Abstract
The sample strategy employed in statistical parameter estimation issues has a major impact on the accuracy of the parameter estimates. Ranked set sampling (RSS) is a highly helpful technique for gathering data when it is difficult or impossible to quantify the units in a population. A bounded power logarithmic distribution (PLD) has been proposed recently, and it may be used to describe many real-world bounded data sets. In the current work, the three parameters of the PLD are estimated using the RSS technique. A number of conventional estimators using maximum likelihood, minimum spacing absolute log-distance, minimum spacing square distance, Anderson-Darling, minimum spacing absolute distance, maximum product of spacings, least squares, Cramer-von-Mises, minimum spacing square log distance, and minimum spacing Linex distance are investigated. The different estimates via RSS are compared with their simple random sampling (SRS) counterparts. We found that the maximum product spacing estimate appears to be the best option based on our simulation results for the SRS and RSS data sets. Estimates generated from SRS data sets are less efficient than those derived from RSS data sets. The usefulness of the RSS estimators is also investigated by means of a real data example.
Similar content being viewed by others
Introduction
In many domains, data analysis has been made simpler, and the margin of error has decreased with the discovery of new probability distributions. New unit distributions are typically created by converting certain well-known continuous distributions, which are more adaptable than the originals, without the need to include extra parameters. A number of distributions has been developed for the purpose of modeling data sets in numerous field, such as finance, risk management, engineering, actuarial sciences, biology, and economics. These distributions include the unit-Birnbaum-Saunders distribution1, the unit Weibull distribution2, the unit Gompertz distribution3, the unit-inverse Gaussian distribution4, the unit Burr-XII distribution5, the unit-Chen distribution6, the unit half-logistic geometric distribution7, the unit exponentiated Frechet distribution8, the unit exponentiated Lomax distribution9, the unit inverse exponentiated Weibull distribution10, the unit power Burr X distribution11, among others.
The power logarithmic distribution (PLD), which combines logarithmic and power function distributions, was introduced by Abd El-Bar et al.12. They mentioned that, compared to the power function distribution, the PLD is more flexible. The probability density function (PDF) of the PLD, with shape parameter \(a > 0\) and scale parameters \(b > 0\) and \(c > 0\), is given by:
where \(\omega \equiv (a,b,c)\) is the set of parameters. The cumulative distribution function (CDF) of the PLD is:
According to PDF (1), the distributions mentioned below are considered as submodels of the PLD:
-
For \(c = 0\), the PDF (1) provides the power function distribution with parameter a.
-
For \(a = 0\), the PDF (1) provides the logarithmic distribution with parameters b and c.
-
For \(a + 1 = \vartheta\) and \(c = 1\), the PDF (1) provides the Log-Lindley distribution with parameters \(\vartheta\) and b.
-
For \(a + 1 = \vartheta\), \(b = 0\), and \(c = 1\), the PDF (1) provides the transformed gamma distribution with parameter \(\vartheta\).
The hazard rate function (HF) of the PLD is:
The PDF and HF plots of the PLD are represented in Fig. 1. From Fig. 1, we can see that the PDF plot takes various shapes, such as growing, decreasing, constant, skewed to the right or left, and upside-down bathtub-shaped. The HF plots can be increasing, U-shaped, bathtub or j-shaped.
The study of economical sampling techniques is one of the major and fascinating areas of statistics. The field’s motivation stems from its exceptional ability to streamline the process of gathering data, particularly in situations when gathering relevant data is costly or time-consuming. In order to obtain accurate and cost-effective findings, researchers have developed a variety of sampling techniques over the past few decades. Ranked set sampling (RSS) is a useful technique for attaining observational economy in terms of the precision attained per sample unit. In the beginning, McIntyre13 presented the idea of RSS as a method for improving the sample mean’s accuracy as a population mean estimate. Ranking can be done without actually quantifying the observations by using expert opinion, visual examination, or any other method. Takahasi and Wakimoto14 provided the mathematical framework for RSS. Dell and Clutter15 demonstrated that, even in the presence of ranking errors, RSS outperforms simple random sampling (SRS). The RSS is extensively used in the fields of environmental monitoring16, entomology17, engineering applications18, forestry19, and information theory20.
The following is a description of the RSS design: Initially, \(s^2\) randomly selected units are taken from the population and divided into s groups of s units each. Without using any measures, the s units in each set are ranked. The unit that ranks lowest among the first s units is selected for actual quantification. The unit that ranks second lowest among the second set of s units is measured. The procedure is carried out again until the largest unit is determined from the sth group of s units. Hence, \(X_{\left( h \right) h} = {\text { }}\left( {{X_{\left( 1 \right) 1}}, {X_{\left( 2 \right) 2}},{\text { }}.{\text { }}.{\text { }}.{\text { }},{X_{\left( s \right) s}}} \right)\), \(h = 1, \ldots , s\), represents the one-cycle RSS. The process can be repeated l times to produce a sample of size \({s^ \cdot } = sl\) if a larger number of samples is needed. The l- cycle RSS is represented as \(X_{\left( h \right) hv} = {\text { }}\left( {{X_{\left( 1 \right) 11}}, {X_{\left( 2 \right) 22}},{\text { }}.{\text { }}.{\text { }}.{\text { }},{X_{\left( s \right) sl}}} \right)\), \(h = 1, \ldots , s\) and \(v = 1, \ldots , l\). In the present work, we write \({X_{hv}}\) instead of \({X_{(h)hv}}\). Wolfe21 mentioned that set sizes (s) larger than five would undoubtedly result in an excessive number of ranking errors and so could not likely considerably increase the efficacy of the RSS. Suppose that \({X_{hv}}\) represents the order statistics of the hth sample, with \(h = 1, \ldots , s\) in the vth cycle. Assuming perfect ranking, the PDF of \({X_{hv}}\), is given by
The issue of RSS-based estimation for a variety of parametric models has been the subject of several studies recently. The location-scale family distributions’ parameter estimator was examined by Stokes22. Bhoj23 investigated the scale and location parameter estimates for the extreme value distribution. Abu-Dayyeh et al.24 used SRS, RSS and a modification of RSS to investigate various estimators for the location and scale parameters of the logistic distribution. Under RSS, median RSS (MRSS), and multistage MRSS in case of imperfect ranking, Lesitha and Yageen25 investigated the scale parameter of a log-logistic distribution. Inference of the log-logistic distribution parameters, based on moving extremes RSS, was discussed by He et al.26. Using RSS and SRS, Yousef and Al-Subh27 obtained the maximum likelihood estimators (MLEs), moment estimators, and regression estimators of the Gumbel distribution parameters. Regarding SRS, RSS, MRSS, and extreme RSS (ERSS), Qian et al.28 derived a number of estimators for the Pareto distribution parameters in the case where one parameter is known and both are unknown. The MLEs for the generalized Rayleigh distribution parameters were derived by Esemen and Gurler29, using SRS, RSS, MRSS and ERSS. In the framework of SRS, RSS, MRSS, and ERSS, Samuh et al.30 presented the MLEs of the parameters pertaining to the new Weibull-Pareto distribution. Yang et al.31 explored the Fisher information matrix of the log-extended exponential-geometric distribution parameters based on SRS, RSS, MRSS, and ERSS. Al-Omari et al.32 investigated the generalized quasi-Lindley distribution parameters using the following estimators: MLEs, maximum product of spacings (MXPS) estimators, weighted least squares estimators, least squares estimators (LSEs), Cramer-von-Mises (CRM) estimators, and Anderson-Darling (AD) estimators based on RSS. Further, Al-Omari et al.33 considered similar procedures discussed as in Al-Omari et al.32 to examine estimators of the x-gamma distribution. Under stratified RSS, Bhushan and Kumar34 examined the effectiveness of combined and separate log type class population mean estimators. The suggested estimators’ mean square error and bias expressions were determined. The efficiency criteria were provided and a theoretical comparison between the proposed and current estimators was conducted. For more recent studies, see35,36,37,38,39,40,41,42.
The statistical literature proposes different estimation techniques since parameter estimation is important in real-world applications. Parameter estimation frequently involves the use of conventional estimation techniques like the LSE and MLE approaches. Both of them have advantages and disadvantages, but the most often used estimation technique is the ML method. The parameters of the PLD may be estimated using eight other methods of estimation in addition to the widely used MLE and LSE. These eight methods are AD, minimum spacing absolute distance (SPAD), MXPS, minimum spacing absolute log distance (SPALoD), minimum spacing square distance (MSSD), CRM, minimum spacing square log distance (MSSLD), and minimum spacing Linex distance (MSLND). It is difficult to compare the theoretical performance of different techniques, hence, extensive simulation studies are carried out under various sample sizes and parameter values to assess the performance of different estimators. Using a simulation scheme, the various PLD estimators based on the RSS design are then contrasted with those offered by the SRS approach. In this regard, six evaluation criteria are employed to assess the effectiveness of the estimating techniques. As far as the authors are aware, no attempt has been made to compare all of these estimators under RSS for the PLD. This fact served as the novelty and motivation for this study as we compare all of these estimators under RSS for the PLD.
The following sections provide a rough outline of the article. The various estimation methods for the PLD under RSS are provided in “Estimation methods based on RSS”. Several PLD estimators under SRS are given in “Estimation methods based on SRS”. The Monte Carlo simulation analysis that compares the effectiveness of the RSS-based estimators is examined in “Numerical simulation”. In “Real data analysis”, data analysis on milk production is conducted to demonstrate the practical applicability of the recommended estimate techniques. Some closing thoughts are included in “Concluding remarks”.
Estimation methods based on RSS
This section discusses ten different estimators for the PLD based on RSS. The suggested estimators are the MLE, AD estimator (ADE), CRM estimator (CRME), MXPS estimator (MXPSE), LSE, SPAD estimator (SPADE), SPALoD estimator (SPALoDE), MSSD estimator (MSSDE), MSSLD estimator (MSSLDE), and MSLND estimator (MSLNDE).
Maximum likelihood method
In the following, the MLEs \({{\hat{a}}^{{\text {ML}}}}\) of a, \({{\hat{b}}^{{\text {ML}}}}\) of b, and \({{\hat{c}}^{{\text {ML}}}}\) of c for the PLD are obtained based on RSS. To get these estimators let \({X_{hv}} = \left\{ {{X_{hv}},\,\,h = 1,\ldots ,s,v = 1,\ldots ,l} \right\}\) be an RSS of size \({s^ \cdot } = sl\) with PDF (1) and CDF (2), where l is cycles count and s is the set size. The likelihood function (LF) of the PLD is obtained by inserting Eqs. (1) and (2) into Eq. (3) as follows:
where \(\Lambda ({x_{vh}},\omega ) = (a + 1)(b - c\ln ({x_{vh}})).\) The log-LF of the PLD, denoted by \({L^{RSS}},\) is as follows:
The MLEs \({{\hat{a}}^{{\text {ML}}}},{{\hat{b}}^{{\text {ML}}}},{{\hat{c}}^{{\text {ML}}}}\) of a, b, c are obtained by maximizing (4) with respect to a, b, and c, as follows:
and
where \({\Lambda '_a}({x_{vh}},\omega ) = \frac{{\partial {\Lambda _a}({x_{vh}},\omega )}}{{\partial a}} = (b - c\ln ({x_{vh}})).\)
We can get the MLEs \({{\hat{a}}^{{\text {ML}}}},{{\hat{b}}^{{\text {ML}}}},{{\hat{c}}^{{\text {ML}}}}\) of a, b, c by setting Eqs. (5)–(7) equal to zero and solving them simultaneously. Note that nonlinear optimization techniques such as the quasi-Newton algorithm are often more effective for solving these equations.
Anderson–Darling method
The class of minimal distance approaches includes the AD method. In this subsection, the ADEs of a, b, c, say \({{\hat{a}}^{{\text {AD}}}}\), \({{\hat{b}}^{{\text {AD}}}}\), \({{\hat{c}}^{{\text {AD}}}}\) of the PLD are obtained using RSS.
Suppose that \({X_{(1:{s^ \cdot })}},{X_{(2:{s^ \cdot })}},\ldots ,{X_{({s^ \cdot }:{s^ \cdot })}}\) are ordered RSS items taken from the PLD with sample size \({s^ \cdot } = sl,\) where s is set size and l is the cycle number. The ADEs \({{\hat{a}}^{{\text {AD}}}},{{\hat{b}}^{{\text {AD}}}},{{\hat{c}}^{{\text {AD}}}}\) of a, b, and c are derived by minimizing the function
where \({\bar{F}}\left( {.\left| \omega \right. } \right)\) is the survival function. Alternatively, the ADEs \({{\hat{a}}^{{\text {AD}}}},{{\hat{b}}^{{\text {AD}}}}\), and \({{\hat{c}}^{{\text {AD}}}}\) of the PLD can be obtained by solving the subsequent non-linear equations in place of Eq. (8):
and
where
and
Also, \({\zeta _1}\left( {{x_{({s^ \cdot } - i + 1:{s^ \cdot })}}\left| \omega \right. } \right) ,\) \({\zeta _2}\left( {{x_{({s^ \cdot } - i + 1:{s^ \cdot })}}\left| \omega \right. } \right) ,\) and \({\zeta _3}\left( {{x_{({s^ \cdot } - i + 1:{s^ \cdot })}}\left| \omega \right. } \right)\) have the same expressions as (9), (10) and (11) by replacing the ordered sample \({x_{(i:{s^ \cdot })}}\) by the ordered sample \({x_{({s^ \cdot } - i + 1:{s^ \cdot })}}\).
Cramer–von-Mises method
The CRM method is a member of the minimal distance method class. This subsection provides the CRME of the parameter a, denoted by \({{\hat{a}}^{{\text {CR}}}}\), the CRME of parameter b, denoted by \({{\hat{b}}^{{\text {CR}}}}\), and the CRME of parameter c, denoted by \({{\hat{c}}^{{\text {CR}}}}\), of the PLD using RSS.
Let \({X_{(1:{s^ \cdot })}},{X_{(2:{s^ \cdot })}},\ldots ,{X_{({s^ \cdot }:{s^ \cdot })}}\) are ordered RSS items taken from the PLD with sample size \({s^ \cdot } = sl,\) where s is set size and l is the cycle number. The CRMEs \({{\hat{a}}^{{\text {CR}}}},{{\hat{b}}^{{\text {CR}}}},{{\hat{c}}^{{\text {CR}}}}\) of a, b, and c are derived by minimizing the function
Rather than using Eq. (12), these estimators can be obtained by solving the non-linear equations
and
where \({\zeta _1}\left( {{x_{(i:{s^ \cdot })}}\left| \omega \right. } \right) ,\) \({\zeta _2}\left( {{x_{(i:{s^ \cdot })}}\left| \omega \right. } \right) ,\) and \({\zeta _3}\left( {{x_{(i:{s^ \cdot })}}\left| \omega \right. } \right)\) are given in Eqs. (9), (10), and (11).
Maximum product of spacings method
This subsection provides the MXPSE of the parameter a, denoted by \({{\hat{a}}^{{\text {MP}}}}\), the MXPSE of parameter b, denoted by \({{\hat{b}}^{{\text {MP}}}}\), and the MXPSE of parameter c, denoted by \({{\hat{c}}^{{\text {MP}}}}\), of the PLD using RSS.
Let \({X_{(1:{s^ \cdot })}},{X_{(2:{s^ \cdot })}},\ldots ,{X_{({s^ \cdot }:{s^ \cdot })}}\) are ordered RSS items from the PLD with sample size \({s^ \cdot } = sl,\) where s is set size and l is the cycle number. The uniform spacings are defined as the differences
where \(F\left( {{x_{(0:{s^ \cdot })}}\left| \omega \right. } \right) = 0,\,\,\,\,F\left( {{x_{({s^ \cdot } + 1:{s^ \cdot })}}\left| \omega \right. } \right) = 1,\) such that \(\sum \limits _{i = 1}^{{s^ \cdot } + 1} {{D_i}(\omega )} = 1.\)
The MXPSEs \({{\hat{a}}^{{\text {MP}}}},{{\hat{b}}^{{\text {MP}}}},{{\hat{c}}^{{\text {MP}}}}\) of a, b, and c are found by maximizing the geometric mean of the spacing, which is obtained by maximizing the following function
The MXPSEs \({{\hat{a}}^{{\text {MP}}}},{{\hat{b}}^{{\text {MP}}}},{{\hat{c}}^{{\text {MP}}}}\) are provided by solving numerically the equations
and
where \({\zeta _1}\left( {.\left| \omega \right. } \right) ,\) \({\zeta _2}\left( {.\left| \omega \right. } \right) ,\) and \({\zeta _3}\left( {.\left| \omega \right. } \right)\) are given in Eqs. (9), (10), and (11).
Least squares method
Here, the LSE of the parameter a, denoted by \({{\hat{a}}^{{\text {LS}}}}\), the LSE of parameter b, denoted by \({{\hat{b}}^{{\text {LS}}}}\), and the LSE of parameter c, denoted by \({{\hat{c}}^{{\text {LS}}}}\), of the PLD are covered using RSS.
Suppose that \({X_{(1:{s^ \cdot })}},{X_{(2:{s^ \cdot })}},\ldots ,{X_{({s^ \cdot }:{s^ \cdot })}}\) are ordered RSS items from the PLD with sample size \({s^ \cdot } = sl,\) where s is set size and l is the cycle number. The LSEs \({{\hat{a}}^{{\text {LS}}}},{{\hat{b}}^{{\text {LS}}}}\) and \({{\hat{c}}^{{\text {LS}}}}\) , are obtained after minimizing the function
with respect to a, b, and c. The LSEs \({{\hat{a}}^{{\text {LS}}}},{{\hat{b}}^{{\text {LS}}}}\), and \({{\hat{c}}^{{\text {LS}}}}\) can be produced by solving the following non-linear equations
and
where \({\zeta _1}\left( {.\left| \omega \right. } \right) ,\) \({\zeta _2}\left( {.\left| \omega \right. } \right) ,\) and \({\zeta _3}\left( {.\left| \omega \right. } \right)\) are given in (9), (10), and (11).
Minimum spacing absolute distance
In the following, we obtain the SPADE of the parameter a, denoted by \({{\hat{a}}^{{\text {SPA}}}}\), the SPADE of parameter b, denoted by \({{\hat{b}}^{{\text {SPA}}}}\), and the SPADE of parameter c, denoted by \({{\hat{c}}^{{\text {SPA}}}}\), of the PLD using RSS.
Suppose that \({X_{(1:{s^ \cdot })}},{X_{(2:{s^ \cdot })}},\ldots ,{X_{({s^ \cdot }:{s^ \cdot })}}\) are ordered RSS items from the PLD with sample size \({s^ \cdot } = sl,\) where s is set size and l is the cycle number. The SPADEs \({{\hat{a}}^{{\text {SPA}}}},{{\hat{b}}^{{\text {SPA}}}}\), and \({{\hat{c}}^{{\text {SPA}}}}\) are obtained after minimizing the following function with respect to a, b, and c:
The SPADEs \({{\hat{a}}^{{\text {SPA}}}},{{\hat{b}}^{{\text {SPA}}}}\), and \({{\hat{c}}^{{\text {SPA}}}}\) are obtained by solving the nonlinear equations
and
numerically, where \({\zeta _1}\left( {.\left| \omega \right. } \right) ,\) \({\zeta _2}\left( {.\left| \omega \right. } \right) ,\) and \({\zeta _3}\left( {.\left| \omega \right. } \right)\) are given in Eqs. (9), (10), and (11).
Minimum spacing absolute-log distance
This subsection provides the SPALoDE of the unknown parameter a, represented by \({{\hat{a}}^{{\text {LD}}}}\), the SPALoDE of the unknown parameter b, represented by \({{\hat{b}}^{{\text {LD}}}}\), and the SPALoDE of the unknown parameter c, represented by \({{\hat{c}}^{{\text {LD}}}}\) of the PLD based on the RSS method.
Let \({X_{(1:{s^ \cdot })}},{X_{(2:{s^ \cdot })}},\ldots ,{X_{({s^ \cdot }:{s^ \cdot })}}\) are ordered RSS items from the PLD with sample size \({s^ \cdot } = sl,\) where s is set size and l is the cycle number. The SPALoDEs \({{\hat{a}}^{{\text {LD}}}},{{\hat{b}}^{{\text {LD}}}}\), and \({{\hat{c}}^{{\text {LD}}}}\) are obtained after minimizing the following function with respect to a, b, and c:
The following nonlinear equations can be numerically solved to obtain \({{\hat{a}}^{{\text {LD}}}},{{\hat{b}}^{{\text {LD}}}}\), and \({{\hat{c}}^{{\text {LD}}}}\) rather of using Eq. (15),
and
where \({\zeta _1}\left( {.\left| \omega \right. } \right) ,\) \({\zeta _2}\left( {.\left| \omega \right. } \right) ,\) and \({\zeta _3}\left( {.\left| \omega \right. } \right)\) are given in Eqs. (9), (10), and (11).
Minimum spacing square distance
In this subsection we are concerned with the MSSDE of parameter a, represented by \({{\hat{a}}^{{\text {SD}}}}\), the MSSDE of parameter b, represented by \({{\hat{b}}^{{\text {SD}}}}\), and the MSSDE of the parameter c, represented by \({{\hat{c}}^{{\text {SD}}}}\) of the PLD based on RSS.
Let \({X_{(1:{s^ \cdot })}},{X_{(2:{s^ \cdot })}},\ldots ,{X_{({s^ \cdot }:{s^ \cdot })}}\) are ordered RSS items from the PLD with sample size \({s^ \cdot } = sl,\) where s is set size and l is the cycle number. The MSSDEs \({{\hat{a}}^{{\text {SD}}}},{{\hat{b}}^{{\text {SD}}}}\), and \({{\hat{c}}^{{\text {SD}}}}\) are obtained after minimizing the following function with respect to a, b, and c:
The following nonlinear equations can be numerically solved to obtain \({{\hat{a}}^{{\text {SD}}}},{{\hat{b}}^{{\text {SD}}}}\), and \({{\hat{c}}^{{\text {SD}}}}\) rather of using Eq. (16),
and
where \({\zeta _1}\left( {.\left| \omega \right. } \right) ,\) \({\zeta _2}\left( {.\left| \omega \right. } \right) ,\) and \({\zeta _3}\left( {.\left| \omega \right. } \right)\) are given in Eqs. (9), (10), and (11).
Minimum spacing square-log distance
Here, the MSSLDE of parameter a, represented by \({{\hat{a}}^{{\text {SLo}}}}\), the MSSLDE of parameter b, represented by \({{\hat{b}}^{{\text {SLo}}}}\), and the MSSLDE of the parameter c, represented by \({{\hat{c}}^{{\text {SLo}}}}\) of the PLD are determined based on RSS.
Let \({X_{(1:{s^ \cdot })}},{X_{(2:{s^ \cdot })}},\ldots ,{X_{({s^ \cdot }:{s^ \cdot })}}\) are ordered RSS items from the PLD with sample size \({s^ \cdot } = sl,\) where s is set size and l is the cycle number. The MSSLDEs \({{\hat{a}}^{{\text {SLo}}}},{{\hat{b}}^{{\text {SLo}}}}\), and \({{\hat{c}}^{{\text {SLo}}}}\) are obtained after minimizing the following function with respect to a, b, and c:
The following nonlinear equations can be numerically solved to obtain \({{\hat{a}}^{{\text {SLo}}}}\), \({{\hat{b}}^{{\text {SLo}}}}\), and \({{\hat{c}}^{{\text {SLo}}}}\) rather of using Eq. (17),
and
where \({\zeta _1}\left( {.\left| \omega \right. } \right) ,\) \({\zeta _2}\left( {.\left| \omega \right. } \right) ,\) and \({\zeta _3}\left( {.\left| \omega \right. } \right)\) are given in Eqs. (9), (10), and (11).
Minimum spacing Linex distance
This subsection provies the MSLNDE of parameter a, say \({{\hat{a}}^{{\text {SLx}}}}\), the MSLNDE of parameter b, say \({{\hat{b}}^{{\text {SLx}}}}\), and the MSLNDE of the parameter c, say \({{\hat{c}}^{{\text {SLx}}}}\) of the PLD based on RSS.
Let \({X_{(1:{s^ \cdot })}},{X_{(2:{s^ \cdot })}},\ldots ,{X_{({s^ \cdot }:{s^ \cdot })}}\) are ordered RSS items from the PLD with sample size \({s^ \cdot } = sl,\) where s is set size and l is the cycle number. The MSLNDEs \({{\hat{a}}^{{\text {SLx}}}},{{\hat{b}}^{{\text {SLx}}}}\), and \({{\hat{c}}^{{\text {SLx}}}}\) are yielded after minimizing the following function with respect to a, b, and c:
The following nonlinear equations can be numerically solved to obtain \({{\hat{a}}^{{\text {SLx}}}}\), \({{\hat{b}}^{{\text {SLx}}}}\), and \({{\hat{c}}^{{\text {SLx}}}}\) rather of using Eq. (18),
and
where \({\zeta _1}\left( {.\left| \omega \right. } \right) ,\) \({\zeta _2}\left( {.\left| \omega \right. } \right) ,\) and \({\zeta _3}\left( {.\left| \omega \right. } \right)\) are given in (9), (10), and (11).
Estimation methods based on SRS
This section provides the MLE, ADE, CRME, MXPS, LSE, SPADE, SPALoDE, MSSDE, MSSLDE, and MSLNDE for the parameters a, b and c for the PLD based on SRS.
Maximum likelihood estimators
Here, the MLEs \({{{\tilde{a}}}^{{\text {ML}}}}\) of a, \({{{\tilde{b}}}^{{\text {ML}}}}\) of b, and \({{{\tilde{c}}}^{{\text {ML}}}}\) of c for the PLD are obtained based on SRS. To get these estimators, suppose that \({x_1},{x_2},\ldots ,{x_{{s^ \cdot }}}\) is an observed SRS of size \({s^ \cdot }\) from the PLD with PDF (1). The log-LF of a, b and c, is given by:
When differentiating \({\ell ^{SRS}}\) with respect to a, b and c, we obtain the following equations:
and
Using the statistical software Mathematica, the nonlinear equations (19)–(21) may be solved numerically after setting them equal to zero, to obtain the MLEs \({{{\tilde{a}}}^{{\text {ML}}}},{{{\tilde{b}}}^{{\text {ML}}}},\) and \({{{\tilde{c}}}^{{\text {ML}}}}\) of a, b, and c, respectively.
Anderson–Darling estimators
In this subsection, the ADE of parameter a, say \({{{\tilde{a}}}^{{\text {AD}}}}\), ADE of parameter b, say \({{{\tilde{b}}}^{{\text {AD}}}}\), and the ADE of parameter c, say \({{{\tilde{c}}}^{{\text {AD}}}}\) of the PLD are obtained using SRS.
Let \({X_{(1)}},{X_{(2)}},\ldots ,{X_{({s^ \cdot })}}\) be ordered items from SRS following PLD with sample size \({s^ \cdot }\). Thus, the ADEs \({{{\tilde{a}}}^{{\text {AD}}}},{{{\tilde{b}}}^{{\text {AD}}}},{{{\tilde{c}}}^{{\text {AD}}}}\) of a, b, and c are derived by minimizing the function
Alternatively, the ADEs of the PLD can be obtained by solving the subsequent non-linear equations in place of Eq. (22):
and
where \({\zeta _1}\left( {.\left| \omega \right. } \right) ,\) \({\zeta _2}\left( {.\left| \omega \right. } \right) ,\) and \({\zeta _3}\left( {.\left| \omega \right. } \right)\) are given in Eqs. (9), (10), and (11) with ordered sample \({x_{(j)}}\) and \({x_{({s^ \cdot } - j + 1)}}.\)
Cramer–von-Mises estimators
Here, we get the CRME of the parameter a, denoted by \({{{\tilde{a}}}^{{\text {CR}}}}\), the CRME of parameter b, denoted by \({{{\tilde{b}}}^{{\text {CR}}}}\), and the CRME of parameter c, denoted by \({{{\tilde{c}}}^{{\text {CR}}}}\), of the PLD are covered using SRS method.
Let \({X_{(1)}},{X_{(2)}},\ldots ,{X_{({s^ \cdot })}}\) are ordered SRS items taken from the PLD with sample size \({s^ \cdot }\). The CRMEs \({{{\tilde{a}}}^{{\text {CR}}}}\), \({{{\tilde{b}}}^{{\text {CR}}}}\), and \({{{\tilde{c}}}^{{\text {CR}}}}\) of a, b, and c are derived by minimizing the function
The following non-linear equations can be solved to get the CRMEs instead of using Eq. (23)
and
where \({\zeta _1}\left( {.\left| \omega \right. } \right) ,\) \({\zeta _2}\left( {.\left| \omega \right. } \right) ,\) and \({\zeta _3}\left( {.\left| \omega \right. } \right)\) are given in Eqs. (9), (10), and (11) with ordered sample \({x_{(j)}}\).
Maximum product of spacings estimators
This subsection presents the MXPSE of the PLD using SRS for parameter a, indicated by \({{{\tilde{a}}}^{{\text {MP}}}}\), the MXPSE of parameter b, indicated by \({{{\tilde{b}}}^{{\text {MP}}}}\), and the MXPSE of parameter c, indicated by \({{{\tilde{c}}}^{{\text {MP}}}}\).
Let \({X_{(1)}},{X_{(2)}},\ldots ,{X_{({s^ \cdot })}}\) are ordered SRS items taken from the PLD with sample size \({s^ \cdot }\). The MXPSEs \({{{\tilde{a}}}^{{\text {MP}}}},{{{\tilde{b}}}^{{\text {MP}}}}\), and \({{{\tilde{c}}}^{{\text {MP}}}}\) of a, b, and c are found by maximizing the geometric mean of the spacing, which is obtained by maximizing
where \(D_j^ * (\omega ) = F\left( {{x_{(j)}}\left| \omega \right. } \right) - F\left( {{x_{(j - 1)}}\left| \omega \right. } \right) ,\,\,\,\,\,j = 1,2,\ldots ,{s^ \cdot } + 1,\) \(F\left( {{x_{(0)}}\left| \omega \right. } \right) = 0,\,\,\,\,F\left( {{x_{({s^ \cdot } + 1)}}\left| \omega \right. } \right) = 1,\) such that \(\sum \limits _{j = 1}^{{s^ \cdot } + 1} {D_j^ * (\omega )} = 1.\) The MXPSEs \({{{\tilde{a}}}^{{\text {MP}}}},{{{\tilde{b}}}^{{\text {MP}}}},{{{\tilde{c}}}^{{\text {MP}}}}\) are provided by numerically solving the equations
and
where \({\zeta _1}\left( {.\left| \omega \right. } \right) ,\) \({\zeta _2}\left( {.\left| \omega \right. } \right) ,\) and \({\zeta _3}\left( {.\left| \omega \right. } \right)\) are given in Eqs. (9), (10), and (11).
Least squares estimators
Here, we use the SRS method to produce the LSE of the parameter a, denoted by \({{{\tilde{a}}}^{{\text {LS}}}}\), the LSE of parameter b, denoted by \({{{\tilde{b}}}^{{\text {LS}}}}\), and the LSE of parameter c, denoted by \({{{\tilde{c}}}^{{\text {LS}}}}\), of the PLD.
Let \({X_{(1)}},{X_{(2)}},\ldots ,{X_{({s^ \cdot })}}\) be ordered SRS items taken from the PLD with sample size \({s^ \cdot }\). The LSEs \({{{\tilde{a}}}^{{\text {LS}}}},{{{\tilde{b}}}^{{\text {LS}}}}\), and \({{{\tilde{c}}}^{{\text {LS}}}}\) are obtained after minimizing the following function with respect to the unknown parameters a, b, and c:
Alternately, the LSEs \({{{\tilde{a}}}^{{\text {LS}}}},{{{\tilde{b}}}^{{\text {LS}}}}\), and \({{{\tilde{c}}}^{{\text {LS}}}}\) are acquired by minimizing the following equations:
and
where \({\zeta _1}\left( {.\left| \omega \right. } \right) ,\) \({\zeta _2}\left( {.\left| \omega \right. } \right) ,\) and \({\zeta _3}\left( {.\left| \omega \right. } \right)\) are given in (9), (10), and (11).
Minimum spacing absolute distance estimators
This subsection provides the SPADE of the unknown parameter a, represented by \({{{\tilde{a}}}^{{\text {SPA}}}}\), the SPADE of the unknown parameter b, represented by \({{{\tilde{b}}}^{{\text {SPA}}}}\), and the SPADE of the unknown parameter c, represented by \({{{\tilde{c}}}^{{\text {SPA}}}}\) of the PLD based on the SRS method.
Let \({X_{(1)}},{X_{(2)}},\ldots ,{X_{({s^ \cdot })}}\) be ordered SRS items taken from the PLD with sample size \({s^ \cdot }\). The SPADEs \({{{\tilde{a}}}^{{\text {SPA}}}},{{{\tilde{b}}}^{{\text {SPA}}}}\), and \({{{\tilde{c}}}^{{\text {SPA}}}}\) are obtained after minimizing the following function with respect to a, b, and c:
Alternately, the following nonlinear equations can be solved numerically to yield the SPADEs \({{{\tilde{a}}}^{{\text {SPA}}}},{{{\tilde{b}}}^{{\text {SPA}}}}\), and \({{{\tilde{c}}}^{{\text {SPA}}}}\)
and
where \({\zeta _1}\left( {.\left| \omega \right. } \right) ,\) \({\zeta _2}\left( {.\left| \omega \right. } \right) ,\) and \({\zeta _3}\left( {.\left| \omega \right. } \right)\) are given in Eqs. (9), (10), and (11).
Minimum spacing absolute-log distance estimators
Here, we determine the SPALoDE of the unknown parameter a, represented by \({{{\tilde{a}}}^{{\text {LD}}}}\), the SPALoDE of the unknown parameter b, represented by \({{{\tilde{b}}}^{{\text {LD}}}}\), and the SPALoDE of the unknown parameter c, represented by \({{{\tilde{c}}}^{{\text {LD}}}}\) of the PLD based on the SRS technique.
Let \({X_{(1)}},{X_{(2)}},\ldots ,{X_{({s^ \cdot })}}\) are ordered SRS items taken from the PLD with sample size \({s^ \cdot }\). The SPALoDEs \({{{\tilde{a}}}^{{\text {LD}}}},{{{\tilde{b}}}^{{\text {LD}}}}\), and \({{{\tilde{c}}}^{{\text {LD}}}}\) are obtained after minimizing the following function with respect to a, b, and c:
The following nonlinear equations can be numerically solved to obtain \({{{\tilde{a}}}^{{\text {LD}}}},{{{\tilde{b}}}^{{\text {LD}}}}\), and \({{{\tilde{c}}}^{{\text {LD}}}}\) rather of using Eq. (24),
and
where \({\zeta _1}\left( {.\left| \omega \right. } \right) ,\) \({\zeta _2}\left( {.\left| \omega \right. } \right) ,\) and \({\zeta _3}\left( {.\left| \omega \right. } \right)\) are given in Eqs. (9), (10), and (11).
Minimum spacing square distance estimators
Here, we determine the MSSDE of the unknown parameter a, represented by \({{{\tilde{a}}}^{{\text {SD}}}}\), the MSSDE of the unknown parameter b, represented by \({{{\tilde{b}}}^{{\text {SD}}}}\), and the MSSDE of the unknown parameter c, represented by \({{{\tilde{c}}}^{{\text {SD}}}}\) of the PLD based on SRS.
Let \({X_{(1)}},{X_{(2)}},\ldots ,{X_{({s^ \cdot })}}\) are ordered SRS items taken from the PLD with sample size \({s^ \cdot }\). The MSSDEs \({{{\tilde{a}}}^{{\text {SD}}}},{{{\tilde{b}}}^{{\text {SD}}}}\), and \({{{\tilde{c}}}^{{\text {SD}}}}\) are obtained after minimizing the following function with respect to a, b, and c:
The following nonlinear equations can be numerically solved to obtain \({{{\tilde{a}}}^{{\text {SD}}}},{{{\tilde{b}}}^{{\text {SD}}}}\), and \({{{\tilde{c}}}^{{\text {SD}}}}\) rather of use Eq. (25),
and
where \({\zeta _1}\left( {.\left| \omega \right. } \right) ,\) \({\zeta _2}\left( {.\left| \omega \right. } \right) ,\) and \({\zeta _3}\left( {.\left| \omega \right. } \right)\) are given in Eqs. (9), (10), and (11).
Minimum spacing square-log distance estimators
Here, the MSSLDE of parameter a, represented by \({{{\tilde{a}}}^{{\text {SLo}}}}\), the MSSLDE of parameter b, represented by \({{{\tilde{b}}}^{{\text {SLo}}}}\), and the MSSLDE of parameter c, represented by \({{{\tilde{c}}}^{{\text {SLo}}}}\) of the PLD are determined based on SRS.
Let \({X_{(1)}},{X_{(2)}},\ldots ,{X_{({s^ \cdot })}}\) are ordered SRS items taken from the PLD with sample size \({s^ \cdot }\). The MSSLDEs \({{{\tilde{a}}}^{{\text {SLo}}}},{{{\tilde{b}}}^{{\text {SLo}}}}\), and \({{{\tilde{c}}}^{{\text {SLo}}}}\) are obtained after minimizing the following function with respect to a, b, and c:
The following nonlinear equations can be numerically solved to obtain \({{{\tilde{a}}}^{{\text {SLo}}}},{{{\tilde{b}}}^{{\text {SLo}}}}\), and \({{{\tilde{c}}}^{{\text {SLo}}}}\) rather of using Eq. (26),
and
where \({\zeta _1}\left( {.\left| \omega \right. } \right) ,\) \({\zeta _2}\left( {.\left| \omega \right. } \right) ,\) and \({\zeta _3}\left( {.\left| \omega \right. } \right)\) are given in Eqs. (9), (10), and (11).
Minimum spacing Linex distance estimators
This subsection provides the MSLNDE of parameter a, say \({{{\tilde{a}}}^{{\text {SLx}}}}\), the MSLNDE of parameter b, say \({{{\tilde{b}}}^{{\text {SLx}}}}\), and the MSLNDE of parameter c, say \({{{\tilde{c}}}^{{\text {SLx}}}}\) of the PLD based on SRS.
Let \({X_{(1)}},{X_{(2)}},\ldots ,{X_{({s^ \cdot })}}\) are ordered SRS items taken from the PLD with sample size \({s^ \cdot }\). The MSLNDEs \({{{\tilde{a}}}^{{\text {SLx}}}},{{{\tilde{b}}}^{{\text {SLx}}}}\), and \({{{\tilde{c}}}^{{\text {SLx}}}}\) are obtained after minimizing the following function with respect to a, b, and c:
The following nonlinear equations can be numerically solved to obtain \({{\hat{a}}^{{\text {SLx}}}},{{\hat{b}}^{{\text {SLx}}}}\), and \({{\hat{c}}^{{\text {SLx}}}}\) rather of using Eq. (27),
and
where \({\zeta _1}\left( {.\left| \omega \right. } \right) ,\) \({\zeta _2}\left( {.\left| \omega \right. } \right) ,\) and \({\zeta _3}\left( {.\left| \omega \right. } \right)\) are given in Eqs. (9), (10), and (11).
Numerical simulation
The variety of estimation techniques described in this study is examined in this section. By creating random data sets produced from the proposed model, the effectiveness of these methods in identifying model parameters is evaluated. After that, these data sets go through ranking processes, and the estimation techniques are used to identify which one is the best. The simulation operates on the assumption of a flawless ranking, as elaborated below:
-
We compute the corresponding sample sizes \(s^ \cdot = sl\), resulting in \(s^ \cdot = sl = 30, 75, 150, 250, 400\). This allows us to create an RSS from the suggested model with a fixed set size of \(s = 5\) and changing cycle numbers \(l = 6, 15, 30, 50, 80\).
-
We generate SRS from the suggested model using the specified sample sizes \(s^ \cdot = 15, 50, 120, 200, 300,450\).
-
Using the actual parameter values (a, b, c), we derive a set of estimates for each sample size.
-
To assess the efficacy of the estimation methods, six metrics are utilized, comprising:
-
The average of absolute bias (BIAS), computed by the formula: \(|\text {Bias}(\widehat{\pmb \omega })| = \frac{1}{H}\sum _{i=1}^{H}|\widehat{\pmb \omega }-\pmb \omega |\).
-
The mean squared error (MSE), determined as follows: \(\text {MSE} = \frac{1}{H}\sum _{i=1}^{H}(\widehat{\pmb \omega }-\pmb \omega )^2\).
-
The mean absolute relative error (MRE), evaluated using the expression: \(\text {MRE} = \frac{1}{H}\sum _{i=1}^{H}|\widehat{\pmb \omega }-\pmb \omega |/\pmb \omega\).
-
The average absolute difference, denoted as \(D_{\text {abs}}\), calculated by: \(D_{\text {abs}} = \frac{1}{s^ \cdot H}\sum _{i=1}^{H}\sum _{j=1}^{s^ \cdot }|F(x_{ij}; \pmb \omega )-F(x_{ij};\widehat{ \pmb \omega })|\), where \(F(x;\pmb \omega )=F(x)\) and \(x_{ij}\) represent values obtained at the i-th iteration sample and j-th component of this sample.
-
The maximum absolute difference, represented by \(D_{\text {max}}\), obtained from: \(D_{\text {max}} = \frac{1}{H}\sum _{i=1}^{H}\max \limits _{j=1,\ldots ,n} |F(x_{ij}; \pmb \omega )-F(x_{ij};\widehat{ \pmb \omega })|\).
-
The average squared absolute error (ASAE), computed as: \(\text {ASAE} = \frac{1}{H}\sum _{i=1}^{H}\frac{|x_{(i)}-\hat{x}_{(i)}|}{x_{(n)}-x_{(1)}}\), where \(x_{(i)}\) denotes the ascending ordered observations, and \(\pmb \omega =(a,b,c)\).
-
-
The metrics delineated in the preceding step function as impartial standards for appraising the precision and dependability of the estimated parameters. Employing these assessment criteria facilitates a thorough evaluation of the efficacy of the estimation methods. This evaluative procedure yields significant insights into the effectiveness and suitability of these methods for the specific model in question.
-
This approach can be repeated several times to provide a solid and trustworthy assessment of the estimation methods. By ensuring consistency and clarity in the performance findings, this repeated assessment improves our comprehension of how successful these strategies are in parameter estimation for the model.
-
The assessment metrics related to RSS and SRS are shown in Suppl Tables 1–10 (see Suppl Appendix). These tables provide a thorough summary of the outcomes attained. The numbers in these tables represent the relative effectiveness of each strategy out of all the estimation techniques that were looked at. Reduced values indicate better performance than the examined estimation methods. These tables are crucial for evaluating the relative merits and efficacy of the various estimation methods.
-
The MSE ratio of SRS to RSS is shown in Suppl Table 11, which facilitates the evaluation of the MSE performance of different sampling techniques and provides information on their efficiency.
-
Suppl Tables 12 and 13 for SRS and RSS (see Suppl Appendix), respectively, give comprehensive rankings, including partial and total ranks. These ranking tables thoroughly analyze each estimation technique’s relative efficacy and performance, facilitating a greater comprehension of its advantages and disadvantages.
After a meticulous examination of the simulation outcomes and the rankings depicted in the tables, several deductions emerge:
-
Notably, our model estimates demonstrate consistency for both SRS and RSS data sets. This consistency implies that the estimates progressively approach to the true parameter values as the sample size expands.
-
Every metric used shows a similar trend: a decline with increasing sample size. This trend implies that more accurate and precise parameter estimations are produced with larger sample numbers.
-
Our simulation findings for the SRS and RSS data sets suggest that MXPSE is the best technique when assessing the precision of our calculations.
-
Estimates from RSS data sets show more efficiency than estimates from SRS data sets, as seen in Suppl Table 11. This result suggests that RSS is a more effective sampling technique, producing estimates with a lower MSE.
Real data analysis
This section emphasizes the usefulness of the suggested estimation techniques by thoroughly elaborating on a real data set. This analysis clarifies how these estimation methods may be applied to real data, demonstrating their usefulness and applicability in real-world research and decision-making scenarios. The data set under consideration features the total milk production during the initial birth of 107 cows from the SINDI. This data set was investigated by Abd El-Bar et al.12, and its values are as follows: 0.4365, 0.4260, 0.5140, 0.6907, 0.7471, 0.2605, 0.6196, 0.8781, 0.4990, 0.6058, 0.6891, 0.5770, 0.5394, 0.1479, 0.2356, 0.6012, 0.1525, 0.5483, 0.6927, 0.7261, 0.3323, 0.0671, 0.2361, 0.4800, 0.5707, 0.7131, 0.5853, 0.6768, 0.5350, 0.4151, 0.6789, 0.4576, 0.3259, 0.2303, 0.7687, 0.4371, 0.3383, 0.6114, 0.3480, 0.4564, 0.7804, 0.3406, 0.4823, 0.5912, 0.5744, 0.5481, 0.1131, 0.7290, 0.0168, 0.5529, 0.4530, 0.3891, 0.4752, 0.3134, 0.3175, 0.1167, 0.6750, 0.5113, 0.5447, 0.4143, 0.5627, 0.5150, 0.0776, 0.3945, 0.4553, 0.4470, 0.5285, 0.5232, 0.6465, 0.0650, 0.8492, 0.8147, 0.3627, 0.3906, 0.4438, 0.4612, 0.3188, 0.2160, 0.6707, 0.6220, 0.5629, 0.4675, 0.6844, 0.3413, 0.4332, 0.0854, 0.3821, 0.4694, 0.3635, 0.4111, 0.5349, 0.3751, 0.1546, 0.4517, 0.2681, 0.4049, 0.5553, 0.5878, 0.4741, 0.3598, 0.7629, 0.5941, 0.6174, 0.6860, 0.0609, 0.6488, 0.2747.
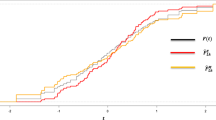
Results of the descriptive analysis can be found in Table 1. A variety of graphical representations is shown in Fig. 2, including histograms, quantile-quantile (Q-Q) plots, violin plots, box plots, total time on test (TTT) plots, and kernel density plots. The probability-probability (P-P) plot, estimated CDF, estimated survival function, and a histogram with the estimated PDF, are given in Fig. 3. The SRS and RSS estimates obtained from the PLD are shown in Tables 2 and 3, respectively. Several goodness-of-fit statistics, namely from the Anderson–Darling test (AT), the Cramer–von-Mises test (WT), and the Kolmogorov–Smirnov test (KST), are used to assess the models, see Table 4. These values (the smaller, the better) demonstrate that RSS is better than SRS for various estimation techniques. Moreover, it is evident from Figs. 4 and 5 how well the models fit the data.
Concluding remarks
The accuracy of parameter estimators is considerably influenced by the sampling technique used in statistical parameter estimation problems. In the current work, the parameter estimates of the PLD are examined using both SRS and RSS approaches. The various estimates obtained by RSS were contrasted with those obtained through SRS. Six metrics were used to evaluate the effectiveness of the estimation methods. Based on our simulation findings for the SRS and RSS data sets, the MXPS method seems to be the best choice in terms of accuracy of the estimates. For both the SRS and RSS data sets, our model estimates show consistency. It may be inferred from this consistency that as the sample size increases, the estimates gradually become closer to the actual parameter values. Compared to estimates obtained from RSS data sets, those created from SRS data sets are less efficient.
Data availability
Any data that supports the findings of this study is included in the article.
References
Mazucheli, J., Menezes, A. F. & Dey, S. The unit-Birnbaum–Saunders distribution with applications. Chil. J. Stat. 9, 47–57 (2018).
Mazucheli, J., Menezes, A. F. B. & Ghitany, M. E. The unit-Weibull distribution and associated inference. J. Appl. Probab. Stat. 13, 1–22 (2018).
Mazucheli, J., Menezes, A. F. & Dey, S. Unit-Gompertz distribution with applications. Statistica 79, 25–43 (2019).
Ghitany, M. E., Mazucheli, J., Menezes, A. F. B. & Alqallaf, F. The unit-inverse Gaussian distribution: A new alternative to two parameter distributions on the unit interval. Commun. Stat. Theory Methods 48, 3423–3438 (2019).
Korkmaz, M.C., & Chesneau, C. On the unit Burr-XII distribution with the quantile regression modeling and applications. Comput. Appl. Math. https://doi.org/10.1007/s40314-021-01418-5.14 (2021)
Korkmaz, M. C., Emrah, A., Chesneau, C. & Yousof, H. M. On the Unit-Chen distribution with associated quantile regression and applications. Math. Slov. 72, 765–786. https://doi.org/10.1515/ms-2022-0052 (2022).
Ramadan, A. T., Tolba, A. H. & El-Desouky, B. S. A unit half-logistic geometric distribution and its application in insurance. Axioms 11, 676 (2022).
Abubakari, A. G., Luguterah, A. & Nasiru, S. Unit exponentiated Frechet distribution: Actuarial measures, quantile regression and applications. J. Indian Soc. Probab. Stat. 23, 387–424 (2022).
Fayomi, A., Hassan, A. S. & Almetwally, E. A. Inference and quantile regression for the unit exponentiated Lomax distribution. PLoS ONE 18(7), e0288635. https://doi.org/10.1371/journal.pone.0288635 (2023).
Hassan, A. S. & Alharbi, R. S. Different estimation methods for the unit inverse exponentiated Weibull distribution. Commun. Stat. Appl. Methods 30(2), 191–213 (2023).
Fayomi, A., Hassan, A. S., Baaqeel, H. M. & Almetwally, E. M. Bayesian inference and data analysis of the unit-power Burr X distribution. Axioms 2023(12), 297. https://doi.org/10.3390/axioms12030297 (2023).
Abd El-Bar, A. M. T., Lima, M. C. S. & Ahsanullah, M. Some inferences based on a mixture of power function and continuous logarithmic distribution. J. Taibah Univ. Sci. 14(1), 1116–1126. https://doi.org/10.1080/16583655.2020.1804140 (2020).
McIntyre, G. A. A method of unbiased selective sampling, using ranked sets. Aust. J. Agric. Res. 3, 385–390 (1952).
Takahasi, K. & Wakimoto, K. On unbiased estimates of the population mean based on the sample stratified by means of ordering. Ann. Inst. Stat. Math. 20(1), 1–31 (1968).
Dell, T. R. & Clutter, J. L. Ranked set sampling theory with order statistics background. Biometrics 28(2), 545–555 (1972).
Kvam, P. H. Ranked set sampling based on binary water quality data with covariates. J. Agric. Biol. Environ. Stat. 8, 271–279 (2003).
Howard, R.W., Jones, S.C., Mauldin, J.K. et al. Abundance, distribution, and colony size estimates for Reticulitermes spp. (Isopter: Rhinotermitidae) in Southern Mississippi. Environ. Entomol. 11, 1290–1293 (1982).
Mahmood, T. On developing linear profile methodologies: A ranked set approach with engineering application. J. Eng. Res. 8, 203–225 (2020).
Halls, L. K. & Dell, T. R. Trial of ranked-set sampling for forage yields. Forest Sci. 12, 22–26 (1966).
Kocyigit, E. G. & Kadilar, C. Information theory approach to ranked set sampling and new sub-ratio estimators. Commun. Stat.-Theory Methods 53(4), 1331–1353 (2024).
Wolfe, D.A. Ranked set sampling: Its relevance and impact on statistical inference. Int. Sch. Res. Not. Probab. Stat. 1–32 (2012)
Stokes, L. Parametric ranked set sampling. Ann. Inst. Stat. Math. 47, 465–482. https://doi.org/10.1007/BF00773396 (1995).
Bhoj, D. S. Estimation of parameters of the extreme value distribution using ranked set sampling. Commun. Stat.-Theory Methods 26(3), 653–667 (1997).
Abu-Dayyeh, W. A., Al-Subh, S. A. & Muttlak, H. A. Logistic parameters estimation using simple random sampling and ranked set sampling data. Appl. Math. Comput. 150(2), 543–554 (2004).
Lesitha, Z. G. & Yageen Thomas, P. Estimation of the scale parameter of a log-logistic distribution. Metrika 76, 427–448. https://doi.org/10.1007/s00184-012-0397-5 (2013).
He, X.-F., Chen, W.-X. & Yang, R. Log-logistic parameters estimation using moving extremes ranked set sampling design. Appl. Math. J. Chin. Univ. 36, 99–113. https://doi.org/10.1007/s11766-021-3720-y (2021).
Yousef, O. M., & Al-Subh, S. A. Estimation of Gumbel parameters under ranked set sampling. J. Mod. Appl. Stat. Methods 13, 24. https://doi.org/10.56801/10.56801/v13.i.741 (2014)
Qian, W. S., Chen, W. X. & He, X. F. Parameter estimation for the Pareto distribution based on ranked set sampling. Stat. Pap.https://doi.org/10.1007/s00362-019-01102-1 (2019).
Esemen, M. & Gurler, S. Parameter estimation of generalized Rayleigh distribution based on ranked set sample. J. Stat. Comput. Simul. 88(4), 615–628 (2018).
Samuh, A. I., Al-Omari, M. H. & Koyuncu, N. Estimation of the parameters of the new Weibull-Pareto distribution using ranked set sampling. Statistica 80, 103–123 (2020).
Yang, R., Chen, W., Yao, D. Long, C., Dong,Y., & Shen, B. The efficiency of ranked set sampling design for parameter estimation for the log-extended exponential-geometric distribution. Iran J. Sci. Technol. Trans. Sci.https://doi.org/10.1007/s40995-020-00855-x (2020).
Al-Omari, A. I., Benchiha, S. & Almanjahie, I. M. Efficient estimation of the generalized quasi Lindley distribution parameters under ranked set sampling and applications. Math. Prob. Eng. 2021, 1214. https://doi.org/10.1155/2021/9982397 (2021).
Al-Omari, A. I., Benchiha, S. & Almanjahie, I. M. Efficient estimation of two-parameter xgamma distribution parameters using ranked set sampling design. Mathematics 10, 3170. https://doi.org/10.3390/math1017317 (2022).
Bhushan, S. & Kumar, A. On some efficient logarithmic type estimators under stratified ranked set sampling. Afr. Mat. 35(2), 40. https://doi.org/10.1007/s13370-024-01180-x (2024).
Hassan, A. S., Elshaarawy, R. S. & Nagy, H. F. Parameter estimation of exponentiated exponential distribution under selective ranked set sampling. Stat. Transit. New Ser. 23(4), 37–58 (2022).
Nagy, H. F., Al-Omari, A. I., Hassan, A. S. & Alomani, G. A. Improved estimation of the inverted Kumaraswamy distribution parameters based on ranked set sampling with an application to real data. Mathematics 10, 4102. https://doi.org/10.3390/math10214102 (2022).
Yousef, M. M., Hassan, A. S., Al-Nefaie, A. H., Almetwally, E. M. & Almongy, H. M. Bayesian estimation using MCMC method of system reliability for inverted Topp–Leone distribution based on ranked set sampling. Mathematics 10, 3122. https://doi.org/10.3390/math1017312 (2022).
Hassan, A. S., Alsadat, N., Elgarhy, M., Chesneau, C. & Elmorsy, R. M. Different classical estimation methods using ranked set sampling and data analysis for the inverse power Cauchy distribution. J. Radiat. Res. Appl. Sci. 16(4), 100685. https://doi.org/10.1016/j.jrras.2023.100685 (2023).
Alsadat, N., Hassan, A. S., Gemeay, A. M., Chesneau, C. & Elgarhy, M. Different estimation methods for the generalized unit half-logistic geometric distribution: Using ranked set sampling. AIP Adv. 13, 085230. https://doi.org/10.1063/5.0169140 (2023).
Hassan, A. S., Elshaarawy, R. S. & Nagy, H. F. Reliability analysis of exponentiated exponential distribution for neoteric and ranked sampling designs with applications. Stat. Optim. Inf. Comput. 11, 580–594 (2023).
Alyami, S. A. et al. Estimation methods based on ranked set sampling for the arctan uniform distribution with application. AIMS Math. 9(4), 10304–10332 (2024).
Rather, K. U. I., Kocyigit, E. G., Onyango, R. & Kadilar, C. Improved regression in ratio type estimators based on robust M-estimation. Plos one 17(12), e0278868. https://doi.org/10.1371/journal.pone.0278868 (2022).
Acknowledgements
This research is supported by researchers Supporting Project number (RSPD2024R548), King Saud University, Riyadh, Saudi Arabia. The authors thank four anonymous reviewers for their valuable feedback and suggestions, which were important and helpful to improve the paper.
Funding
Open Access funding enabled and organized by Projekt DEAL. This research was supported within the project RSPD2024R548, King Saud University, Riyadh, Saudi Arabia. We acknowledge financial support from the Open Access Publication Fund of the University of Hamburg.
Author information
Authors and Affiliations
Contributions
Najwan Alsadat: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Writing - Original Draft, Visualization Amal S. Hassan: Conceptualization, Formal analysis, Investigation, Writing - Original Draft, Writing - Review & Editing Mohammed Elgarhy: Validation, Data Curation, Writing - Original Draft, Writing - Review & Editing, Visualization Arne Johannssen: Conceptualization, Investigation, Writing - Review & Editing, Project administration Ahmed M. Gemeay: Writing - Review & Editing, Supervision, Project administration.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Alsadat, N., Hassan, A.S., Elgarhy, M. et al. Estimation methods based on ranked set sampling for the power logarithmic distribution. Sci Rep 14, 17652 (2024). https://doi.org/10.1038/s41598-024-67693-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-67693-4
- Springer Nature Limited