Abstract
Leprosy is a chronic infectious and neurological disease that is caused by infection of Mycobacterium leprae (M. leprae). A recent genome-wide association study indicated a suggestive association of LRRK2 genetic variant rs1873613 with leprosy in Chinese population. To validate this association and further identify potential causal variants of LRRK2 with leprosy, we genotyped 13 LRRK2 variants in 548 leprosy patients and 1078 healthy individuals from Yunnan Province and (re-)analyzed 3225 Han Chinese across China. Variants rs1427267, rs3761863, rs1873613, rs732374 and rs7298930 were significantly associated with leprosy per se and/or paucibacillary leprosy (PB). Haplotype A-G-A-C-A was significantly associated with leprosy per se (P=0.018) and PB (P=0.020). Overexpression of the protective allele (Thr2397) of rs3761863 in HEK293 cells led to a significantly increased nuclear factor of activated T-cells’ activity compared with allele Met2397 after lipopolysaccharides stimulation. Allele Thr2397 could attenuate 1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine-induced autophagic activity in U251 cells. These data suggest that the protective effect of LRRK2 variant p.M2397T on leprosy might be mediated by increasing immune response and decreasing neurotoxicity after M. leprae loading. Our findings confirm that LRRK2 is a susceptible gene to leprosy in Han Chinese population.
Similar content being viewed by others
Introduction
Leprosy is a chronic infectious and neurological disease that is caused by Mycobacterium leprae (M. leprae) infection. The bacterium can affect human peripheral nerve and skin with consequent nerve damage and/or disabilities.1 Over decades, the prevalence of leprosy has been reduced gradually; however, it still affects approximately 200 000 people annually according to the World Health Organization report.2 Up to now, leprosy remains a public health problem, and the molecular underpinnings of M. leprae infection and leprosy onset have not been fully elucidated.
Development of leprosy after M. leprae infection depends on host genetic background. Among those people who were potentially exposed to M. leprae, <5% of them develop disease.3, 4 In addition, most of leprosy patients living in endemic areas always have poor nutrition conditions, suggesting that environment and host genetic background have key roles in leprosy susceptibility. During the past years, accumulating evidence showed that host genetic background can influence the infection of M. leprae and its clinical manifestation, such as TLR1,5 HLA-DRB1,6 NOD2,6, 7 ILs,8, 9 TNF,10 VDR,11 MRC1,12 IFNG,13, 14 FCN2, MBL2 and CFH.15, 16 These innate and adaptive immune response relevant genes had a complex interplay with pathogen and host during disease onset.
Human leucine-rich repeat kinase 2 (LRRK2, OMIM 609007) gene encodes dardarin (PARK8), which is involved in the interferon-gamma response and host response to pathogens.17 This gene is located in the 12q12 chromosomal region and contains 51 exons. LRRK2 is a multi-domain protein with 2527 amino acids, includes enzymatic activity domains (a GTPase and a kinase domain), a leucine-rich repeat (LRR) domain and a C-terminal WD40 repeat domain.18 Recently, single-nucleotide polymorphism (SNP) rs1873613 of the LRRK2 gene was identified as a risk factor for leprosy in Han Chinese in a genome-wide association study (GWAS).6 In previous studies, LRRK2 mutations were recognized as the most common cause of hereditary Parkinsonism,19, 20 which is a neurodegenerative movement disorder characterized by the presence of intracytoplasmic lewy bodies and lewy neuritis.21 Genetic polymorphisms of LRRK2 were also reported to be associated with susceptibility to Crohn’s disease22 and cancer.23 In addition, LRRK2 might have a key role in autophagy,24 and overexpression of mutant LRRK2 in human neuroblastoma cells caused autophagy activation and significantly decreased neurite length.25 LRRK2 could act as a regulator to control the nuclear factor of activated T-cells (NFAT) activity in the immune system26 and regulated microglial inflammatory responses.27
In this study, we genotyped 13 SNPs of the LRRK2 gene (including the GWAS hit rs1873613)6 in 527 Han Chinese with leprosy and 1078 healthy subjects from Southwest China to investigate whether this gene is involved in leprosy and to identify the potential causal variant(s). We found that five variants (rs1427267, rs3761863 (p.M2397T), rs1873613, rs732374 and rs7298930) were significantly associated with leprosy per se and/or paucibacillary leprosy (PB) patients. Functional characterization showed that polymorphism p.M2397T (rs3761863) can increase the NFAT activity after lipopolysaccharides (LPS) stimulation in HEK293 cells and attenuate 1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine (MPTP)-induced autophagic activity in U251 cells. Our results confirmed that LRRK2 is a susceptibility gene to leprosy in Han Chinese population.
Results
Association of LRRK2 SNPs and haplotypes with leprosy per se and PB patients
The minor allele frequency (MAF) for SNPs (except for six rare/pathogenic SNPs, which had a frequency of 0.0–0.04) analyzed in this study ranged from 28.2% to 49.4% (Tables 1 and 2). Considering a MAF of 0.282 as observed in our samples, the power to detect an odds ratio (OR) value as low as 1.6 for risk allele was expected to be >97% while the power for MAF of 0.494 was expected to be >95%.
With the exception of rs732374 (P=0.010) and rs7298930 (P=0.027), none of the analyzed tag SNPs and the reported GWAS hit SNP had a deviation from Hardy–Weinberg equilibrium (HWE) in the control group. Note that we also observed a marginally significant HWE P-value for rs732374 (P=0.05) in Han Chinese from Eastern China in our previous study.28 The deviation from HWE was unlikely caused by genotyping errors, as we found no problem during the double check of the original genotyping data, and we could consistently validate the genotyping results by sequencing 2% randomly selected individuals. There was no significant difference (P>0.05) when we compared genotype frequency of these two SNPs in the control population with HapMap CHB data (82 Han Chinese) or with 3225 Han Chinese reported in our recent studies.28, 29 The reported pathogenic LRRK2 mutations were also found in some subjects, albeit with a very low frequency (Table 3).
We failed to validate the association between the GWAS-reported SNP rs1873613 with leprosy per se. However, when we subdivided leprosy into PB and multibacillary leprosy (MB) populations according to their clinical expression, genotype GG of rs1873613 had a marginal significant difference (P=0.045) between the PB group and control group, and allele G had a lower frequency in the PB group (26.4%) compared with the control group (31.8%; OR=0.769, P=0.020). Among the six tag SNPs, three of them appeared to have a protective role against leprosy per se (rs3761863, genotype GG: OR=0.713, P=0.038; rs732374, genotype AA: OR=0.651, P=0.020; and rs7298930, genotype CC: OR=0.647, P=0.005) and the PB group (rs3761863, genotype GG: OR=0.584, P=0.014; rs732374, genotype AA: OR=0.592, P=0.026; and rs7298930, genotype CC: OR=0.558, P=0.005) at the genotypic level. However, tag SNP rs1427267 conferred a risk to leprosy per se (genotype AA: OR=1.403, P=0.026; genotype AG: OR=1.394, P=0.015), MB group (genotype AG: OR=1.512, P=0.015) and the PB group (genotype AA: OR=1.585, P=0.020) at the genotypic level. This pattern was robust when these SNPs were analyzed at the allelic level. SNPs rs732374 and rs7298930 appeared to have a protective role against leprosy per se (A allele of rs732374: OR =0.804, P=0.008; and C allele of rs7298930: OR=0.811, P=0.006) and the PB group (A allele of rs732374: OR=0.767, P=0.018; and C allele of rs7298930: OR=0.739, P=0.003). SNP rs3761863 was associated with the PB group (G allele: OR=0.775, P=0.013). Moreover, SNP rs1427267 conferred a risk to leprosy per se (A allele: OR=1.179, P=0.030) and the PB group (A allele: OR=1.274, P=0.016). When we aggregated the Han Chinese samples that were reported in our previous studies28, 29 and this study (n=4303) together to increase the statistical power, the above associations with leprosy per se were further confirmed at the genotypic level (Table 2).
We performed linkage disequilibrium analysis to test whether these tag SNPs in the case and control groups were linked together. As shown in Figure 1, both populations had a similar linkage disequilibrium structure, and this result was consistent with the observation in our recent studies of Han Chinese from other parts of China.28, 29 Note that rs1427267 was linked with rs7298930 (r2=0.81 in case population and r2=0.83 in control population), so we excluded SNP rs7298930 in the following analysis.
A total of 22 haplotypes in the cases and 23 haplotypes in the controls (order of SNPs: rs1873613–rs732374–rs1427267–rs7307310–rs3761863) were reconstructed based on five tag SNPs. Among them, seven haplotypes were observed as the most common haplotype. The overall haplotype test was performed to show the global difference in haplotype frequencies between the case (also grouped into PB and MB) and control populations. There were significant differences for the two groups (Chi-square test: case vs control, P=0.031; MB vs control, P=0.221; PB vs control, P=0.033). In particular, haplotype A-G-A-C-A (consists of non-protective allele of each SNPs) posed a risk effect on leprosy per se (OR=1.282, P=0.018) and the PB group (OR=1.369, P=0.020). Haplotype A-G-G-C-A had a marginally significant protective effect on leprosy per se (OR=0.685, P=0.046; Table 4).
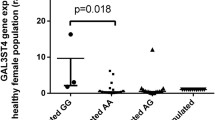
We also measured the serological difference of LRRK2 between cases and controls. However, we found no significant difference of serum LRRK2 concentrations between the cases and controls (t-test, P>0.05; Supplementary Figure S1).
LRRK2 allele Thr2397 increases NFAT activity in HEK-293 cell stimulated by LPS
To understand the underpinning of the protective role of SNP rs3761863 (p.Met2397Thr) on PB leprosy, we performed an evolutionary comparison to evaluate the conservation of this position in vertebrates and characterized different alleles of variant p.M2397T on NFAT activity in HEK293 cells treated with and without LPS. LRRK2 variant p.M2397T was relatively conserved among the primates and rodentia (Supplementary Figure S2). We made a variety of constructs, including full-length LRRK2 (wild type (WT)) and variants bearing pathogenic mutations p.G2019S, p.M2397T and deletion of WD40 domain (ΔWD40). As shown in Figure 2, after 12 h treatment with LPS (1 μg ml−1), the reported Parkinson’s disease (PD)-associated pathogenic mutation p.G2019S and the truncated LRRK2 mutant ΔWD40 had no observed effect on regulating NFAT activity compared with the WT LRRK2. In contrast, variant p.M2397T significantly increased NFAT activity (t-test, P<0.05) upon LPS treatment compared with the WT.
Functional assays of LRRK2 allele p.Thr2397. (a) Immunoblot analysis of total cell lysates of transfected HEK293 cells showing overexpressed LRRK2 protein. (b) NFAT luciferase assay of HEK293 cells transfected with equal amount of empty vector or mutant LRRK2 and stimulated with and without LPS (1 μg ml−1). (c) Immunoblot analysis of total U251cell lysates with stable expression of mutant LRRK2 induced by doxycycline (1 μg ml−1) treatment. (d) Immunoblot analysis of total U251 cell lysates with stable expression of mutant LRRK2 treated with or without MPTP (500 μM) in the presence of doxycycline (top), and densitometry of the results presented as the ratio of LCB-II/LCB-I (below). Data were representative of three independent experiments.
Stable expression of LRRK2 allele Thr2397 attenuates MPTP-induced autophagy in U251 cells
MPTP is a neurotoxic drug that induces autophagy in neuroblastoma cells.30 We tested whether the stable expression of LRRK2 p.M2397T variant could regulate autophagy in U251 cells with and without MPTP treatment. Consistent with previous study that LRRK2 p.G2019S variant could cause a high-level basal autophagy in cultured fibroblasts,31 we found that p.G2019S and p.M2397T of LRRK2 had a similar level of basal autophagy based on the quantification of LC3B-I to LC3B-II conversion. However, after treatment with MPTP (500 μM), the control (without transfection), PLVX (vector), WT LRRK2 and p.G2019S variant groups had a higher autophagic activity than those without MPTP treatment. Variants Thr2397 and ΔWD40 of LRRK2 salvaged autophagy in cells after MPTP treatment (Figure 2).
Discussion
Recently, Zhang et al.6 carried out the first leprosy GWAS in Han Chinese population, and rs1873613 of the LRRK2 gene was identified to be associated with leprosy. The association between this variant and leprosy was said to be validated in Indian patients.32 Previous investigations also showed that overexpression of mutant LRRK2 (for example, p.G2019S, p.R1441C, p.I2020T, p.Y1699C) in cultured cells caused neuron damage,33 autophagy,34 inflammation26, 27 and mitochondrial dysfunction.35 All these observations suggested that LRRK2 had important roles in cellular activity and might explain why LRRK2 was involved in the pathogenesis of PD, Crohn’s disease, cancer and/or leprosy.36 Intriguingly, we found that the LRRK2 gene had experienced positive selection both in hominidae and primate lineages during the evolution (ω2>ω1 and P-value <0.05; Supplementary Method and Supplementary Table S1) by branch model test, which might account for the adaptation to its biological functions and/or host diseases in primates. Moreover, these vertebrate species that were reported to be infected by M. leprae, including primates (human, chimpanzee, gorilla and monkey), rodentia (guinea pig, mouse and rat), hedgehog, tree shrew and nine-banded armadillo,4, 37, 38 were clustered together in their respective clades in the neighbor-joining tree of LRRK2 amino-acid sequences (Supplementary Figure S2). Based on the clustering pattern of the tree, we speculated that those species which were marked with a ‘?’ might be infected by M. leprae. Taken all these lines of evidence together, it seemed that LRRK2 gene is likely to be involved in leprosy.
In this study, we aimed to define the potential role of LRRK2 in leprosy. We first attempted to validate and identify LRRK2 genetic variants that were associated with leprosy, followed by functional characterization of risk allele at the cellular level. Besides the previously reported GWAS-hit SNP rs1873613,6 additional SNPs were genotyped to have a better characterization of the association of LRRK2 variants with leprosy. The analyzed case and control populations had no potential population stratification and sampling bias based on our previous analysis of matrilineal genetic components,39 which could serve as a good basis for genetic association analysis. In contrast with the GWAS report,6 we failed to replicate the reported association of rs1873613 with leprosy per se, but we discerned an association of this SNP with PB leprosy. This result might be explained by the weak linkage disequilibrium of rs1873613 with potential causal variant and/or a large chromosomal distance from the LRRK2 gene. The complexity of leprosy clinical outcomes and different regional samples might also account for the incomplete consistency between our study and the GWAS report.6 Note that rs1873613 had a positive signal as expression quantitative trait loci for the nearby SLC2A13 gene according to the information provided by the expression quantitative trait loci database Genevar (www.sanger.ac.uk/resources/software/genevar/). Further analysis of the SLC2A13 gene would be worthwhile to clarify the role of rs1873613 in leprosy.
Intriguingly, four LRRK2 SNPs (rs1427267, rs3761863, rs732374 and rs7298930) were identified to be strongly associated with leprosy per se and PB at both the genotypic and allelic levels. Similarly, we observed significant associations of LRRK2 haplotypes that were composed of SNPs rs1873613–rs732374–rs1427267–rs7307310–rs3761863 with leprosy per se and/or PB (Table 4). These results reinforced the notion that LRRK2 genetic variants confer susceptibility to leprosy.
Among these leprosy-associated SNPs, the non-synonymous SNP rs3761863 (p.Met2397Thr) was previously identified to be associated with Crohn’s disease22 and was reported to affect the amount and stability of LRRK2 protein in vitro.26 However, we observed no effect of this variant on the serum LRRK2 level between the case and control populations (Supplementary Figure S1). We next characterized the putative role of p.M2397T compared with the PD-associated pathogenic mutation p.G2019S. We demonstrated that overexpression of allele p.T2397 had a significantly increased NFAT activity after LPS stimulation in HEK293 cells, whereas mutant p.S2019 had no effect on NFAT activity (Figure 2). This result was reasonable as the kinase function of LRRK2 did not participate in NFAT regulation.26
There is increasing evidence that demonstrates the active role of LRRK2 in regulating macroautophagy, chaperone-mediated autophagy and mitophagy.40, 41, 42 Therefore, we investigated whether LRRK2 mutations could affect the progress of autophagy in the presence of MPTP. MPTP caused a destruction of dopaminergic neurons in the nigrostriatal pathway,43 and this degeneration of neurons was associated with MPTP-induced autophagy.30, 44 We observed that Thr2397-LRRK2 and ΔWD40-LRRK2 could inhibit the process of MPTP-induced autophagy. In contrast, the PD-pathogenic mutation p.G2019S could increase the highest ratio of the LC3-II conversion. This result suggested that variant p.M2397T might have an opposite role with p.G2019S in regulating autophagy. Variant p.M2397T is located within the WD40 domain of the LRRK2 protein. Deletion of WD40 domain could eliminate LRRK2 dimer complex formation and kinase activity.45 Moreover, in vivo loss-of-function study of LRRK2 in zebrafish showed that deletion of WD40 domain caused brain dopaminergic neurons loss and axon tract disorganization.46 We found that the WD40 domain had a protective role in MPTP-induced neurotoxicity. In addition, p.M2397T had a similar role as the WD40 domain during this process, suggesting that individuals with allele T2397 might have different susceptibility to autophagy. Collectively, we speculate that the protective effect of allele T2397 on leprosy might be mediated by an increasing immune response and decreasing neurotoxicity after M. leprae loading.
A limitation of this study is the lack of direct relevance between autophagy and M. leprae infection affected by LRRK2 genetic polymorphisms. It may provide more evidence to work on the early onset leprosy patients with Met2397 or Thr2397, and assess the autophagy and bacteria index at the lesion, because the role of LRRK2 and autophagy in bacterial clearance and disease clinical manifestations may be most important early in infection. Unfortunately, we do not have access to these samples at the moment. Another limitation of the study is that there is a possibility of rare LRRK2 mutations in leprosy even though we analyzed several reported pathogenic mutations.
In summary, we showed that the GWAS top hit SNP rs1873613 of the LRRK2 gene was not associated with leprosy per se but was associated with PB in our samples. We identified additional tag and functional SNPs that were associated with leprosy per se and its subtypes. Functional assays showed that LRRK2 variant p.M2397T affected NFAT activity in immune response and MPTP-induced autophagy. Further experiments are needed to explore this association and to understand the LRRK2 functional change in leprosy.
Materials and Methods
Ethics statement
Written informed consents conforming to the tenets of the Declaration of Helsinki were obtained from each participant prior to the study. The institutional review board of the Kunming Institute of Zoology approved this study.
Study subjects
We performed a case–control study in a population from Yuxi Prefecture, Yunnan Province of Southwest China. This study was carried out in 1626 samples, including 548 leprosy patients (onset age from 2 to 67 years, mean age: 24.9±12.5 years; male/female ratio=399/149; multibacillary: paucibacillary=295: 252, one individual without a clear clinical classification) and 1078 healthy control subjects from the same geographic area (age from 4 to 91 years, mean age: 39.9±17.6 years; male/female ratio=590: 488). The diagnosis of leprosy patients was based on clinical and histopathological features, as well as the bacteriological index if available, as described in our recent epidemiological study for leprosy in this region.47 Additional 3225 Han Chinese (776 samples from Sichuan Province, 966 samples from Hunan Province and 1483 samples from Shanghai) reported in our recent studies were included in this study for comparison.28, 29 All healthy individuals had no history of leprosy, HIV infection and tuberculosis. These samples had been analyzed for other susceptibility genes to leprosy in our previous studies.14, 15, 16, 39
LRRK2 SNP selection and genotyping
Genomic DNA was extracted from whole blood by using the AxyPrep Blood Genomic DNA Miniprep Kit (AP-MN-BL-GDNA-250, Axygen, Union City, CA, USA). Thirteen SNPs of the LRRK2 gene were selected and analyzed in this study, following a similar strategy described in our previous studies.28, 29 Among them, 6 tag SNPs (rs732374, rs4473003, rs1427267, rs7298930, rs7307310, rs3761863) captured 57 SNPs (totally 67 SNPs) of the LRRK2 gene according to the international HapMap project database of CHB (http://hapmap.ncbi.nlm.nih.gov/, Phase 3, CHB). Six rare variants (MAF<0.05; rs34594498 (p.A419V), rs33939927 (p.R1441C), rs35801418 (p.Y1699C), rs34637584 (p.G2019S), rs35870237 (p.I2020T), rs34778348 (p.G2385R)) were chosen, because these pathogenic mutations led to aberrant LRRK2 function and were regarded as the genetic cause of PD.18 We included these reported (rare) pathogenic LRRK2 mutations in the analysis to discern whether these mutations may have a role in leprosy. Finally, the GWAS-hit SNP rs1873613 of leprosy6 (which is located at 66.4 Kb upstream of the LRRK2 gene) was also included in the analysis.
Two genotyping methods were employed in this study. SNPs rs1427267, rs3761863 and rs1873613 were detected by PCR restriction fragment length polymorphism. Briefly, PCR was performed in a 20-μl reaction volume with 50 ng of genomic DNA, 10 pmol of each specific primer (Supplementary Table S2), 0.2 mM of each dNTP, 2 mM of MgCl2 and 0.5 U Taq DNA polymerase (TaKaRa Biotechnology Co. Ltd, Dalian, China). PCR products were digested by specific restriction enzymes NmuCI, TaaI and SwaI (New England Biolabs, Beverly, MA, USA), respectively. Each SNP and genotypic profiles were determined by 2% agarose gel electrophoresis. The other 10 SNPs (rs732374, rs7307310, rs7298930, rs4473003, rs34594498, rs33939927, rs35801418, rs34637584, rs35870237 and 34778348) were detected using multiplex PCR and the SNaPshot technique (ABI PRISM SNaPshot Multiplex System, Foster City, CA, USA) as described in our recent study.28 All SNPs had a call rate of >98%. Further analysis of SNPs rs1427267, rs3761863 and rs1873613 that were initially genotyped by restriction fragment length polymorphism using SNaPshot confirmed the genotyping results. Direct sequencing of 2% of samples validated 100% correctness of genotyping.
Statistical analysis for LRRK2 genetic association
Deviation from HWE was assessed for each SNP by Chi-square tests. Cases and controls were compared according to the frequencies of genotypes and alleles. Linkage disequilibrium structure was determined by using Haploview.48 Haplotype construction and frequency were estimated by using the Phase software (Seattle, WA, USA).49 The global difference in haplotype frequency between cases and controls was estimated by Chi-square test. Potential association of SNP with leprosy (including leprosy subtype) was estimated using unconditional logistic regression model, with an adjustment for sex. All analyses were performed using SPSS 16.0 (SPSS Inc., Chicago, IL, USA). Power calculations were performed using the Quanto software (Los Angeles, CA, USA).50
Quantification of plasma LRRK2 level
We randomly chose 229 leprosy samples (including 112 MB patients and 117 PB patients) and 57 healthy controls to measure plasma LRRK2 level by using the Enzyme-Linked Immunosorbent Assay Kit (R&D systems, Minneapolis, MN, USA) according to the manufacturer’s instructions.
Functional assay for LRRK2 variant p.M2397T (rs3761863)
We performed cellular assay to characterize the potential effect of the LRRK2 variant p.M2397T (rs3761863). Human LRRK2 cDNA and a truncated mutant, LRRK2-delWD40 (ΔWD40, deletion of residues 2010–2527 of LRRK2, which represents the WD40 domain), were amplified by two sets of primers (Supplementary Table S3) to introduce restriction endonuclease sites for XhoI and NotI, and PCR products were cloned into pCMV-c-Myc vector (Clontech, Mountain View, CA, USA), respectively. We generated two mutants (p.G2019S and p.M2397T) based on the WT LRRK2 by using site-directed mutagenesis PCR methods. All mutants were verified by sequencing.
To generate a stable and inducible cell line expressing WT or mutant LRRK2 upon doxycycline (Dox, Sigma, St Louis, MO, USA) induction, WT or mutant LRRK2 was subcloned into pLVX-tight-puro (Clontech) by using two sets of primers to introduce restriction endonuclease sites (NotI and MluI) and the c-Myc tag (Supplementary Table S3). HEK293 cells were transfected with LRRK2 vector, packaging plasmid psPAX2 (Addgene, Cambridge, MA, USA) and envelop plasmid PMD2.G (Addgene) with a ratio of 3:2:1, respectively. The lentivirus was collected from cell supernatant at 48 h after transfection. Human U251 cells grown in a 12-well plate were sequentially infected with 500 μl lentivirus supernatant in the presence of 1 μg ml−1 Polybrene (Sigma) and were selected by puromycin (1 μg ml−1) and G418 (500 μg ml−1) for 2 weeks.
HEK293 cells were plated in 24-well plates at a density of 1 × 104 cells, cultured overnight and then were transfected with 0.2 μg of pNFAT-TA-Luc (Clontech) reporter vector and 0.3 μg of expression constructs (LRRK2-cMyc and its mutants) by using Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA). As an internal control, cells were also transfected with 0.02 μg Renilla luciferase construct. The transfected cells were left untreated or treated with LPS (1 μg ml−1, Sigma) for 12 h. Cells were lysed and subjected to a luciferase reporter assay (Promega, Madison, WI, USA) following the manufacturer’s instructions. U251 cells with stable expression of WT and mutant LRRK2 were treated with or without MPTP (500 μM) for 24 h, and cell lysates were prepared using protein lysis buffer (Beyotime, Shanghai, China). Western blots for target protein were performed using the standard method. In brief, a total of 30 μg protein was separated by 10–15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and was transferred to a polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA, USA). The membrane was blocked with 5% (w/v) skim milk for 2 h at room temperature. The membrane was incubated with primary antibodies against c-Myc (Sigma, 1:1000), LC3B (Cell Signaling Technology, Danvers, MA, USA, 1:1000) and β-actin (EnoGene Biotech Co. Ltd, New York, NY, USA, 1:10000) overnight at 4 °C, and after three washes with TBST buffer, the membrane was incubated with a peroxidase-conjugated anti-rabbit immunoglobulin G (KPL, Milford, MA, USA, 1:5000) for 1 h at room temperature. The epitope was visualized using an ECL Western Blot Detection Kit (Millipore, Billerica, MA, USA).
References
Britton WJ, Lockwood DN . Leprosy. Lancet 2004; 363: 1209–1219.
WHO. Leprosy update 2011 Wkly Epidemiol Rec 2011; 86: 389–399.
Scollard DM, Adams LB, Gillis TP, Krahenbuhl JL, Truman RW, Williams DL . The continuing challenges of leprosy. Clin Microbiol Rev 2006; 19: 338–381.
Worobec SM . Treatment of leprosy/Hansen's disease in the early 21st century. Dermatol Ther 2009; 22: 518–537.
Johnson CM, Lyle EA, Omueti KO, Stepensky VA, Yegin O, Alpsoy E et al. Cutting edge: a common polymorphism impairs cell surface trafficking and functional responses of TLR1 but protects against leprosy. J Immunol 2007; 178: 7520–7524.
Zhang FR, Huang W, Chen SM, Sun LD, Liu H, Li Y et al. Genomewide association study of leprosy. N Engl J Med 2009; 361: 2609–2618.
Misch EA, Berrington WR, Vary JC Jr ., Hawn TR . Leprosy and the human genome. Microbiol Mol Biol Rev 2010; 74: 589–620.
Yang D, Song H, Xu W, Long H, Shi C, Jing Z et al. Interleukin 4-590T/C polymorphism and susceptibility to leprosy. Genet Test Mol Biomarkers 2011; 15: 877–881.
Sousa AL, Fava VM, Sampaio LH, Martelli CM, Costa MB, Mira MT et al. Genetic and immunological evidence implicates interleukin 6 as a susceptibility gene for leprosy type 2 reaction. J Infect Dis 2012; 205: 1417–1424.
Cardoso CC, Pereira AC, Brito-de-Souza VN, Duraes SM, Ribeiro-Alves M, Nery JA et al. TNF -308G>A single nucleotide polymorphism is associated with leprosy among Brazilians: a genetic epidemiology assessment, meta-analysis, and functional study. J Infect Dis 2011; 204 :: 1256–63.
Sapkota BR, Macdonald M, Berrington WR, Misch EA, Ranjit C, Siddiqui MR et al. Association of TNF, MBL, and VDR polymorphisms with leprosy phenotypes. Hum Immunol 2010; 71: 992–998.
Alter A, de Léséleuc L, Van Thuc N, Thai VH, Huong NT, Ba NN et al. Genetic and functional analysis of common MRC1 exon 7 polymorphisms in leprosy susceptibility. Hum Genet 2010; 127: 337–348.
Cardoso CC, Pereira AC, Brito-de-Souza VN, Dias-Baptista IM, Maniero VC, Venturini J et al. IFNG +874 T>A single nucleotide polymorphism is associated with leprosy among Brazilians. Hum Genet 2010; 128: 481–490.
Wang D, Feng J-Q, Li Y-Y, Zhang D-F, Li X-A, Li QW et al. Genetic variants of the MRC1 gene and the IFNG gene are associated with leprosy in Han Chinese from Southwest China. Hum Genet 2012; 131: 1251–1260.
Zhang D-F, Huang X-Q, Wang D, Li Y-Y, Yao Y-G . Genetic variants of complement genes ficolin-2, mannose-binding lectin and complement factor H are associated with leprosy in Han Chinese from Southwest China. Hum Genet 2013; 132: 629–640.
Zhang D-F, Wang D, Li Y-Y, Yao Y-G . Mapping genetic variants in the CFH gene for association with leprosy in Han Chinese. Genes Immun 2014; 15: 506–510.
Gardet A, Benita Y, Li C, Sands BE, Ballester I, Stevens C et al. LRRK2 is involved in the IFN-gamma response and host response to pathogens. J Immunol 2010; 185: 5577–5585.
Mata IF, Wedemeyer WJ, Farrer MJ, Taylor JP, Gallo KA . LRRK2 in Parkinson's disease: protein domains and functional insights. Trends Neurosci 2006; 29: 286–293.
Paisán-Ruíz C, Jain S, Evans EW, Gilks WP, Simón J, van der Brug M et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron 2004; 44: 595–600.
Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 2004; 44: 601–607.
Recchia A, Debetto P, Negro A, Guidolin D, Skaper SD, Giusti P . Alpha-synuclein and Parkinson's disease. FASEB J 2004; 18: 617–626.
Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet 2008; 40: 955–962.
Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G et al. Patterns of somatic mutation in human cancer genomes. Nature 2007; 446: 153–158.
Tong Y, Giaime E, Yamaguchi H, Ichimura T, Liu Y, Si H et al. Loss of leucine-rich repeat kinase 2 causes age-dependent bi-phasic alterations of the autophagy pathway. Mol Neurodegener 2012; 7: 2.
Plowey ED, Cherra SJ 3rd, Liu YJ, Chu CT . Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J Neurochem 2008; 105: 1048–1056.
Liu Z, Lee J, Krummey S, Lu W, Cai H, Lenardo MJ . The kinase LRRK2 is a regulator of the transcription factor NFAT that modulates the severity of inflammatory bowel disease. Nat Immunol 2011; 12: 1063–1070.
Moehle MS, Webber PJ, Tse T, Sukar N, Standaert DG, DeSilva TM et al. LRRK2 inhibition attenuates microglial inflammatory responses. J Neurosci 2012; 32: 1602–1611.
Bi R, Zhao L, Zhang C, Lu W, Feng J-Q, Wang Y et al. No association of the LRRK2 genetic variants with Alzheimer's disease in Han Chinese individuals. Neurobiol Aging 2014; 35: 444. e5–e9.
Li X, Zhang W, Zhang C, Gong W, Tang J, Yi Z et al. No association between genetic variants of the LRRK2 gene and schizophrenia in Han Chinese. Neurosci Lett 2014; 566: 210–215.
Zhu JH, Horbinski C, Guo F, Watkins S, Uchiyama Y, Chu CT . Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am J Pathol 2007; 170: 75–86.
Bravo-San Pedro JM, Niso-Sántano M, Gómez-Sánchez R, Pizarro-Estrella E, Aiastui-Pujana A, Gorostidi A et al. The LRRK2 G2019S mutant exacerbates basal autophagy through activation of the MEK/ERK pathway. Cell Mol Life Sci 2013; 70: 121–136.
Marcinek P, Jha AN, Shinde V, Sundaramoorthy A, Rajkumar R, Suryadevara NC et al. LRRK2 and RIPK2 variants in the NOD 2-mediated signaling pathway are associated with susceptibility to Mycobacterium leprae in Indian populations. PLoS One 2013; 8: e73103.
Smith WW, Pei Z, Jiang H, Moore DJ, Liang Y, West AB et al. Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration. Proc Natl Acad Sci USA 2005; 102: 18676–18681.
Alegre-Abarrategui J, Christian H, Lufino MM, Mutihac R, Venda LL, Ansorge O et al. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum Mol Genet 2009; 18: 4022–4034.
Saha S, Guillily MD, Ferree A, Lanceta J, Chan D, Ghosh J et al. LRRK2 modulates vulnerability to mitochondrial dysfunction in Caenorhabditis elegans. J Neurosci 2009; 29: 9210–9218.
Lewis PA, Manzoni C . LRRK2 and human disease: a complicated question or a question of complexes? Sci Signal 2012; 5: pe2.
Watson SR, Morrison NE, Collins FM . Delayed hypersensitivity responses in mice and guinea pigs to Mycobacterium leprae Mycobacterium vaccae, and Mycobacterium nonchromogenicum cytoplasmic proteins. Infect Immun 1979; 25: 229–236.
Wang HY, Liu JH, Ye SZ, Yu LC, Shi MQ, Sang HG . Preliminary observations on experimental leprosy in tupaias (Tupaia belangeri yunalis. Leprosy Rev 1990; 61: 12–18.
Wang D, Su L-Y, Zhang A-M, Li YY, Li XA, Chen LL et al. Mitochondrial DNA copy number, but not haplogroup, confers a genetic susceptibility to leprosy in Han Chinese from Southwest China. PLoS One 2012; 7: e38848.
Orenstein SJ, Kuo SH, Tasset I, Arias E, Koga H, Fernandez-Carasa I et al. Interplay of LRRK2 with chaperone-mediated autophagy. Nat Neurosci 2013; 16: 394–406.
Manzoni C, Mamais A, Dihanich S, Abeti R, Soutar MP, Plun-Favreau H et al. Inhibition of LRRK2 kinase activity stimulates macroautophagy. Biochim Biophys Acta 2013; 1833: 2900–2910.
Zhu Y, Wang C, Yu M, Cui J, Liu L, Xu Z . ULK1 and JNK are involved in mitophagy incurred by LRRK2 G2019S expression. Protein Cell 2013; 4: 711–721.
Bandopadhyay R, de Belleroche J . Pathogenesis of Parkinson's disease: emerging role of molecular chaperones. Trends Mol Med 2010; 16: 27–36.
Wong AS, Lee RH, Cheung AY, Yeung PK, Chung SK, Cheung ZH et al. Cdk5-mediated phosphorylation of endophilin B1 is required for induced autophagy in models of Parkinson's disease. Nat Cell Biol 2011; 13: 568–579.
Jorgensen ND, Peng Y, Ho CC, Rideout HJ, Petrey D, Liu P et al. The WD40 domain is required for LRRK2 neurotoxicity. PLoS One 2009; 4: e8463.
Sheng D, Qu D, Kwok KH, Ng SS, Lim AY, Aw SS et al. Deletion of the WD40 domain of LRRK2 in Zebrafish causes Parkinsonism-like loss of neurons and locomotive defect. PLoS Genet 2010; 6: e1000914.
Li Y-Y, Li X-A, He L, Wang D, Chen WY, Chen L et al. Trends in new leprosy case detection over 57 years (1952-2008) in Yuxi, Yunnan Province of Southwest China. Leprosy Rev 2011; 82: 6–16.
Barrett JC, Fry B, Maller J, Daly MJ . Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21: 263–265.
Stephens M, Smith NJ, Donnelly P . A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 2001; 68: 978–989.
Gauderman WJ . Sample size requirements for matched case-control studies of gene-environment interaction. Stat Med 2002; 21: 35–50.
Acknowledgements
We are grateful to all the participants in this study. We thank Mr Jia-Qi Feng and Dr Yue-Mei Feng for technical assistance. This study was supported by the National Natural Science Foundation of China (31271346 and 30925021) and the Ministry of Science and Technology of China (2011CB910902). The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict interest.
Additional information
Supplementary Information accompanies this paper on Genes and Immunity website
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, D., Xu, L., Lv, L. et al. Association of the LRRK2 genetic polymorphisms with leprosy in Han Chinese from Southwest China. Genes Immun 16, 112–119 (2015). https://doi.org/10.1038/gene.2014.72
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2014.72
- Springer Nature Limited
This article is cited by
-
LRRK2 Biology from structure to dysfunction: research progresses, but the themes remain the same
Molecular Neurodegeneration (2019)
-
LRRK2 is involved in the pathogenesis of system lupus erythematosus through promoting pathogenic antibody production
Journal of Translational Medicine (2019)
-
The LRRK2 signalling system
Cell and Tissue Research (2018)
-
Human genetics of mycobacterial disease
Mammalian Genome (2018)
-
Selective LRRK2 kinase inhibition reduces phosphorylation of endogenous Rab10 and Rab12 in human peripheral mononuclear blood cells
Scientific Reports (2017)