Abstract
Objectives
To investigate the association between the functional Fc gamma receptor 3 A (FCGR3A) V158F and FCGR2A R131H polymorphisms and rituximab therapy in patients with autoimmune diseases.
Methods
We searched the Medline, Embase, and Cochrane databases for relevant articles. We conducted a meta-analysis of the association between FCGR3A V158F and FCGR2A R131H polymorphisms and responsiveness to rituximab in patients with autoimmune diseases.
Results
Eleven studies, consisting of 661 responders and 267 non-responders for FCGR3A V158F polymorphism and 156 responders and 89 non-responders for FCGR2A R131H polymorphism, were included. The meta-analysis revealed a significant association between the FCGR3A V allele and responsiveness to rituximab (odds ratio [OR] = 1.600, 95% confidence interval [CI] = 1.268–2.018, P < 0.001). Furthermore, associations were found using the dominant and homozygous contrast models. Subgroup analysis showed an association between the FCGR3A V allele and responsiveness to rituximab in European, RA, ITP, small (<50) and large (≥50) groups, and short- (≤6 months) and long-term follow-up periods (≥6 months). These associations were also found in recessive, dominant or homozygous contrast models. Meta-analysis revealed no association between the FCGR2A R allele and responsiveness to rituximab (OR = 1.243, 95% CI = 0.825–1.873, P = 0.229).
Conclusions
We demonstrated that the FCGR3A F158V polymorphism is associated with better responsiveness to rituximab therapy in patients with autoimmune diseases, indicating that individuals carrying the FCGR3A V allele will likely respond better to rituximab. However, FCGR2A R131H polymorphism was not associated with better response to rituximab.
Similar content being viewed by others
Introduction
A wide range of autoimmune diseases impact about 5% of people worldwide. Their absence of self-tolerance results in immune-mediated tissue damage and is one of their defining traits [1]. Autoimmune disorders have a variety of biochemical and clinical signs, but they all share the same pathological mechanisms, such as abnormal B-cell regulation. The necessity of biological treatments that suppress B cells in individuals with intractable autoimmune disease has been shown by the crucial role of B cells in illness. Using antibody-dependent cell-mediated cytotoxicity, the hybrid monoclonal antibody, rituximab, mainly attacks B cells. (ADCC) (4). The Fc gamma receptors (FCGR) on natural killer cells, particularly FcRIIIa, initiate ADCC [2]. Although rituximab is one of the most successful treatments for autoimmune illness, it does not work for everyone [3,4,5]. The causes of this lack of response are unknown. It would be possible to foresee a patient’s response to rituximab, which would remove needless medication, lower expenses for treatment, and significantly improve patient care.
In the identification of immunological complexes, FCGRs play a vital function [6]. Their biochemical reaction has been associated with mutations in the genes encoding FCGRs, which change their preference for the Fc region and include FCGR2A and FCGR3A. A single nucleotide substitution at position 596 results in either valine (V158) or phenylalanine (F158) at position 158, which is the source of the FCGR3A V158F (rs396991) variant. (F158V) [7]. High IgG binding of the FCGR3A V158 variant is associated with a more potent immune response, which is mediated by complement-dependent cytotoxicity, cellular cytotoxicity, and apoptosis [7]. A similar change is made by the FCGR2A R131H (rs1801274) mutation, which changes the amino acid from arginine (R) to histidine at position 131 (H) (R131H). IgG and H131R interact differentially; H131 has a stronger attraction for IgG compared to R131 [8]. Therefore, genomic differences unique to biological agents that regulate the action of FCGR3A and FCGR2A may affect the biological effectiveness [8]. Numerous studies have looked into the relationship between FCGR3A V158F and FCGR2A R131H SNPs and the reactivity to rituximab therapy, because of their significance in the pathogenesis of autoimmune disorders [9,10,11,12,13,14,15,16,17,18,19]. However, the results of these inquiries remain unclear. These variations could be caused by clinical heterogeneity, inadequate statistical strength, or insufficient group size. As a result, we carried out a meta-analysis to get around the constraints of each research and clarify the differences [20,21,22]. In individuals with autoimmune disorders, we looked at the associations between rituximab reactivity and the FCGR2A R131H and FCGR3A V158F polymorphisms.
Materials and methods
Identification of eligible studies and data extraction
We considered all studies that examined the association between FCGR3A F158V and FCGR2A R131H polymorphisms and responsiveness to biologics in patients with autoimmune diseases. A literature search was conducted using the Medline, Embase, and Cochrane Library databases. References within individual publications were reviewed to identify additional studies that were not indexed in the electronic databases. The following keywords and terms were searched: “Fc gamma receptor,” “FCGR3A,” “FCGR2A,” “polymorphism,” “rituximab,” and “autoimmune disease.” No language restrictions were applied. A study was included in the analysis if it: (1) was published before November 2022, (2) presented original data (ensuring independence among the studies), and (3) provided sufficient data to calculate odds ratios (ORs). The exclusion criteria were as follows: (1) inclusion of duplicate data, (2) containment of unextractable data, and (3) study of other polymorphisms. Two independent reviewers searched the literature and extracted the data from the original studies. Discrepancies between the reviewers were resolved by consensus. The following information was extracted from each article: author identification, year of publication, country of study, biologic names, follow-up duration, response criteria used, and genotypes or alleles of FCGR3A F158V and FCGR2A R131H polymorphisms in responders and non-responders.
Evaluation of statistical associations
We estimated the overall contrast between the V and F (allelic effect), VV vs. VF + FF (recessive), VV + VF vs. FF (dominant), and VV + FF (homozygote contrast) models of FCGR3A F158V polymorphism in response to biologics. Similarly, we estimated the overall contrast between the H and R, HH vs. HR + RR, HH + HR vs. RR, and HH vs. RR models of FCGR2A R131H polymorphism in response to rituximab. Point estimates of risk, ORs, and 95% confidence intervals (CI) were calculated for each included study. Cochran’s Q-test was used to assess within- and between-study variations, heterogeneity, and the null hypothesis that all studies evaluated the same effect. The heterogeneity effect was quantified using the I2 statistic, which ranges from 0% to 100% and provides an estimate of the total point estimate variability attributable to heterogeneity rather than chance [23]. I2 values of 25%, 50%, and 75% were designated as low, moderate, and high heterogeneity, respectively. The fixed-effects model assumes that a genetic factor has a similar effect on responders across all included studies and that the observed variation among the studies is caused by chance alone [24]. Conversely, the random-effects model assumes that different studies have substantial diversity and assesses both within-study sampling errors and between-study variances [25]. The two models show similar results when the study groups are homogeneous. However, when the groups are heterogeneous, the random-effects model usually provides wider CIs than the fixed-effects model. The random-effects model and is best used when significant between-study heterogeneity is present [25]. Statistical analyses were performed using the Comprehensive Meta-Analysis Program (Biostat, Englewood, NJ, USA).
Exploring study quality, heterogeneity, and publication bias
The Newcastle–Ottawa Scale (NOS) was used to assess the quality of each study was given a score [26]. The highest score is 9, and a score in the range of 5–9 is considered to be of high methodological quality. Although funnel plots are often used to detect publication bias, they require diverse study types with varying sample sizes. Furthermore, the interpretation of the plots are subjective. Therefore, we evaluated publication bias using Egger’s linear regression test [27], which uses a natural logarithm scale of ORs to measure funnel plot asymmetry.
Results
Studies included in the meta-analysis
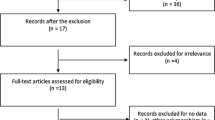
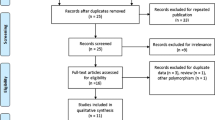
Electronic and manual searches identified 273 eligible studies, of which 17 were selected for full-text review based on the title and abstract details. Six of these 17 studies were excluded because they did not include patients with autoimmune diseases, had no data, or reported other polymorphisms. Finally, 11 studies, consisting of 661 responders and 267 non-responders for the FCGR3A V158F polymorphism and 156 responders and 89 non-responders for FCGR2A R131H polymorphism, that met the inclusion criteria were included [9,10,11,12,13,14,15,16,17,18,19, 28] (Fig. 1). Sixteen separate comparison groups were considered in the meta-analysis. The sample sizes in the studies ranged from 29 to 302. Eleven studies considered FCGR3A F158V polymorphism and five studies investigated the FCGR2A polymorphism. The follow-up period ranged from 3 to 18 months. The quality assessment scores of the included studies ranged from 5 to 7, indicating high quality (Table 1). There was no excluded study based on a low NOS score. The characteristics of these studies are summarized in Table 1.
Association between FCGR3A V158F polymorphism and responsiveness to rituximab
The meta-analysis revealed a significant association between the FCGR3A V allele and responsiveness to rituximab (OR = 1.600, 95% CI = 1.268–2.018, P < 0.001) (Table 2, Fig. 2). Furthermore, associations were found using the dominant and homozygous contrast models (Table 2). Subgroup analysis showed an association between the FCGR3A V allele and responsiveness to rituximab in European, RA, ITP, small (<50) and large (≥50) groups, and short- (≤6 months) and long-term (>6 months) follow-up periods (Table 2, Supplementary Fig.). These associations were also found in recessive, dominant or homozygous contrast models (Table 2).
Association between the FCGR2A R131H polymorphism and responsiveness to rituximab
The meta-analysis revealed no association between the FCGR2A R allele and responsiveness to rituximab in any study participant (OR = 1.243, 95% CI = 0.825–1.873, P = 0.229) (Table 3, Fig. 3). Furthermore, no associations were found between the recessive, dominant, and homozygous contrast models (Table 3). Subgroup analysis showed no association between the FCGR2A R allele and responsiveness to rituximab in European, RA, ITP, AAV, small (<50) and large (≥50) groups, and short- (≤6 months) and long-term (≥6 months) follow-up periods (Table 3, Supplementary Fig.).
Heterogeneity and publication bias
Between-study heterogeneity was found in the subgroup meta-analysis of FCGR3A V158F and FCGR2A R131H polymorphisms (Table 2); however, there was no such heterogeneity in the allelic meta-analysis (Tables 2 and 3). Funnel plots showed symmetry, while Egger’s regression analysis showed no evidence of publication bias for the FCGR polymorphisms addressed (Egger’s regression test P-values > 0.1), indicating no publication bias in this meta-analysis (Fig. 4).
Discussion
Our meta-analysis revealed that patients with the FCGR3A 158 V variation reacted to rituximab more favorably than those with the FCGR3A 158 F variety. However, the rituximab reaction was not correlated with the FCGR2A R131H mutation. To the best of our knowledge, this is the first meta-analysis to compile the data on the relationship between the FCGR3A V158F and FCGR2A R131H polymorphisms and rituximab response in individuals with autoimmune diseases. It also establishes the utility of the FCGR3A F158V polymorphism in predicting rituximab response. Our meta-analysis shows a paucity of reliable pharmacogenetic data on the FCGR2A R131H variation in autoimmune disease patients, though. We concentrated on the FCGR2A and FCGR3A polymorphisms due to their role in ADCC and the abundance of papers looking into their relationship with rituximab response in autoimmune disorders.
Our study’s findings, which further support the idea that genetic variants in FCGR3A, which have been linked to decreased FCGR affinity, are linked to rituximab reactivity, may be explained by FCGR binding and complement interactions. In comparison to the 158 F isoform, the 158 V isoform has a greater propensity for binding to IgG1 and IgG3 [29]. The FCGR3A 158 V variations may enhance the acquisition of the IgG-opsonized pathogen or IgG immune complex, which enters directly into the antigen-processing pathway [30], leading to a more effective display of the arthritogenic peptides. In comparison, the FCGR3A 158 F gene binds to fewer immune complexes and may even suppress inflammation reactions [7, 31]. Individuals with the high affinity V gene are more effectively reduced from peripheral B cells by rituximab. Rituximab insensitivity may be predicted by low FCGR3A mRNA.
The genome q21-q23 risk areas for inflammatory diseases have been found, and the potential genes there are the FCGR2A genes [32, 33]. The susceptibility to biologic treatment may be impacted by alleles linked to autoimmune disease risk [34]. Furthermore, the FCGR2A R131H mutation impacts either H or R at location 131 within the receptor’s second Ig-like region, making it physiologically important [34]. As a consequence, the curative reaction is reduced when the high-affinity H gene enhances the removal of biologics from the bloodstream. The FCGR2A R131H mutation was not linked to a heightened rituximab reaction, according to our meta-analysis. However, because of the small sample size, we could not rule out type II errors. The FCGR3A V158F statistics for rituximab are not unique to autoimmune illness because it is also used to treat cancer [30].
The statistical discrepancy between the FCGR3A V158F variant’s dominant and recessive modes of action is a hot subject in the area of genetic epidemiology. This difference has been seen in numerous studies, with some citing a dominant impact and others a genetic effect. The possible molecular processes that might be causing this disparity have been the subject of a number of theories, though they are still not completely known. Additional hereditary or external variables may alter how the FCGR3A V158F variation affects illness risk. Another reason for the difference between the dominant and recessive theories is that the FCGR3A V158F variation may have complicated and context-dependent effects on the Fc receptor’s functionality. The immunological processes of immune complex uptake and antibody-dependent cellular killing are both mediated by the Fc receptor. Depending on the existence of other immunological components, the specific virus or antigen met, or both, the FCGR3A V158F variant’s impact on these activities may vary.
There are a few limitations on this research. First, because of the small sample size, a publishing bias may have had an impact on the findings of our meta-analysis, especially those of the subgroup analysis. Second, the meta-analysis might have been impacted by variability and influencing variables. The duration of the breaks between exams differed. (from 3 to 18 months). Furthermore, we were unable to take into consideration factors that may have affected the reaction to biologics in this research, including gender, rheumatoid factor status, illness length, and initial DAS28. The meaning of the response phenotype varied between research, and illness seriousness and response phenotype intensity also added to the variety of results. Thirdly, the findings cannot be applied to other groups because the bulk of the data used in this meta-analysis originated from European countries. The proportional impacts of FCGR3A and FCGR2A SNPs on disease risk and treatment reaction may change depending on the gene rates of these polymorphisms, which differ between racial groups.
The results of the current research showed that the FCGR3A V158F variant is related to rituximab response, which implies that people who carry the FCGR3A V gene may respond to rituximab more favorably. In individuals with autoimmune diseases, the FCGR2A R131H mutation was not linked to an improved reaction to rituximab therapy. As a result, identifying the genetic variation in individuals with autoimmune disorders may make it easier to tailor treatments based on the likelihood that they will respond to rituximab. In order to determine the prognostic value of these SNPs in individuals with autoimmune disorders, additional research is necessary.
References
Marrack P, Kappler J, Kotzin BL. Autoimmune disease: why and where it occurs. Nat Med. 2001;7:899–905.
Bruhns P. Properties of mouse and human IgG receptors and their contribution to disease models. Blood J Am Soc Hematol. 2012;119:5640–9.
Lee YH, Bae S-C, Song GG. The efficacy and safety of rituximab for the treatment of active rheumatoid arthritis: a systematic review and meta-analysis of randomized controlled trials. Rheumatol Int. 2011;31:1493–9.
Choi SJ, Ahn SM, Oh JS, Hong S, Lee C-K, Yoo B, et al. Initial preserved renal function as a predictor of favorable renal response to rituximab in refractory or relapsing lupus nephritis: a single-center cohort study in Korea. J Rheum Dis. 2022;29:22–32.
Do H, Pyo JY, Song JJ, Park Y-B, Lee S-W. Implication of serious infections in patients with antineutrophil cytoplasmic antibody-associated vasculitis for the first cycle of rituximab: a pilot study in a single Korean center. J Rheum Dis. 2023;30:45–52.
Ravetch JV, Bolland S. IgG Fc receptors. Annu Rev Immunol. 2001;19:275–90.
Koene HR, Kleijer M, Algra J, Roos D, von dem Borne AE, de Haas, et al. Fc gammaRIIIa-158V/F polymorphism influences the binding of IgG by natural killer cell Fc gammaRIIIa, independently of the Fc gammaRIIIa-48L/R/H phenotype. Blood. 1997;90:1109–14.
Sanders LA, Feldman RG, Voorhorst-Ogink MM, de Haas M, Rijkers GT, Capel PJ, et al. Human immunoglobulin G (IgG) Fc receptor IIA (CD32) polymorphism and IgG2-mediated bacterial phagocytosis by neutrophils. Infect Immun. 1995;63:73–81.
Jiménez Morales A, Maldonado‐Montoro M, Martinez de la Plata JE, Pérez Ramírez C, Daddaoua A, Alarcón Payer C, et al. FCGR2A/FCGR3A gene polymorphisms and clinical variables as predictors of response to tocilizumab and rituximab in patients with rheumatoid arthritis. J Clin Pharmacol. 2019;59:517–31.
Ellithy HN, Ahmed SH, Shahin GH, Matter MM, Talatt M. The impact of Fc gamma receptor IIa and IIIa gene polymorphisms on the therapeutic response of rituximab in Egyptian adult immune thrombocytopenic purpura. Hematology. 2018;23:169–74.
Pál I, Szamosi S, Hodosi K, Szekanecz Z, Váróczy L. Effect of Fcγ-receptor 3a (FCGR3A) gene polymorphisms on rituximab therapy in Hungarian patients with rheumatoid arthritis. RMD Open. 2017;3:e000485.
Cartin‐Ceba R, Indrakanti D, Specks U, Stone JH, Hoffman GS, Kallenberg CG, et al. The pharmacogenomic association of Fcγ receptors and cytochrome P450 enzymes with response to rituximab or cyclophosphamide treatment in antineutrophil cytoplasmic antibody–associated vasculitis. Arthritis Rheumatol. 2017;69:169–75.
Stork AC, Notermans NC, van den Berg LH, Schellevis RD, Niermeijer J-MF, Nederend M, et al. Fcγ receptor IIIA genotype is associated with rituximab response in antimyelin-associated glycoprotein neuropathy. J Neurol Neurosurg Psychiatry. 2014;85:918–20.
Quartuccio L, Fabris M, Pontarini E, Salvin S, Zabotti A, Benucci M, et al. The 158VV Fcgamma receptor 3A genotype is associated with response to rituximab in rheumatoid arthritis: results of an Italian multicentre study. Ann Rheum Dis. 2014;73:716–21.
Zhu Y, Zhuang Y, Yang G-H, Qiang X-F, Yang L, Shen Y-F. Polymorphisms of FcγRIIA, FcγRIIIA and FcγRIIB in patients with immune thrombocytopenia and their clinical significance. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2013;21:135–9.
Kastbom A, Cöster L, Ärlestig L, Chatzidionysiou A, van Vollenhoven RF, Padyukov L, et al. Influence of FCGR3A genotype on the therapeutic response to rituximab in rheumatoid arthritis: an observational cohort study. BMJ Open. 2012;2;e001524.
Ruyssen-Witrand A, Rouanet S, Combe B, Dougados M, Le Loët X, Sibilia J, et al. Fcγ receptor type IIIA polymorphism influences treatment outcomes in patients with rheumatoid arthritis treated with rituximab. Ann Rheum Dis. 2012;71:875–7.
Robledo G, Márquez A, Dávila-Fajardo CL, Ortego-Centeno N, Rubio JLC, Garrido EDR, et al. Association of the FCGR3A-158F/V gene polymorphism with the response to rituximab treatment in Spanish systemic autoimmune disease patients. DNA Cell Biol. 2012;31:1671–7.
Cooper N, Stasi R, Cunningham‐Rundles S, Cesarman E, McFarland JG, Bussel JB. Platelet‐associated antibodies, cellular immunity and FCGR3a genotype influence the response to rituximab in immune thrombocytopenia. Br J Haematol. 2012;158:539–47.
Lee YH, Song GG. The gut microbiome and osteoarthritis: a two-sample mendelian randomization study. J Rheum Dis. 2021;28:94–100.
Lee YH. An overview of meta-analysis for clinicians. Korean J Intern Med. 2018;33:277.
Lee Y-H, Bae S-C, Song G-G. Omega-3 polyunsaturated fatty acids and the treatment of rheumatoid arthritis: a meta-analysis. Arch Med Res. 2012;43:356–62.
Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58.
Egger M, Smith GD, Phillips AN. Meta-analysis: principles and procedures. BMJ. 1997;315:1533–7.
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88.
Wells G, Shea B, O’connell D, Peterson J, Welch V, Losos M, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. 2013. http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp.
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34.
Louis E, El Ghoul Z, Vermeire S, Dall’Ozzo S, Rutgeerts P, Paintaud G, et al. Association between polymorphism in IgG Fc receptor IIIa coding gene and biological response to infliximab in Crohn’s disease. Alimentary Pharmacol Ther. 2004;19:511–9.
Arend WP. The innate immune system in rheumatoid arthritis. Arthritis Rheum. 2001;44:2224–34.
Liu D, Tian Y, Sun D, Sun H, Jin Y, Dong M. The FCGR3A polymorphism predicts the response to rituximab-based therapy in patients with non-Hodgkin lymphoma: a meta-analysis. Ann Hematol. 2016;95:1483–90.
Lee YH, Ji JD, Song GG. Associations between FCGR3A polymorphisms and susceptibility to rheumatoid arthritis: a metaanalysis. J Rheumatol. 2008;35:2129–35.
Choi SJ, Rho YH, Ji JD, Song GG, Lee YH. Genome scan meta-analysis of rheumatoid arthritis. Rheumatology. 2006;45:166–70.
Raychaudhuri S, Thomson BP, Remmers EF, Eyre S, Hinks A, Guiducci C, et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet. 2009;41:1313–8.
Su K, Wu J, Edberg JC, McKenzie SE, Kimberly RP. Genomic organization of classical human low-affinity Fcgamma receptor genes. Genes Immun. 2002;3:S51–6.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lee, Y.H., Song, G.G. Association between functional FCGR3A F158V and FCGR2A R131H polymorphisms and responsiveness to rituximab in patients with autoimmune diseases: a meta-analysis. Pharmacogenomics J 23, 210–216 (2023). https://doi.org/10.1038/s41397-023-00308-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41397-023-00308-9
- Springer Nature Limited