinek M. 2018. Cas9 versus Cas12a/Cpf1: Structure–function comparisons and implications for genome editing. Wiley Interdiscip. Rev. RNA. e1481. https://doi.org/10.1002/wrna.1481
Kelley M.L., Strezoska Z., He K., Vermeulen A., Smith A. 2016. Versatility of chemically synthesized guide RNAs for CRISPR-Cas9 genome editing. J. Biotechnol.233, 74–83. https://doi.org/10.1016/j.jbiotec.2016.06.011
Andersson M., Turesson H., Olsson N., Fält A.S., Ohlsson P., Gonzalez M.N., Samuelsson M., Hofvander P. 2018. Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol. Plant.164, 378‒384. https://doi.org/10.1111/ppl.12731
Strohkendl I., Saifuddin F.A., Rybarski J.R., Finkelstein I.J., Russell R. 2018. Kinetic basis for DNA target specificity of CRISPR-Cas12a. Mol. Cell.71, 816‒824.e3. https://doi.org/10.1016/j.molcel.2018.06.043
Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., Cradick T.J., Marraffini L.A., Bao G., Zhang F. 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol.31, 827‒832. https://doi.org/10.1038/nbt.2647
Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B.L., Xavier R.J., Root D.E. 2014. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014. 32, 1262‒1267. https://doi.org/10.1038/nbt.3026
Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R., Virgin H.W., Listgarten J., Root D.E. 2016. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol.34, 184‒191. https://doi.org/10.1038/nbt.3437
Abadi S., Yan W.X., Amar D., Mayrose I. 2017. A machine learning approach for predicting CRISPR-Cas9 cleavage efficiencies and patterns underlying its mechanism of action. PLoS Comput. Biol.13, e1005807. https://doi.org/10.1371/journal.pcbi.1005807
Kim H.K., Min S., Song M., Jung S., Choi J.W., Kim Y., Lee S., Yoon S., Kim H.H. 2018. Deep learning improves prediction of CRISPR-Cpf1 guide RNA activity. Nat. Biotechnol.36, 239‒241. https://doi.org/10.1038/nbt.4061
Labuhn M., Adams F.F., Ng M., Knoess S., Schambach A., Charpentier E.M., Schwarzer A., Mateo J.L., Klusmann J.H., Heckl D. 2018. Refined sgRNA efficacy prediction improves large- and small-scale CRISPR-Cas9 applications. Nucleic Acids Res.46, 1375‒1385. https://doi.org/10.1093/nar/gkx1268
Listgarten J., Weinstein M., Kleinstiver B.P., Sousa A.A., Joung J.K., Crawford J., Gao K., Hoang L., Elibol M., Doench J.G., Fusi N. 2018. Prediction of off-target activities for the end-to-end design of CRISPR guide RNAs. Nat. Biomed. Eng.2, 38‒47. https://doi.org/10.1038/s41551-017-0178-6
Xue L., Tang B., Chen W., Luo J. 2019. Prediction of CRISPR sgRNA activity using a deep convolutional neural network. J. Chem. Inf. Model.59, 615‒624. https://doi.org/10.1021/acs.jcim.8b00368
Minkenberg B., Zhang J., Xie K., Yang Y. 2019. CRISPR-PLANT v2: An online resource for highly specific guide RNA spacers based on improved off-target analysis. Plant Biotechnol. J.17, 5‒8. https://doi.org/10.1111/pbi.13025
Zhu H., Liang C. 2019. CRISPR-DT: designing gRNAs for the CRISPR-Cpf1 system with improved target efficiency and specificity. Bioinformatics.35, 2783–2789. https://doi.org/10.1093/bioinformatics/bty1061
Graf R., Li X., Chu V.T., Rajewsky K. 2019. sgRNA sequence motifs blocking efficient CRISPR/Cas9-mediated gene editing. Cell Rep.26, 1098‒1103.e3. https://doi.org/10.1016/j.celrep.2019.01.024
Wong N., Liu W., Wang X. 2015. WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/ Cas9 system. Genome Biol.16, 218. https://doi.org/10.1186/s13059-015-0784-0
Garafutdinov R.R., Baimiev An.Kh., Maleev G.V., Alekseev Ya.I., Zubov V.V., Chemeris D.A., Kir’yanova O.Yu., Gubaidullin I.M., Matniyazov R.T., Sakhabutdinova A.R., Nikonorov Yu.M., Kuluev B.R., Baymiev Al.Kh., Chemeris A.V. 2019. Diversity of primers for PCR and principles of their selection. Biomika. 11, 23–70. https://doi.org/10.31301/2221-6197.bmcs.2019-04
Hahn F., Nekrasov V. 2019. CRISPR/Cas precision: Do we need to worry about off-targeting in plants? Plant Cell Rep.38, 437‒441. https://doi.org/10.1007/s00299-018-2355-9
Tang X., Liu G., Zhou J., Ren Q., You Q., Tian L., Xin X., Zhong Z., Liu B., Zheng X., Zhang D., Malzahn A., Gong Z., Qi Y., Zhang T., Zhang Y. 2018. A large-scale whole-genome sequencing analysis reveals highly specific genome editing by both Cas9 and Cpf1 (Cas12a) nucleases in rice. Genome Biol.19, 84. https://doi.org/10.1186/s13059-018-1458-5
Ossowski S., Schneeberger K., Lucas-Lledó J.I., Warthmann N., Clark R.M., Shaw R.G., Weigel D., Lynch M. 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana.Science.327, 92‒94. https://doi.org/10.1126/science.1180677
Wolt J.D., Wang K., Sashital D., Lawrence-Dill C.J. 2016. Achieving plant CRISPR targeting that limits off-target effects. Plant Genome.9 (3). https://doi.org/10.3835/plantgenome2016.05.0047
Svitashev S., Schwartz C., Lenderts B., Young J.K., Mark Cigan A. 2016. Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat. Commun.7, 13274. https://doi.org/10.1038/ncomms13274
Liang Z., Chen K., Li T., Zhang Y., Wang Y., Zhao Q., Liu J., Zhang H., Liu C., Ran Y., Gao C. 2017. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun.8, 14261. https://doi.org/10.1038/ncomms14261
Khromov A.V., Makhotenko A.V., Snigir’ E.V., Makarova S.S., Makarov V.V., Suprunova T.P., Miroshnichenko D.N., Kalinina N.O., Dolgov S.V., Tal’yanskii M.E. 2018. Delivery of CRISPR/Cas9 ribonucleoprotein complex to apical meristem cells for DNA-free editing of potato Solanum tuberosum genome. Biotekhmologiya.34, 51‒58.
Miroshnichenko D.N., Shul’ga O.A., Timerbaev V.R., Dolgov S.V. 2019. Production of nontransgenic plants with an edited genome: Achievements, problems, and prospects. Biotekhnologiya.35, 3‒26. https://doi.org/10.21519/0234-2758-2019-35-1-3-26
Oliveros J.C., Franch M., Tabas-Madrid D., San-León D., Montoliu L., Cubas P., Pazos F. 2016. Breaking-Cas-interactive design of guide RNAs for CRISPR-Cas experiments for ENSEMBL genomes. Nucleic Acids Res.44, 267‒271. https://doi.org/10.1093/nar/gkw407
Stemmer M., Thumberger T., Del Sol Keyer M., Wittbrodt J., Mateo J.L. 2015. CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One.24, e0124633.
Brazelton V.A. Jr., Zarecor S., Wright D.A., Wang Y., Liu J., Chen K., Yang B., Lawrence-Dill C.J. 2015. A quick guide to CRISPR sgRNA design tools. GM Crops Food.6, 266–276. https://doi.org/10.1080/21645698.2015.1137690
Montague T.G., Cruz J.M., Gagnon J.A., Church G.M., Valen E. 2014. CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res.42, 401–407. https://doi.org/10.1093/nar/gku410
Labun K., Montague T.G., Gagnon J.A., Thyme S.B., Valen E. 2016. CHOPCHOP v2: A web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res.44, 272‒276. https://doi.org/10.1093/nar/gkw398
Heigwer F., Zhan T., Breinig M., Winter J., Brugemann D., Leible S., Boutros M. 2016. CRISPR library designer (CLD): Software for multispecies design of single guide RNA libraries. Genome Biol.17, 55. https://doi.org/10.1186/s13059-016-0915-2
Haeussler M., Schönig K., Eckert H., Eschstruth A., Mianné J., Renaud J.B., Schneider-Maunoury S., Shkumatava A., Teboul L., Kent J., Joly J.S.,Concordet J.P. 2016. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol.17, 148. https://doi.org/10.1186/s13059-016-1012-2
Concordet J.P., Haeussler M. 2018. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res.46, 242‒245. https://doi.org/10.1093/nar/gky354
Naito Y., Hino K., Bono H., Ui-Tei K. 2015. CRISPRdirect: Software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics.31, 1120–1123. https://doi.org/10.1093/bioinformatics/btu743
Xie X., Ma X., Zhu Q., Zeng D., Li G., Liu Y.-G. 2017. CRISPR-GE: A convenient software toolkit for CRISPR-based genome editing. Mol. Plant.10, 1246‒1249. https://doi.org/10.1016/j.molp.2017.06.004
Prykhozhij S.V., Rajan V., Gaston D., Berman J.N. 2015. CRISPR Multitargeter: A web tool to find common and unique CRISPR single guide RNA targets in a set of similar sequences. PLoS One.10, e0119372. https://doi.org/10.1371/journal.pone.0119372
Prykhozhij S.V., Rajan V., Gaston D., Berman J.N. 2015. Correction: CRISPR MultiTargeter: A Web Tool to find common and unique CRISPR single guide RNA targets in a set of similar sequences. PLoS One.10, e0138634. https://doi.org/10.1371/journal.pone.0138634
Lei Y., Lu L., Liu H.Y., Li S., Xing F., Chen L.L. 2014. CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant.7, 1494–1496. https://doi.org/10.1093/mp/ssu044
Liu H., Ding Y., Zhou Y., Jin W., Xie K., Chen L.L. 2017. CRISPR-P 2.0: An improved CRISPR-Cas9 tool for genome editing in plants. Mol. Plant.10, 530–532. https://doi.org/10.1016/j.molp.2017.01.003
Sun J., Liu H., Liu J., Cheng S., Peng Y., Zhang Q., Yan J., Liu H.J., Chen L.L. 2019. CRISPR-Local: A local single-guide RNA (sgRNA) design tool for non-reference plant genomes. Bioinformatics.35, 2501–2503. https://doi.org/10.1093/bioinformatics/bty970
Xie K., Zhang J., Yang Y. 2014. Genome-wide prediction of highly specific guide RNA spacers for the CRISPR-Cas9 mediated genome editing in model plants and major crops. Mol. Plant.7, 923‒926. https://doi.org/10.1093/mp/ssu009
Yan M., Zhou S.R., Xue H.W. 2015. CRISPR Primer Designer: Design primers for knockout and chromosome imaging CRISPR-Cas system. J. Integr. Plant Biol.57, 613‒617. https://doi.org/10.1111/jipb.12295
Bae S., Park J., Kim J.S. 2014. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics.30, 1473–1475. https://doi.org/10.1093/bioinformatics/btu048
Bae S., Kweon J., Kim H.S., Kim J.S. 2014. Microhomology-based choice of Cas9 nuclease target sites. Nat. Methods.11, 705–706. https://doi.org/10.1038/nmeth.3015
Park J., Bae S., Kim J.S. 2015. Cas-Designer: A web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics.31, 4014–4016. https://doi.org/10.1093/bioinformatics/btv537
Park J., Kim J.S., Bae S. 2016. Cas-Database: Web-based genome-wide guide RNA library design for gene knockout screens using CRISPR-Cas9. Bioinformatics.32, 2017–2023. https://doi.org/10.1093/bioinformatics/btw103
Hwang G.H., Park J., Lim K., Kim S., Yu J., Yu E., Kim S.T., Eils R., Kim J.S., Bae S. 2018. Web-based design and analysis tools for CRISPR base editing. BMC Bioinformatics.19, 542. https://doi.org/10.1186/s12859-018-2585-4
Park J., Bae S. 2018. Cpf1-Database: Web-based genome-wide guide RNA library design for gene knockout screens using CRISPR-Cpf1. Bioinformatics.34, 1077‒1079. https://doi.org/10.1093/bioinformatics/btx695
Zhu H., Misel L., Graham M., Robinson M.L., Liang C. 2016. CT-Finder: A web service for CRISPR optimal target prediction and visualization. Sci. Rep.23, 25516. https://doi.org/10.1038/srep25516
Zhu H., Richmond E., Liang C. 2018. CRISPR-RT: A web service for designing CRISPR-C2c2 crRNA with improved target specificity. Bioinformatics.34, 117‒119. https://doi.org/10.1093/bioinformatics/btx580
Hough S.H., Kancleris K., Brody L., Humphryes-Kirilov N., Wolanski J., Dunaway K., Ajetunmobi A., Dillard V. 2017. Guide Picker is a comprehensive design tool for visualizing and selecting guides for CRISPR experiments. BMC Bioinformatics.18, 167. https://doi.org/10.1186/s12859-017-1581-4
Heigwer F., Kerr G., Boutros M. 2014. E-CRISP: Fast CRISPR target site identification. Nat. Methods.11, 122–123. https://doi.org/10.1038/nmeth.2812
O’Brien A., Bailey T.L. 2014. GT-Scan: Identifying unique genomic targets. Bioinformatics.30, 2673–2675. https://doi.org/10.1093/bioinformatics/btu354
Rastogi A., Murik O., Bowler C., Tirichine L. 2016. PhytoCRISP-Ex: A web-based and stand-alone application to find specific target sequences for CRISPR/CAS editing. BMC Bioinformatics.17, 261. https://doi.org/10.1186/s12859-016-1143-1
Liu H., Wei Z., Dominguez A., Li Y., Wang X., Qi L.S. 2015. CRISPR-ERA: A comprehensive design tool for CRISPR-mediated gene editing, repression and activation. Bioinformatics.31, 3676–3678. https://doi.org/10.1093/bioinformatics/btv423
O’Brien A.R., Wilson L.O.W., Burgio G., Bauer D.C. 2019. Unlocking HDR-mediated nucleotide editing by identifying high-efficiency target sites using machine learning. Sci. Rep.9, 2788. https://doi.org/10.1038/s41598-019-39142-0
Merritt B.B., Cheung L.C. 2019. GRIBCG: A software for selection of sgRNAs in the design of balancer chromosomes. BMC Bioinformatics.20, 122. https://doi.org/10.1186/s12859-019-2712-x
Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.D., Fernandez J.P., Mis E.K., Khokha M.K., Giraldez A.J. 2015. CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods.12, 982‒988. https://doi.org/10.1038/nmeth.3543
Dandage R., Després P.C., Yachie N., Landry C.R. 2019. beditor: A computational workflow for designing libraries of guide RNAs for CRISPR-mediated base editing. Genetics.212, 377‒385, https://doi.org/10.1534/genetics.119.302089
Chari R., Mali P., Moosburner M., Church G.M. 2015. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat. Methods.12, 823‒826. https://doi.org/10.1038/nmeth.3473
Horlbeck M.A., Witkowsky L.B., Guglielmi B., Replogle J.M., Gilbert L.A., Villalta J.E., Torigoe S.E., Tjian R., Weissman J.S. 2016. Nucleosomes impede Cas9 access to DNA in vivo and in vitro.eLife.5, pii: e12677. https://doi.org/10.7554/eLife.12677
Jensen K.T., Fløe L., Petersen T.S., Huang J., Xu F., Bolund L., Luo Y., Lin L. 2017. Chromatin accessibility and guide sequence secondary structure affect CRISPR-Cas9 gene editing efficiency. FEBS Lett.591, 1892‒1901. https://doi.org/10.1002/1873-3468.12707
Yarrington R.M., Verma S., Schwartz S., Trautman J.K., Carroll D. 2018. Nucleosomes inhibit target cleavage by CRISPR-Cas9 in vivo.Proc. Natl. Acad. Sci. U. S. A.115, 9351‒9358. https://doi.org/10.1073/pnas.1810062115
Feng C., Yuan J., Wang R., Liu Y., Birchler J.A., Han F. 2016. Efficient targeted genome modification in maize using CRISPR/Cas9 system. J. Genet. Genomics.43, 37‒43. https://doi.org/10.1016/j.jgg.2015.10.002
Singh R., Kuscu C., Quinlan A., Qi Y., Adli M. 2015. Cas9-chromatin binding information enables more accurate CRISPR off-target prediction. Nucleic Acids Res.43, e118. https://doi.org/10.1093/nar/gkv575
Zhang S., Li X., Lin Q., Wong K.C. 2019. Synergizing CRISPR/Cas9 off-target predictions for ensemble insights and practical applications. Bioinformatics.35, 1108‒1115. https://doi.org/10.1093/bioinformatics/bty748
Zhang D., Hurst T., Duan D., Chen S.J. 2019. Unified energetics analysis unravels SpCas9 cleavage activity for optimal gRNA design. Proc. Natl. Acad. Sci. U. S. A.116, 8693‒8698. https://doi.org/10.1073/pnas.1820523116
Funding
This work was supported by the Russian Foundation for Basic Research, project nos. 18-04-00118 А, 19-016-00117 А, 19-016-00139 А, and by the governmental grants, nos. АААА-А16-116020350028-4, AAAA-A19-119021190011-0.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The present article does not contain any studies with human or animals as objects.
Conflict of interest. The authors declare no conflict of interests.
Additional information
Translated by M. Bibov
Abbreviations: gRNA, guide RNA; Cas9, CRISPR associated protein 9; Cpf1, CRISPR from Prevotella and Francisella 1; CRISPR, Clustered Regularly Interspaced Short Palindromic Repeats; PAM, Protospacer Adjacent Motif.
Rights and permissions
About this article
Cite this article
Gerashchenkov, G.A., Rozhnova, N.A., Kuluev, B.R. et al. Design of Guide RNA for CRISPR/Cas Plant Genome Editing. Mol Biol 54, 24–42 (2020). https://doi.org/10.1134/S0026893320010069
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893320010069