Abstract
The aim of this study was to determine whether the functional HLA-G 14 bp insertion (I)/deletion (D) and +3142 G/C polymorphisms are associated with susceptibility to systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA). A meta-analysis was conducted on the associations between the HLA-G 14 bp I/D, and +3142 G/C polymorphisms and SLE or RA using; (1) allele contrast, (2) the recessive model, (3) the dominant model, and (4) additive model. A total of 14 comparison studies from 11 articles met our inclusion criteria, comprising eight on SLE (1,284 patients and 1,885 controls) and four on RA (820 patients and 772 controls), and three studies investigated response to methotrexate (MTX) in RA according to the HLA-G 14 bp I/D polymorphisms (249 responders and 205 nonresponders). Meta-analysis revealed an association between the II + ID genotype of the HLA-G 14 bp I/D polymorphism in overall group (OR = 1.205, 95 % CI = 1.036–1.403, P = 0.016). Ethnicity-specific meta-analysis showed no association between the II + ID genotype and SLE in the South American, European, and Asian population. Meta-analysis revealed a significant association between SLE and the HLA-G +3142 G allele in all study subjects (OR = 1.367, 95 % CI = 1.158–1.613, P = 2.2 × 10−5) and in the South American group (OR = 1.531, 95 % CI = 1.242–1.888, P = 6.7 × 10−5). However, no association between HLA-G 14 bp I/D, and +3142 G/C polymorphisms and RA was found (OR for HLA-G I allele = 1.013, 95 % CI = 0.879–1.167, P = 0.859; OR for +3142 G allele = 0.980, 95 % CI = 0.742–1.294, P = 0.888). Furthermore, HLA-G 14 bp I/D polymorphism was not found to be associated with response to MTX in RA. This meta-analysis demonstrates that the HLA-G 14 bp I/D polymorphism is associated with susceptibility to SLE, and HLA-G +3142 G/C polymorphisms are associated with susceptibility to SLE in South Americans.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Systemic lupus erythematosus (SLE) is a prototypic human autoimmune disease and a disorder of generalized autoimmunity. It is characterized by intense inflammation and multiple organ damage. Rheumatoid arthritis (RA) is a chronic inflammatory disease that predominantly involves synovial joints and affects up to 1 % of adults worldwide. Although the etiologies of SLE and RA are not fully understood, it is clear that they both have genetic components [1, 2].
HLA-G is a nonclassical major HLA class Ib molecule and has an immunomodulatory properties by suppressing the cytotoxicity of natural killer (NK) cells, CD4+, CD8+ lymphocytes, and dendritic cell functions [3, 4]. The HLA-G gene, which is located on chromosome 6p21.31, contains a 14 bp insertion (I)/deletion (D) (rs1704) and a +3142 G/C (rs1063320) polymorphisms in the 3′ untranslated region (3′UTR) of HLA-G. HLA-G expression is influenced by the HLA-G 14 bp I/D and +3142 G/C polymorphisms. HLA-G 14 bp I/D polymorphisms affect the HLA-G function by influencing the stability and splicing pattern of HLA-G mRNA isoform [5]. The HLA-G +3142 G allele has a binding site with higher affinity for microRNAs, resulting in less expression of HLA-G [6].
The functional HLA-G 14 bp I/D and +3142 G/C polymorphisms have been associated with SLE and RA in some reports, but by no means in all [7–17]. The reasons for this disparity may be small sample sizes, low statistical power, and/or clinical heterogeneity. Therefore, in order to overcome the limitations of individual studies, resolve inconsistencies, and reduce the likelihood that random errors are responsible for false-positive or false-negative associations [18–20], we turned to meta-analysis. The aim of the present study was to determine using meta-analysis whether the functional HLA-G 14 bp I/D and +3142 G/C polymorphisms are susceptible to SLE or RA.
Methods
Identification of eligible studies and data extraction
Published studies that examined the associations between HLA-G polymorphisms and SLE or RA were searched from the literature by using the MEDLINE and EMBASE citation databases, including articles in which HLA-G polymorphisms were analyzed in SLE or RA. Different combinations of keywords, such as “HLA-G,” “polymorphism,” “systemic lupus erythematosus,” “SLE,” “rheumatoid arthritis,” and “RA” were entered as Medical Subject Heading (MeSH) components and as text words. The references cited in the obtained papers were also examined to identify additional studies not indexed by MEDLINE and EMBASE. Genetic association studies that determined the distributions of HLA-G 14 bp I/D or +3142 G/C polymorphism in patients and in controls were included. Overall, the inclusion criteria for the searched studies were as follows: (1) case control study design; (2) original data; and (3) sufficient genotype or allele data to calculate odds ratios (ORs). No language restriction was applied. The exclusion criteria for the searched studies were as follows: (1) overlapping data; (2) inability to ascertain the number of null and wild genotypes or alleles; and (3) studies of family members, because our analysis was based on linkage considerations. Two independent reviewers extracted the data on the methods and results from the original studies for meta-analysis. Any discrepancy between the reviewers was resolved by consensus between the two or by intervention of a third reviewer. The following information was extracted from each study: author name, year of publication, ethnicity of the study population, demographics, number of cases and controls, and the genotype and allele frequencies of each HLA-G 14 bp I/D and +3142 G/C polymorphisms. For analysis of association between HLA-G 14 bp I/D polymorphism and response to MTX in RA, dose of methotrexate (MTX), length of follow-up period, response criteria used, and genotypes of the HLA-G 14 bp I/D polymorphism in responders and nonresponders were additionally extracted.
Evaluations of statistical associations
Allele frequencies at HLA-G 14 bp I/D and +3142 G/C polymorphisms were determined by the allele counting method. Chi-square test was used to ascertain whether the observed frequencies of genotypes in the controls conformed to the Hardy–Weinberg equilibrium (HWE). Meta-analyses were performed using (1) allelic contrast and (2) recessive, (3) dominant, and (4) additive models. Point estimates of risks, ORs, and 95 % confidence intervals (CI) were estimated for each study. Cochran’s Q statistic was used to assess within- and between-study variations and heterogeneities. This heterogeneity test assesses the null hypothesis that all studies evaluated the same effect. The effect of heterogeneity was quantified using I 2, which ranges from 0 to 100 %, and represents the proportion of between-study variability attributable to heterogeneity rather than to chance [21]. I 2 values of 25, 50, and 75 % were nominally considered low, moderate, and high estimates, respectively. The fixed-effects model assumes that a genetic factor has a similar effect on disease susceptibility across all investigated studies, and that the observed variations among the studies are caused by chance alone [22]. However, the random-effects model assumes that different studies show substantial diversity and assesses both within-study sampling errors and between-study variances [23]. When the study groups are homogeneous, the two models are similar. In contrast, when the groups are not homogeneous, the random-effects model usually provides wider CIs than the fixed-effects model. The random-effects model is best used in the presence of significant between-study heterogeneity [23]. Statistical manipulations were performed using the Comprehensive Meta-Analysis program (Biostat, Englewood, NJ, USA). The powers of the studies were computed as the probabilities of detecting associations between the HLA-G 14 bp I/D or +3142 G/C polymorphism and SLE or RA at a significance level of 0.05, assuming an OR of about 1.5 (small effect size, convention w = 0.1) [24]. Power analysis was performed using the statistical software G*Power (http://www.psycho.uni-duesseldorf.de/aap/projects/gpower).
Exploring publication bias
While funnel plots are often used to detect publication bias, funnel plotting requires diverse study types of varying sample sizes and involves subjective judgments. Accordingly, we evaluated any publication bias by using Egger’s linear regression test [25], which measures funnel plot asymmetry using a natural logarithm scale of ORs.
Results
Studies included in the meta-analysis
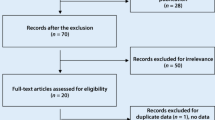
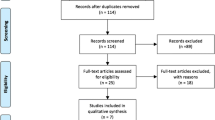
Fifty-eight studies were identified by electronic and manual searches, and 16 were selected for full-text review based on the title and abstract details. Five studies were excluded because they discussed HLA-G polymorphisms other than those considered in this study, showed data duplication, or were review article. Thus, only 11 studies met our inclusion criteria [7–17], and three of which contained data on two different groups [8, 11, 14] that were then analyzed separately (Fig. 1). Finally, a total of 14 separate comparisons were considered in this meta-analysis. These studies comprised eight on SLE consisting of 1,284 patients and 1,885 controls, and four on RA consisting of 820 patients and 772 controls, and three studies investigated response to MTX in RA according to the HLA-G 14 bp I/D polymorphism (249 responders and 205 nonresponders). Ethnicity-specific meta-analysis was conducted on the South American, European, and Asian populations. The statistical powers of these studies ranged between 26.4 and 72.2 %, and none of the studies had a statistical power exceeding 80 % (Table 1). Selected details of the individual studies are summarized in Table 1.
Meta-analysis of HLA-G 14 bp I/D, and +3142 G/C polymorphisms and SLE susceptibility
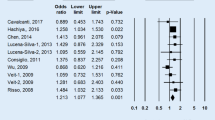
A summary of meta-analyses findings concerning associations between the HLA-G polymorphisms and SLE is provided in Table 2. Meta-analysis of HLA-G 14 bp I/D polymorphism in all study subjects revealed no association between SLE and the HLA-G 14 bp I allele (OR = 1.137, 95 % CI = 0.953–1.356, P = 0.154) (Table 2). Stratification by ethnicity indicated no association between the HLA-G 14 bp I allele and SLE in the South American, European, and Asian population (Table 2). A similar pattern was noted for the HLA-G 14 bp I allele by the analysis using the recessive model and homozygote contrast (Table 2). However, meta-analysis under dominant model revealed an association between the II + ID genotype of the HLA-G 14 bp I/D polymorphism in overall group (OR = 1.205, 95 % CI = 1.036–1.403, P = 0.016) (Table 2, Fig. 2). Ethnicity-specific meta-analysis showed no association between the II + ID genotype and SLE in the South American, European, and Asian population (Table 2). Meta-analysis of HLA-G +3142 G/C polymorphism in all study subjects revealed a significant association between SLE and the HLA-G +3142 G allele (OR = 1.367, 95 % CI = 1.158–1.613, P = 2.2 × 10−5) (Table 2). Stratification by ethnicity indicated an association between the HLA-G +3142 G allele and SLE in the South American group (OR = 1.531, 95 % CI = 1.242–1.888, P = 6.7 × 10−5) (Table 2). Furthermore, analysis using the recessive, dominant models, and homozygote showed the same pattern for the HLA-G +3142 G allele (Table 2; Fig. 2).
Meta-analysis of HLA-G 14 bp I/D, and +3142 G/C polymorphisms and RA susceptibility
No association between HLA-G 14 bp I/D, and +3142 G/C polymorphisms and RA was found using the allele contrast (OR for HLA-G I allele = 1.013, 95 % CI = 0.879–1.167, P = 0.859; OR for +3142 G allele = 0.980, 95 % CI = 0.742–1.294, P = 0.888), recessive, dominant, or additive models in all study subjects or South American, respectively (Table 3; Fig. 3).
Association between HLA-G 14 bp I/D polymorphism and response to MTX treatment in RA
We investigated the possible effects of HLA-G 14 bp I/D polymorphism on MTX response in RA. Meta-analysis showed no association between the HLA-G 14 bp I allele and response to MTX therapy (OR = 0.940, 95 % CI = 0.720–1.228, P = 0.651) (Table 4). Furthermore, no association was found between HLA-G 14 bp I/D polymorphism and a response to MTX treatment using recessive, dominant, or additive models (Table 4; Fig. 4).
Heterogeneity and publication bias
In SLE, some between-study heterogeneity was found in the meta-analyses of the HLA-G 14 bp I/D polymorphism, but no heterogeneity was found during meta-analyses of the HLA-G +3142 G/C polymorphism. In RA, no between-study heterogeneity was found during meta-analyses of the HLA-G 14 bp I/D and +3142 G/C polymorphisms (Table 3). Deviation from the HWE among controls implies potential bias during control selection or genotyping errors. However, the distributions of genotypes in normal control groups were consistent with the HWE in all studies on SLE and RA. Egger’s regression test showed no evidence of publication bias in the meta-analyses (Egger’s regression test P values >0.1).
Discussion
HLA-G plays a key role in immunosuppression by inhibiting the cytotoxic activity of NK and cytotoxic T cells, CD4 + T cell all proliferative response, and inhibiting their maturation and function of antigen-presenting cells [3, 4]. Considering HLA-G importance in immune modulation, the link between HLA-G expression and autoimmune diseases [12], and HLA-G gene located at a strong linkage region of SLE and RA [1, 2], HLA-G gene has been considered as a candidate gene in the context of SLE and RA.
The present study addressed the association between the HLA-G 14 bp I/D and +3142 G/C polymorphisms and susceptibility to SLE and RA. We found an association between SLE and the HLA-G 14 bp I/D, and +3142 G/C polymorphisms. The results of our study are consistent of an immunosuppressive role of HLA-G in SLE. However, we found no association between RA and the HLA-G polymorphisms. This discrepancy may reflect the different role of these polymorphisms in different autoimmune disease or underpowered studies of small sample sizes of subgroup analysis.
SLE is a prototypic of autoimmune disease and is characterized by multiple organ involvement, polyclonal B cell activation, and autoantibody production. The HLA-G expression may be a mechanism of protection against autoimmune responses by downregulation of inflammatory processes and involvement in immune tolerance [26]. The HLA-G 14 bp I/D and +3142 G/C polymorphisms may have been functionally implicated in SLE [5, 6], and in the present study, we found an overrepresentation of the HLA-G 14 bp I allele and +3142 G allele in SLE patients versus controls. Both the HLA-G 14 bp I and +3142 G alleles have been known to be associated with lower expression of HLA-G [5, 6].
RA is a chronic inflammatory autoimmune disease characterized by synovial cell proliferation and T lymphocyte accumulation within a synovial tissue and is associated with immune dysregulation. Our results showed that the HLA-G 14 bp I/D and +3142 G/C polymorphisms are associated with susceptibility to SLE, but not to RA. This finding suggests that the HLA-G 14 bp I/D and +3142 G/C polymorphisms have disease-dependent functionality or that the analysis in RA was underpowered. In studies undertaken to identify genetic factors with small effects on risk, sample size is a critical consideration. Thus, we are unable to rule out the possibility that RA is associated with the HLA-G polymorphisms due to a lack of statistical power. The studies on the effect of HLA-G 14 bp I/D polymorphisms on response to MTX in RA have produced contradictory results. For example, one study showed increased response to MTX in patients with HLA-G 14 bp D allele [15], but two later studies failed to replicate this finding [16, 17]. In the present study, no relation was found between HLA-G 14 bp I/D polymorphism and response to MTX therapy.
The present study has some limitations that require consideration. First, our ethnic-specific meta-analysis included data from South American, European, and Asian patients, and thus, our results are applicable to only these ethnic groups. The relative importance of HLA-G polymorphisms during the development of SLE and RA may depend on ethnicity. However, we were unable to perform ethnicity-specific meta-analysis in various populations due to limited data. Second, the HLA-G polymorphism may be associated with clinical manifestations in addition to disease susceptibility. We could not stratify and analyze factors, such as sex, or clinical or environmental variables, because of a lack of data. Third, our results should be interpreted with caution due to the limited number of studies included, which restricted further subgroup analyses. There were only three studies included in each meta-analysis of the HLA-G +3142 G/C polymorphism and RA, and HLA-G 14 bp I/D polymorphism and response to MTX treatment in RA. Thus, this study was two small to reach a conclusive result. Fourth, Although Egger’s regression test showed no evidence of publication bias in the meta-analyses, publication bias is inevitable when there were fewer than nine literatures. Only a total eight studies on SLE, four studies on RA, and three studies investigated response to methotrexate (MTX) in RA according to the HLA-G 14 bp I/D polymorphisms in this meta-analysis. As a result, Egger’s regression for publication bias sensitivity is not high, and the publication bias exists by default in the meta-analysis.
In conclusion, this meta-analysis demonstrates that the HLA-G 14 bp I/D polymorphism is associated with susceptibility to SLE, and HLA-G +3142 G/C polymorphism is associated with susceptibility to SLE in South Americans. However, no association was found between the HLA-G 14 bp I/D, and +3142 G/C polymorphisms and susceptibility to RA. Given the important roles of HLA-G in immunologic process, larger-scale studies in different ethnic populations are required to explore the roles of polymorphisms of the HLA-G gene in the pathogeneses of SLE.
References
Lee YH, Nath SK (2005) Systemic lupus erythematosus susceptibility loci defined by genome scan meta-analysis. Hum Genet 118(3–4):434–443
Choi SJ, Rho YH, Ji JD, Song GG, Lee YH (2006) Genome scan meta-analysis of rheumatoid arthritis. Rheumatology (Oxford) 45(2):166–170
Dorling A, Monk NJ, Lechler RI (2000) HLA-G inhibits the transendothelial migration of human NK cells. Eur J Immunol 30(2):586–593
Bainbridge DR, Ellis SA, Sargent IL (2000) HLA-G suppresses proliferation of CD4(+) T-lymphocytes. J Reprod Immunol 48(1):17–26
Rousseau P, Le Discorde M, Mouillot G, Marcou C, Carosella ED, Moreau P (2003) The 14 bp deletion-insertion polymorphism in the 3′ UT region of the HLA-G gene influences HLA-G mRNA stability. Hum Immunol 64(11):1005–1010
Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L et al (2007) Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet 81(4):829–834
Chen T, Lin J, Jin S, Ma S, Yu L, Huang K et al (2014) Association of three polymorphisms in the 3′ untranslated region of the HLA-G gene with systemic lupus erythematosus in a population from Yunnan. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 31(2):228–232
Lucena-Silva N, de Souza VS, Gomes RG, Fantinatti A, Muniz YC, de Albuquerque RS et al (2013) HLA-G 3′ untranslated region polymorphisms are associated with systemic lupus erythematosus in 2 Brazilian populations. J Rheumatol 40(7):1104–1113
Consiglio CR, Veit TD, Monticielo OA, Mucenic T, Xavier RM, Brenol JC et al (2011) Association of the HLA-G gene +3142C > G polymorphism with systemic lupus erythematosus. Tissue Antigens 77(6):540–545
Wu FX, Wu LJ, Luo XY, Tang Z, Yang MH, Xie CM et al (2009) Lack of association between HLA-G 14-bp polymorphism and systemic lupus erythematosus in a Han Chinese population. Lupus 18(14):1259–1266
Veit TD, Cordero EA, Mucenic T, Monticielo OA, Brenol JC, Xavier RM et al (2009) Association of the HLA-G 14 bp polymorphism with systemic lupus erythematosus. Lupus 18(5):424–430
Rizzo R, Hviid TV, Govoni M, Padovan M, Rubini M, Melchiorri L et al (2008) HLA-G genotype and HLA-G expression in systemic lupus erythematosus: HLA-G as a putative susceptibility gene in systemic lupus erythematosus. Tissue Antigens 71(6):520–529
Catamo E, Addobbati C, Segat L, Sotero Fragoso T, Domingues Barbosa A, Tavares Dantas A et al (2014) HLA-G gene polymorphisms associated with susceptibility to rheumatoid arthritis disease and its severity in Brazilian patients. Tissue Antigens 84(3):308–315
Veit TD, de Lima CP, Cavalheiro LC, Callegari-Jacques SM, Brenol CV, Brenol JC et al (2014) HLA-G +3142 polymorphism as a susceptibility marker in two rheumatoid arthritis populations in Brazil. Tissue Antigens 83(4):260–266
Rizzo R, Rubini M, Govoni M, Padovan M, Melchiorri L, Stignani M et al (2006) HLA-G 14-bp polymorphism regulates the methotrexate response in rheumatoid arthritis. Pharmacogenet Genomics 16(9):615–623
Stamp LK, O’Donnell JL, Chapman PT, Barclay ML, Kennedy MA, Frampton CM et al (2009) Lack of association between HLA-G 14 bp insertion/deletion polymorphism and response to long-term therapy with methotrexate response in rheumatoid arthritis. Ann Rheum Dis 68(1):154–155
Kooloos WM, Wessels JA, van der Straaten T, Allaart CF, Huizinga TW, Guchelaar HJ (2010) Functional polymorphisms and methotrexate treatment outcome in recent-onset rheumatoid arthritis. Pharmacogenomics 11(2):163–175
Lee YH, Rho YH, Choi SJ, Ji JD, Song GG (2007) PADI4 polymorphisms and rheumatoid arthritis susceptibility: a meta-analysis. Rheumatol Int 27(9):827–833
Lee YH, Bae SC, Choi SJ, Ji JD, Song GG (2011) Associations between vitamin D receptor polymorphisms and susceptibility to rheumatoid arthritis and systemic lupus erythematosus: a meta-analysis. Mol Biol Rep 38(6):3643–3651
Lee YH, Bae SC, Choi SJ, Ji JD, Song GG (2012) The association between the PTPN22 C1858T polymorphism and rheumatoid arthritis: a meta-analysis update. Mol Biol Rep 39(4):3453–3460
Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21(11):1539–1558
Egger M, Smith GD, Phillips AN (1997) Meta-analysis: principles and procedures. BMJ 315(7121):1533–1537
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7(3):177–188
Faul F, Erdfelder E, Buchner A, Lang AG (2009) Statistical power analyses using G*Power 3.1: tests for correlation and regression analyses. Behav Res Methods 41(4):1149–1160
Egger M, Smith GD, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315(7109):629–634
Carosella ED, Moreau P, Aractingi S, Rouas-Freiss N (2001) HLA-G: a shield against inflammatory aggression. Trends Immunol 22(10):553–555
Acknowledgments
This study was supported in part by a Grant of the Korea Healthcare technology R&D Project, Ministry for Health and Welfare, Republic of Korea (HI13C2124).
Conflict of interest
The authors have no financial or nonfinancial conflict of interest to declare.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lee, Y.H., Bae, SC. & Song, G.G. Meta-analysis of associations between functional HLA-G polymorphisms and susceptibility to systemic lupus erythematosus and rheumatoid arthritis. Rheumatol Int 35, 953–961 (2015). https://doi.org/10.1007/s00296-014-3155-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00296-014-3155-3