Abstract
Leprosy is a chronic infectious disease caused by Mycobacterium leprae, a low virulence mycobacterium, and the outcome of disease is dependent on the host genetics for either susceptibility per se or severity. The IFNG gene codes for interferon-γ (IFN-γ), a cytokine that plays a key role in host defense against intracellular pathogens. Indeed, single nucleotide polymorphisms (SNPs) in IFNG have been evaluated in several genetic epidemiological studies, and the SNP +874T>A, the +874T allele, more specifically, has been associated with protection against infectious diseases, especially tuberculosis. Here, we evaluated the association of the IFNG locus with leprosy enrolling 2,125 Brazilian subjects. First, we conducted a case–control study with subjects recruited from the state of São Paulo, using the +874 T>A (rs2430561), +2109 A>G (rs1861494) and rs2069727 SNPs. Then, a second study including 1,370 individuals from Rio de Janeiro was conducted. Results of the case–control studies have shown a protective effect for +874T carriers (ORadjusted = 0.75; p = 0.005 for both studies combined), which was corroborated when these studies were compared with literature data. No association was found between the SNP +874T>A and the quantitative Mitsuda response. Nevertheless, the spontaneous IFN-γ release by peripheral blood mononuclear cells was higher among +874T carriers. The results shown here along with a previously reported meta-analysis of tuberculosis studies indicate that the SNP +874T>A plays a role in resistance to mycobacterial diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Leprosy is a chronic infectious disease caused by Mycobacterium leprae, which mainly infects macrophages in skin and Schwann cells in peripheral nerves. The clinical manifestations are presented in a wide spectrum ranging from a localized to a disseminated form called tuberculoid (TT) and lepromatous (LL) leprosy, respectively. The intermediate forms are called borderline and classified into three groups according to their proximity to polar forms (Ridley and Jopling 1962). M. leprae is highly adapted to the intracellular environment and exhibits very low genetic diversity indicating a single-clone origin (Monot et al. 2009) and suggesting that variability of clinical presentations are, at least in part, explained by host immune responses. Also, genetics of the human host is responsible for the susceptibility to leprosy per se observed among only a small percentage of the exposed individuals (1–3%). The genetic component of susceptibility to leprosy per se and clinical types was first demonstrated using twin studies (Chakravartti and Vogel 1973), familial aggregation (Shields et al. 1987) and segregation analyses (Abel and Demenais 1988). Recently, several case–control and family-based association analyses have reported the role of genes coding for proteins of the immune system, independently confirming their findings in distinct populations (Moraes et al. 2006). Among these genes are TNF (Santos et al. 2002; Shaw et al. 2001), IL10 (Malhotra et al. 2005; Moraes et al. 2004; Pereira et al. 2009), LTA (Alcais et al. 2007), HLADRB1 (Vanderborght et al. 2007) and MRC1 (Alter et al. 2010). Other studies have explored leprosy reactions as phenotypes, which is much more complex, but indicate that toll-like receptors (TLR) and ninjurin are associated with reversal reaction and nerve injury, respectively (Cardoso et al. 2007; Misch et al. 2008). Moreover, a genome-wide scan performed in a Chinese population enrolling more than 5,000 cases and controls confirmed the participation of 6p21 genes (e.g., HLADRB1) and depicted novel genes (e.g., NOD2, LRKK2) that regulates an entire pathway of basic cellular functions associated with immune responses (Zhang et al. 2009).
Interferon-gamma (IFN-γ) is a cytokine secreted mainly by CD4+ Th1 cells, CD8+ T cells and NK cells whenever these cells are stimulated by IL-12, among other stimuli. IFN-γ, in turn, induces IL-12 production in phagocytes and inhibits IL-4 secretion by Th2 populations, which may further drive Th1 differentiation in vivo (Schroder et al. 2004). This axis of activation plays a critical role in killing intracellular pathogens such as Mycobacterium, Leishmania, Salmonella and Chlamydia species and is considered a key modulator of innate and adaptive immune responses (Agnello and Gadina 2006), since it promotes macrophage microbicidal activity based on oxidative metabolism (Nathan et al. 1983). In leprosy, IFN-γ enables macrophages to effectively inhibit or kill M. leprae (Hagge et al. 2004; Ramasesh et al. 1991). In vitro studies have shown that T lymphocytes obtained from TT patients secrete mainly IFN-γ, while the LL patients release IL-4 preferentially (Sieling et al. 1994). Taken together, these data suggest that IFNG is an important candidate gene for susceptibility to leprosy per se.
The IFNG gene is located in the 12q14 region, and despite the high conservation of its coding region, some polymorphisms have been detected along the IFNG sequence, especially in introns 1 and 3, and in the 3′-UTR region (Vandenbroeck and Goris 2006). The single nucleotide polymorphism (SNP) +874 T>A (rs2430561) has been a target in association studies of diverse infectious diseases, especially those caused by intracellular pathogens. The +874T allele was associated with higher IFN-γ production in blood cells stimulated with purified protein derivative (PPD) from M. tuberculosis (López-Maderuelo et al. 2003) and was also defined as a key allele in TB resistance after a meta-analysis study (Pacheco et al. 2008). Nevertheless, previous studies did not detect association between this SNP and leprosy (Fitness et al. 2004; Franceschi et al. 2009).
Herein, we conducted a population-based study using two Brazilian case–control panels to investigate the role of genetic polymorphisms along the sequence of IFNG in leprosy susceptibility. Further analyses were also performed to assess the effect of these genetic variations in the in vivo intradermic response to M. leprae antigens (lepromin) and in vitro IFN-γ release.
Subjects and methods
Subjects
A total of 2,125 individuals, 1,370 from the metropolitan area of Rio de Janeiro (RJ) and 755 from Bauru (a city in the countryside of the state of São Paulo) were included in this study. Of these, 1,045 were cases (670 from RJ and 375 from Bauru) and 1,080 were controls (700 from RJ and 380 from Bauru). Diagnosis was determined by experienced professionals from the Souza Araújo Clinic at Fiocruz and Lauro de Souza Lima Institute (Bauru). Patients were classified according to the Ridley and Jopling classification system (Ridley and Jopling 1962), and for treatment purposes were also classified as pauci- or multibacillary. All patients were treated as specified by the World Health Organization (WHO). Inclusion criteria for cases recruited were every patient diagnosed with leprosy in Souza Araújo (Fiocruz) and Lauro de Souza Lima Institute (Bauru) who agreed to participate in the study and signed the written informed consent. Patients co-infected with HIV and Mycobacterium tuberculosis were excluded from the study, as well as the carriers of any immunodeficiency. No age restrictions were applied. The healthy control groups were composed of unrelated individuals who lived in the same endemic area as the patients.
In the RJ population, a lepromin challenge was also performed at the time of diagnosis (active disease) and analyzed 28 days after inoculation as described (Moraes et al. 2001). Only borderline tuberculoid patients were included in the analyses of Mitsuda response. All individuals were phenotypically classified as Afro- or Euro descendants after careful inspection of characteristics such as facial morphological features, hair type and skin color of both the individual as well as his/her family. General characteristics of patients and controls recruited in this study are summarized in the Online Resource (Supplementary Table 1).
For the functional analyses, 28 healthy individuals were selected in accordance with their genotype for the +874 T/A SNP. More specifically, a total of 9 volunteers presented the AA genotype, of whom 6 were women and 3 were men (mean age: 40.1 years); 19 carried the +874T allele (TA or TT genotypes), of which 15 were women and 4 were men (mean age: 43.8 years). Written informed consent was obtained from all individuals included in the study as required by the local ethics committees (CEP-FIOCRUZ and Lauro de Souza Lima Institute, Bauru).
Study design
During the course of this work, two independent case–control studies were conducted. First, we compared the frequencies of the IFNG SNPs +874 T>A (rs2430561), +2109 A>G (rs1861494) and rs2069727 between cases and controls recruited from Bauru. The analyses were performed for all the subjects and then among individuals of four subgroups defined according to sex and ethnicity. Haplotype analyses were conducted following this same method. Then, a second study was conducted using a sample selected from RJ and to assess the reproducibility of the results. Considering a minor allele frequency (MAF) of 0.4, the minimal OR values to reach a power of 80% were about 1.65 for Bauru and 1.55 for the RJ population. When MAFs of 0.3 and 0.2 were applied, these values were increased to 1.72 and 1.66 for Bauru and 1.6 and 1.55 for the RJ population, respectively. The two studies were also combined in an overall analysis, in which this power could be reached with OR values of about 1.36 (MAF = 0.4), 1.42 (MAF = 0.3) and 1.37 (MAF = 0.2). Finally, functional analyses were performed in the RJ study assessing the effect of the SNPs in the Mitsuda response of 230 borderline tuberculoid (BT) patients, and a group of individuals from Bauru was used to determine the effect of the SNPs on IFN-γ production.
DNA extraction and genotyping
DNA was extracted from frozen blood samples using a salting-out precipitation method. The SNP +874 T>A was genotyped by PCR-ARMS as described (Pravica et al. 2000). The polymorphism +2109 A>G created a restriction site for the enzyme Aci I and was genotyped by a previously reported PCR–RFLP method (Henri et al. 2002). Briefly, PCR reactions were carried out with 50 ng of genomic DNA, 0.3 μM of each specific primer, 1 U of Taq DNA polymerase (Invitrogen, Brazil), 0.2 mM of each dNTP (Invitrogen, CA, USA) and 1.5 mM of MgCl2 in a final reaction volume of 25 μL. Genotyping of the SNP at +874 included a second primer pair (0.15 μM, each) used as an internal control for amplification. The SNP rs2069727 was genotyped using a real-time SNP genotyping assay (Assay ID: C__2683475_10) according to the manufacturer’s instructions (Applied Biosystems, CA, USA). Conventional and real-time PCR reactions were performed using the MJ Research PTC100 (Hercules, CA, USA) and ABI Prism 7000 (Applied Biosystems, CA, USA) thermal cyclers, respectively. PCR reactions were repeated at least once for all samples in which results were unclear. After that, about 5–10% of the samples remained to be genotyped.
PBMC culture and IFN-γ measurement
Mononuclear cells were separated from 20 mL of venous blood by Ficoll-Hypaque (Sigma, St Louis, MO, USA). The total number of mononuclear cells was determined in a Neubauer chamber and final concentration was adjusted to 2 × 106 mononuclear cells/mL using RPMI-1640 medium containing l-glutamine and 25 mM HEPES buffer (Gibco, Grand Island, NY, USA) supplemented with 10% fetal calf serum (Gibco), penicillin (100 UI/mL) and streptomycin (100 μg/mL) (Gibco). The cell suspension (100 μL/well) was distributed in 96-well, flat-bottom tissue culture plates (Corning, New York, NJ, USA) in the presence of 8 μg/mL phytohemaglutinin (PHA, Gibco), 10 μg/mL of E. coli lipopolysaccharide (LPS, Sigma), 10 μg/mL of M. leprae sonicated antigen (AgSon) and 4 μg/mL of whole M. leprae antigen (8 bacilli:1 mononuclear cell). The antigens were kindly provided by Dr. Patrick Brennan, Department of Microbiology, Immunology and Pathology, Colorado State University, USA. Control cultures were not stimulated. Plates were incubated at 37°C in an atmosphere with 5% CO2 for 5 days and the supernatants were collected and stored at −80°C until the cytokine measurement. The total concentration of IFN-γ in the mononuclear cell culture supernatant was measured by ELISA using IFN-γ Duo-Set (R&D Systems, Minneapolis, USA).
Statistical analyses
Case–control
All statistical analyses were carried out as previously described (Pereira et al. 2009). Briefly, deviations from Hardy–Weinberg equilibrium (HWE) were accessed by Chi-square tests. Cases and controls were compared according to the frequencies of genotypes, alleles and minor allele carriers. Odds ratios (ORs) were obtained through unconditional logistic regression models, controlling for potential confounders such as sex, ethnicity and study place in the combined analysis. A Cochran–Armitage trend test was applied to describe possible allele-dose effects. Haplotype frequencies were estimated by maximum likelihood and compared with the same logistic regression models conducted for the isolated SNPs. Linkage disequilibrium was assessed by the r 2 statistics using frequencies obtained from the control group. All analyses were performed using the R for windows (R Development Core Team 2008), version 2.10.1, with the packages “genetics”, “haplo.stats” and “coin”.
Functional analyses
Mitsuda response was evaluated as a continuous outcome using the Mann–Whitney test to compare the means of the induration (mm) in +874T carriers and non-carriers as a whole or in the subgroups defined according to the presence or absence of a BCG scar and reversal reaction. A Welch Two Sample t test was applied for differences of means of IFN-γ production among carriers and non-carriers of the +874T allele for each stimulus. Results were presented as stripcharts with mean bars. p values <0.05 were considered to be statistically significant. All analyses were performed using R for Windows 2.10.1 and the package “MASS”.
Results
For the case–control study with the sample from Bauru, total counts and frequencies of the three different genotypes, alleles and carriers of the minor allele are shown for each SNP in Table 1. Frequencies of all SNPs were in HWE in both control and patient groups. Comparisons of cases and controls according to the frequencies of the SNPs +874 T>A, +2109 A>G, rs2069727 and leprosy showed protection, after adjustment for sex and ethnicity, with borderline statistical significance (OR = 0.72; p = 0.04 for +874T carriers and OR = 0.69; p = 0.03 for rs2069727C carriers; Table 1).
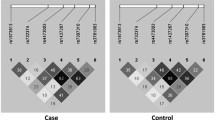
Pairwise linkage disequilibrium (LD) analyses performed using all individuals from the control group showed an r 2 value of 0.9 for the SNPs +874 T>A and rs2069727, indicating that these polymorphisms belong to the same LD bin, while the low r 2 values (r 2 < 0.2) obtained for comparisons with the +2109 A>G polymorphism indicated that this latter SNP was included in another bin. Thus, to exclude redundancy, the +874T>A SNP was selected for haplotype analysis, along with +2109 A>G. Frequencies of each haplotype in cases and controls were also compared by using logistic regression models and a borderline p value was found after adjustment (Table 1).
The results shown so far indicated that the SNP at +874 T>A could be used as a tag for the bin and as a marker for the genetic association of IFNG found in the Bauru population. Therefore, only this SNP was genotyped in the replication study. Data from the study with the sample recruited from Rio de Janeiro are shown in Table 2. As observed in this table, the OR values confirmed that the genotypes carrying the +874T allele presented protection against leprosy (adjusted OR = 0.74; p = 0.03). Furthermore, the OR values showed a more prominent protective effect for the TT genotype and, this allele-dose effect was confirmed by a Cochran–Armitage trend test (p = 0.003). Data obtained from the overall analysis including subjects from both populations increased the power of the study and the p value indicated more accurately that OR values indeed showed a clear protective effect (adjusted OR = 0.75; p = 0.005) in comparisons between +874T carriers and non-carriers (Table 2). The allele-dose effect was also observed in this analysis (p = 0.003).
Results of our logistic regression models showed a statistically significant interaction between the effect of the SNP +874T>A and the covariate ethnicity (p = 0.02 for genotype comparisons in both populations combined). Indeed, the ethnicity-stratified models conducted in the Bauru population suggested a specificity in the effect of the SNPs +874 T>A and rs2069727, as both were prominently associated with protection against leprosy among Afro-descendants (Table 3 and Online Resource, Supplementary Table 2). More specifically, carriers of the +874T and rs2069727C alleles were less likely to develop leprosy (OR = 0.40; p = 0.004 and OR = 0.37; p = 0.006, respectively) in the Afro-descendants group (Online Resource, Supplementary Table 2). Similarly, the haplotype carrying the +874T allele was also associated with protection (adjusted OR = 0.52; p = 0.03) only among Afro-descendants (Online Resource, Supplementary Table 2). The same alleles were not associated with leprosy in the Euro-descendants (Online Resource, Supplementary Table 2). The same pattern was observed in the replication study and in the overall analysis of both studies, as the adjusted OR values indicated protection for the T carriers only among Afro-descendants (OR = 0.59; p = 0.01 in RJ and OR = 0.53; p = 0.0002 for both studies combined). Results of the ethnicity-stratified models are shown in detail in Table 3 and in the Online Resource (Supplementary Tables 2 and 3). No association was found when statistical analyses were stratified by sex (data not shown).
Comparisons of the two patients’ subgroups defined according to the WHO classification did not show any evidence for association between IFNG SNPs and the development of pauci- or multibacillary disease (data not shown).
Finally, we evaluated the functional status of this marker. First, no differences were found when the means of the quantitative Mitsuda response of +874T carriers and non-carriers (Mean 4.47 vs. 5.14; p = 0.43) were compared (Online Resource, Supplementary Fig. 1) even after stratification of the patients according to the BCG status and the presence of reversal reaction (data not shown). Then, we tested the effect of the +874T>A SNP in the IFN-γ release (Fig. 1). In the absence of any stimulus, results of ELISA measurements showed higher IFN-γ levels in the culture supernatants of +874T carriers (+874 TA and TT genotypes) when compared with non-carriers (Mean 0.197 vs. 0.048; p = 0.02). Nevertheless, when the PBMC cultures were stimulated with either LPS, PHA or M. leprae antigens, the IFN-γ levels were not different, regardless of the genotype of the donor (p > 0.05 for comparisons between +874T carriers and non-carriers).
Interferon-γ (IFN-γ) production by peripheral blood mononuclear cells (PBMC) according to the +874T>A genotypes. PBMC were cultured for 5 days with whole M. leprae (8 bacilli per cell), sonicated M. leprae antigen (10 μg/mL), lipopolysaccharide from E. coli (LPS, 10 μg/mL), phytohemagglutinin-M (PHA, 8 μg/mL) or without stimulus. Results were represented for each sample as the IFN-γ concentration in pg/mL. The lines represent the mean value of each group. N = 19 for +874T carriers (+874TA and TT) and 9 for non-carriers (+874AA). IFN-γ levels were significantly higher among +874T carriers when cells were cultivated in the absence of any stimulus (p = 0.02). IFNg = IFN-γ
Discussion
Given the pivotal role of IFNG in the development of cellular immune response, several genetic association studies have been conducted to define a marker associated with susceptibility to intracellular pathogen diseases. To date, the +874T allele has been shown to play a role in infectious diseases such as tuberculosis (Pacheco et al. 2008), while for others, such as leishmaniasis (Matos et al. 2007; Salih et al. 2007) and hepatitis (Liu et al. 2006), results are still controversial. Here, two independent case–control studies were conducted to define the role of IFNG SNPs in leprosy susceptibility along with functional studies.
Results obtained from the Bauru population suggested that the +874T and rs2069727C alleles were associated with protection against leprosy. Moreover, linkage disequilibrium analysis showed that these polymorphisms were in almost perfect LD and, therefore, belonged to the same bin (r 2 > 0.8 cutoff defined by the HapMap Project (Carlson et al. 2004), suggesting that the association observed for both SNPs is genetically redundant. Accordingly, a detailed analysis of HapMap and literature had already reported a strong correlation between these SNPs in ethnically distinct populations (Vandenbroeck and Goris 2006). Thus, we used only the +874T>A and +2109A>G SNPs for haplotype analyses and found borderline p values for haplotypes carrying the +874T allele, reinforcing that the SNP at +2109 did not bring any extra information and was not associated with leprosy.
Moreover, it seems that the +874T>A SNP could capture the information of this locus; we selected this marker as a tag for further analyses in a replication study developed in an independent population to validate our genetic epidemiological findings (Alter et al. 2008; Pacheco and Moraes 2009). This study included a total of 1,370 individuals recruited in Rio de Janeiro and was also based on a case–control design. The +874T allele was also associated with protection against leprosy in this study. Then, we combined both populations and adjusted the analysis using the origin of the population as an additional covariate. Results of this overall analysis reinforced our data, as the +874T allele was significantly associated with protection against leprosy in all comparisons performed.
Stratified analyses were also performed to assess a possible differential effect of the SNPs on the subgroups defined according to non-genetic covariates. Results of the ethnicity-stratified analyses suggested a specific effect, as the +874T allele conferred protection against leprosy only among Afro-descendants. Moreover, the OR values reflected that this association was stronger in this subgroup, suggesting that the effect was probably more specific in this population. This pattern was observed in Bauru, RJ and also in the analysis of both populations combined. Indeed, a lower frequency of the +874T allele among individuals classified as Afro-descendants was detected in this study, replicating previous findings reported for distinct populations (Delaney et al. 2004; Girnita et al. 2006; Govan et al. 2003; Larcombe et al. 2008). Nevertheless, given the clear population admixture observed in Brazil, it is very difficult to define ethnicity according to phenotypic features (Parra et al. 2003) and, therefore, further analyses using population-specific ancestry markers are still necessary to better characterize the structure of the populations included in this study.
The Mitsuda response measures the granulomatous response to intradermally injected heat-killed leprosy bacilli (lepromin) and reflects the ability to develop an immune-inflammatory response against M. leprae antigens. Results of family-based studies have linked the quantitative Mitsuda response to polymorphisms in the NRAMP1 gene (Alcais et al. 2000), while population studies have reported a role for TNF SNPs (Moraes et al. 2001). Here, no differences were found when the quantitative Mitsuda response was compared in +874T carriers and non-carriers even after the stratification of the patients according to the covariates, BCG vaccination and reversal reaction. These results suggest that, although associated with leprosy development, the SNP +874T>A is not associated with the strength of the inflammation in Mitsuda reactions.
We also evaluated the possibility of conducting a meta-analysis and, after a careful literature review, found only two association studies, which had failed to detect an association between this SNP and leprosy susceptibility (Fitness et al. 2004; Franceschi et al. 2009). From these, the Malawian study (Fitness et al. 2004) had to be excluded due to a deviation from the HWE in the control group (Pacheco et al. 2008), which could probably be attributed to an inconsistency in the genotyping method (Pacheco and Moraes 2009). Since there was only one additional study to analyze along with our new data, we could not perform a meta-analysis study in this paper. Given the small sample size (N = 404) used by Franceschi et al. (2009), the lack of association observed could be attributed to low statistical power to detect small differences. However, results of a random effects model combining our data and the study by Franceschi et al. (2009) corroborated the trend observed with our data (data not shown).
The hypothesis to explain an association between the +874T allele and protection against intracellular pathogens suggests that individuals carrying this allele would produce higher IFN-γ levels that could, in turn, mount up an efficient and protective immune response. This hypothesis suggests that certain stimuli, such as purified protein derivative (PPD) from M. tuberculosis or leishmania antigens, induce increased levels of IFN-γ among carriers of the +874T allele (López-Maderuelo et al. 2003; Matos et al. 2007). Nevertheless, in a study conducted in India in which PBMCs were stimulated with PHA, culture filtrates or live M. tuberculosis, no differences were observed in +874T allele versus +874AA allele carriers (Vidyarani et al. 2006). Results obtained from ELISPOT detection of IFN-γ in PBMC cultures from Turkish tuberculosis patients demonstrated that PPD, but not CFP10 and ESAT6 (two recombinant M. tuberculosis proteins), showed an association of the +874TT genotype with higher production (Sallakci et al. 2007). The results concerning mycobacterial antigens indicate that for some stimuli, such as PPD, for example, there is a significant difference in the production of IFN-γ, although for other stimuli, such as recombinant proteins or live M. tuberculosis, no differences are observed. Also, the method chosen for cytokine detection and the cell type used in the assay can also yield differences in the results obtained by different researchers.
Results of our cell cultures were similar to previously reported data, although some differences could be observed. The unstimulated group had a mean value of 51.6 pg/mL, which was similar to the 40.5 pg/mL produced by healthy controls in the study by Cozen et al. (Cozen et al. 2008). The presence of the sonicated M. leprae stimulus increased the IFN-γ levels to 246.48 pg/mL and, in this case, the literature shows a mean value of about 350 pg/mL using the same concentration of M. leprae for 96 h (Lopez Roa et al. 2008). As expected, the PHA stimulus (8 μg/mL) leads to the highest IFN releases (2,062 pg/mL), which can also be compared to the 1,000 pg/mL produced in the presence of a lower concentration (5 μg/mL) of the same stimulus (Castellano et al. 2009). Curiously, our results pointed to a functional effect in which +874T carriers were also high IFN-γ producers, but only in the absence of any stimuli. This result confirmed the functionality of this SNP among Brazilians, as we previously observed for Leishmania antigens (Matos et al. 2007), although the specific enhancement of IFNG +874T allele in the presence of M. leprae antigens was not observed. We were unable to test PPD, recombinant M. leprae proteins, other time points or other IFN-γ quantitation methods (e.g., ELISPOT, flow cytometry), which could help understand the genotype–phenotype correlation in this locus. Further analyses are being conducted to define the functionality of this SNP in response to specific M. leprae antigens. Another explanation can be obtained from results using mathematical models that suggest functional effects of different polymorphisms can neutralize IFNG +874T, leading to a wider range of phenotypes. In the particular case of tuberculosis, combinations between IFNG and HLA SNPs were shown to have distinct effects on host susceptibility. These data could explain the discrepancies, which are often observed between results from populations with distinct genetic backgrounds (Chang et al. 2008).
IFN-γ is the final product of the activation of IL-12/IL-23 axis of the immune host response, which culminates in macrophage killing of intracellular pathogens such as mycobacteria. The syndrome called “Mendelian susceptibility to mycobacterial diseases” (MSMD) reinforces the key role for the type 1 cytokine cascade, since this disorder has been consistently associated with loss of function mutations in genes coding for IL-12p40, STAT1, the β1 chain of the IL-12/IL-23 receptor and the IFN-γ receptor chains (van de Vosse et al. 2009). In all cases, the overall effect observed is a complete inability of the individual to release or respond to IFN-γ, leading to lack of control over infection even when the pathogen is poorly invasive. These data raise the question of the extent to which the immune response of an individual would be affected by mutations with less drastic effects on gene function. It is possible that common polymorphisms in these genes would lead to small effects in the type 1 response functionality and, therefore, be associated with susceptibility/resistance to common diseases. Recently, novel pathways associated with leprosy susceptibility were revealed (Zhang et al. 2009), and IFNG was suggested as a key gene in triggering protective immune responses. Thus, it is likely that IFN-γ production is regulated by SNPs such as +874T>A, contributing to the outcome of the disease, although the exact mechanism that controls the genotype–phenotype relationship through interactions with other polymorphisms is not clearly defined.
Taken together, the data generated in this study and our previously reported meta-analysis of tuberculosis data (Pacheco et al. 2008) have consistently demonstrated that IFNG polymorphisms, such as +874T>A and other SNPs arranged in the same bin, participate in the resistance to both M. leprae (this paper) and M. tuberculosis infection (Pacheco et al. 2008).
References
Abel L, Demenais F (1988) Detection of major genes for susceptibility to leprosy and its subtypes in a Caribbean island: Desirade Island. Am J Hum Genet 42(2):256–266
Agnello D, Gadina M (2006) The biology of cytokines: general principles, properties, and lessons from animal models. In: Vandenbroeck K (ed) Cytokine gene polymorphisms in multifactorial conditions, vol 1, 1st edn. CRC Press Taylor & Francis Group, Boca Raton, pp 3–33
Alcais A, Sanchez FO, Thuc NV, Lap VD, Oberti J, Lagrange PH, Schurr E, Abel L (2000) Granulomatous reaction to intradermal injection of lepromin (Mitsuda reaction) is linked to the human NRAMP1 gene in Vietnamese leprosy sibships. J Infect Dis 181:302–308
Alcais A, Alter A, Antoni G, Orlova M, Nguyen VT, Singh M, Vanderborght PR, Katoch K, Mira MT, Vu HT, Ngyuen TH, Nguyen NB, Moraes M, Mehra N, Schurr E, Abel L (2007) Stepwise replication identifies a low-producing lymphotoxin-alpha allele as a major risk factor for early-onset leprosy. Nat Genet 39:517–522
Alter A, Alcais A, Abel L, Schurr E (2008) Leprosy as a genetic model for susceptibility to common infectious diseases. Hum Genet 123:227–235
Alter A, de Léséleuc L, Van Thuc N, Thai V, Huong N, Ba NN, Cardoso CC, Grant AV, Abel L, Moraes MO, Alcaïs A, Schurr E (2010) Genetic and functional analysis of common MRC1 exon 7 polymorphisms in leprosy susceptibility. Hum Genet 127:337–348
Cardoso CC, Martinez AN, Guimaraes PE, Mendes CT, Pacheco AG, de Oliveira RB, Teles RM, Illarramendi X, Sampaio EP, Sarno EN, Dias-Neto E, Moraes MO (2007) Ninjurin 1 asp110ala single nucleotide polymorphism is associated with protection in leprosy nerve damage. J Neuroimmunol 190:131–138
Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA (2004) Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 74:106–120
Castellano LR, Filho DC, Argiro L, Dessein H, Prata A, Dessein A, Rodrigues V (2009) Th1/Th2 immune responses are associated with active cutaneous leishmaniasis and clinical cure is associated with strong interferon-gamma production. Hum Immunol 70:383–390
Chakravartti M, Vogel F (1973) A twin study on leprosy. In: Becker PE, Lenz W, Vogel F, Wendt GG (eds) Topics in human genetics, vol 1. Georg Thieme, Stuttgart, pp 1–123
Chang ST, Linderman JJ, Kirschner DE (2008) Effect of multiple genetic polymorphisms on antigen presentation and susceptibility to Mycobacterium tuberculosis infection. Infect Immun 76:3221–3232
Cozen W, Gill PS, Salam MT, Nieters A, Masood R, Cockburn MG, Gauderman WJ, Martinez-Maza O, Nathwani BN, Pike MC, Van Den Berg DJ, Hamilton AS, Deapen DM, Mack TM (2008) Interleukin-2, interleukin-12, and interferon-gamma levels and risk of young adult Hodgkin lymphoma. Blood 111:3377–3382
Delaney NL, Esquenazi V, Lucas DP, Zachary AA, Leffell MS (2004) TNF-alpha, TGF-beta, IL-10, IL-6, and INF-gamma alleles among African Americans and Cuban Americans. Report of the ASHI Minority Workshops: Part IV. Hum Immunol 65:1413–1419
Fitness J, Floyd S, Warndorff DK, Sichali L, Mwaungulu L, Crampin AC, Fine PE, Hill AV (2004) Large-scale candidate gene study of leprosy susceptibility in the Karonga district of northern Malawi. Am J Trop Med Hyg 71:330–340
Franceschi DS, Mazini PS, Rudnick CC, Sell AM, Tsuneto LT, Ribas ML, Peixoto PR, Visentainer JE (2009) Influence of TNF and IL10 gene polymorphisms in the immunopathogenesis of leprosy in the south of Brazil. Int J Infect Dis 13:493–498
Girnita DM, Webber SA, Ferrell R, Burckart GJ, Brooks MM, McDade KK, Chinnock R, Canter C, Addonizio L, Bernstein D, Kirklin JK, Girnita AL, Zeevi A (2006) Disparate distribution of 16 candidate single nucleotide polymorphisms among racial and ethnic groups of pediatric heart transplant patients. Transplantation 82:1774–1780
Govan VA, Carrara HR, Sachs JA, Hoffman M, Stanczuk GA, Williamson AL (2003) Ethnic differences in allelic distribution of IFN-g in South African women but no link with cervical cancer. J Carcinog 2:3
Hagge DA, Ray NA, Krahenbuhl JL, Adams LB (2004) An in vitro model for the lepromatous leprosy granuloma: fate of Mycobacterium leprae from target macrophages after interaction with normal and activated effector macrophages. J Immunol 172:7771–7779
Henri S, Stefani F, Parzy D, Eboumbou C, Dessein A, Chevillard C (2002) Description of three new polymorphisms in the intronic and 3’UTR regions of the human interferon gamma gene. Genes Immun 3:1–4
Larcombe LA, Orr PH, Lodge AM, Brown JS, Dembinski IJ, Milligan LC, Larcombe EA, Martin BD, Nickerson PW (2008) Functional gene polymorphisms in Canadian aboriginal populations with high rates of tuberculosis. J Infect Dis 198:1175–1179
Liu M, Cao B, Zhang H, Dai Y, Liu X et al (2006) Association of interferon-gamma gene haplotype in the Chinese population with hepatitis B virus infection. Immunogenetics 58:859–864
Lopez Roa RI, Guerrero Velasquez C, Alvarado Navarro A, Montoya Buelna M, Garcia Niebla C, Fafutis Morris M (2008) Recovery of IFN-gamma levels in PBMCs from lepromatous leprosy patients through the synergistic actions of the cytokines IL-12 and IL-18. Int Immunopharmacol 8:1715–1720
López-Maderuelo D, Arnalich F, Serantes R, González A, Codoceo R (2003) Interferon-gamma and interleukin-10 gene polymorphisms in pulmonary tuberculosis. Am J Respir Crit Care Med 167:970–975
Malhotra D, Darvishi K, Sood S, Sharma S, Grover C, Relhan V, Reddy BS, Bamezai RN (2005) IL-10 promoter single nucleotide polymorphisms are significantly associated with resistance to leprosy. Hum Genet 118:295–300
Matos GI, Covas Cde J, Bittar Rde C, Gomes-Silva A, Marques F, Maniero VC, Amato VS, Oliveira-Neto MP, Mattos Mda S, Pirmez C, Sampaio EP, Moraes MO, Da-Cruz AM (2007) IFNG +874T/A polymorphism is not associated with American tegumentary leishmaniasis susceptibility but can influence Leishmania induced IFN-gamma production. BMC Infect Dis 7:33
Misch EAMM, Ranjit C, Sapkota BR, Wells RD (2008) Human TLR1 Deficiency Is Associated with Impaired Mycobacterial Signaling and Protection from Leprosy Reversal Reaction. PLoS Negl Trop Dis 2:e231
Monot M, Honoré N, Garnier T, Zidane N, Sherafi D, Paniz-Mondolfi A, Matsuoka M, Taylor GM, Donoghue HD, Bouwman A, Mays S, Watson C, Lockwood D, Khamispour A, Dowlati Y, Jianping S, Rea TH, Vera-Cabrera L, Stefani MM, Banu S, Macdonald M, Sapkota BR, Spencer JS, Thomas J, Harshman K, Singh P, Busso P, Gattiker A, Rougemont J, Brennan PJ, Cole ST (2009) Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat Genet 41:1282–1289
Moraes MODN, Suffys PN, Santos AR, Almeida AS, Nery JA, Sampaio EP, Sarno EN (2001) Tumor necrosis factor-alpha promoter polymorphism TNF2 is associated with a stronger delayed-type hypersensitivity reaction in the skin of borderline tuberculoid leprosy patients. Immunogenetics 53:45–47
Moraes MO, Pacheco AG, Schonkeren JJ, Vanderborght PR, Nery JA, Santos AR, Moraes ME, Moraes JR, Ottenhoff TH, Sampaio EP, Huizinga TW, Sarno EN (2004) Interleukin-10 promoter single-nucleotide polymorphisms as markers for disease susceptibility and disease severity in leprosy. Genes Immun 5:592–595
Moraes MO, Cardoso CC, Vanderborght PR, Pacheco AG (2006) Genetics of host response in leprosy. Lepr Rev 77:189–202
Nathan CFMH, Wiebl ME, Rubin BY (1983) Identification of interferon-gamma as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J Exp Med 158:170
Pacheco AG, Moraes MO (2009) Genetic polymorphisms of infectious diseases in case–control studies. Dis Markers 27:173–186
Pacheco AG, Cardoso CC, Moraes MO (2008) IFNG +874T/A, IL10–1082G/A and TNF -308G/A polymorphisms in association with tuberculosis susceptibility: a meta-analysis study. Hum Genet 123:477–484
Parra FC, Amado RC, Lambertucci JR, Rocha J, Antunes CM, Pena SD (2003) Color and genomic ancestry in Brazilians. Proc Natl Acad Sci USA 100:177–182
Pereira AC, Brito-de-Souza VN, Cardoso CC, Dias-Baptista IM, Parelli FP, Venturini J, Villani-Moreno FR, Pacheco AG, Moraes MO (2009) Genetic, epidemiological and biological analysis of interleukin-10 promoter single-nucleotide polymorphisms suggests a definitive role for -819C/T in leprosy susceptibility. Genes Immun 10:174–180
Pravica VPC, Stevens A, Lee JH, Hutchinson IV (2000) A single nucleotide polymorphism in the first intron of the human IFN-gamma gene: absolute correlation with a polymorphic CA microsatellite marker of high IFN-gamma production. Hum Immunol 61:863–866
R Development Core Team (2008) R: a language and environment for statistical computing. Vienna, Austria
Ramasesh NAL, Franzblau SG, Krahenbuhl JL (1991) Effects of activated macrophages on Mycobacterium leprae. Infect Immun 59:2864–2869
Ridley DS, Jopling WH (1962) A classification of leprosy for research purposes. Lepr Rev 33:119–128
Salih MA, Ibrahim ME, Blackwell JM, Miller EN, Khalil EA, ElHassan AM, Musa AM, Mohamed HS (2007) IFNG and IFNGR1 gene polymorphisms and susceptibility to post-kala-azar dermal leishmaniasis in Sudan. Genes Immun 8:75–78
Sallakci N, Coskun M, Berber Z, Gurkan F, Kocamaz H, Uysal G, Bhuju S, Yavuzer U, Singh M, Yegin O (2007) Interferon-gamma gene +874T-A polymorphism is associated with tuberculosis and gamma interferon response. Tuberculosis (Edinb) 87:225–230
Santos AR, Suffys PN, Vanderborght PR, Moraes MO, Vieira LM, Cabello PH, Bakker AM, Matos HJ, Huizinga TW, Ottenhoff TH, Sampaio EP, Sarno EN (2002) Role of tumor necrosis factor-alpha and interleukin-10 promoter gene polymorphisms in leprosy. J Infect Dis 186:1687–1691
Schroder K, Hertzog PJ, Ravasi T, Hume DA (2004) Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol 75:163–189
Shaw MA, Donaldson IJ, Collins A, Peacock CS, Lins-Lainson Z (2001) Association and linkage of leprosy phenotypes with HLA class II and tumour necrosis factor genes. Genes Immun 2:196–204
Shields ED, Russell DA, Pericak-Vance MA (1987) Genetic epidemiology of the susceptibility to leprosy. J Clin Invest 79(4):1139–1143
Sieling PAWX, Gately MK, Oliveros JL, McHugh T et al (1994) IL-12 regulates T helper type 1 cytokine responses in human infectious disease. J Immunol 153:3639–3647
van de Vosse E, van Dissel JT, Ottenhoff TH (2009) Genetic deficiencies of innate immune signalling in human infectious disease. Lancet Infect Dis 9:688–698
Vandenbroeck K, Goris A (2006) The IFNG-IL26-IL22 cytokine gene cluster. In: Vandenbroeck K (ed) Cytokine gene polymorphisms in multifactorial conditions, vol 1. CRC Press Taylor & Francis, Boca Raton, pp 157–174
Vanderborght PR, Pacheco AG, Moraes ME, Antoni G, Romero M, Verville A, Thai VH, Huong NT, Ba NN, Schurr E, Sarno EN, Moraes MO (2007) HLA-DRB1*04 and DRB1*10 are associated with resistance and susceptibility, respectively, in Brazilian and Vietnamese leprosy patients. Genes Immun 8:320–324
Vidyarani M, Selvaraj P, Prabhu Anand S, Jawahar MS, Adhilakshmi AR, Narayanan PR (2006) Interferon gamma (IFNgamma) & interleukin-4 (IL-4) gene variants & cytokine levels in pulmonary tuberculosis. Indian J Med Res 124:403–410
Zhang FRHW, Chen SM, Sun LD, Liu H, Li Y, Cui Y, Yan XX, Yang HT, Yang RD, Chu TS, Zhang C, Zhang L, Han JW, Yu GQ, Quan C, Yu YX, Zhang Z, Shi BQ, Zhang LH, Cheng H, Wang CY, Lin Y, Zheng HF, Fu XA, Zuo XB, Wang Q, Long H, Sun YP, Cheng YL, Tian HQ, Zhou FS, Liu HX, Lu WS, He SM, Du WL, Shen M, Jin QY, Wang Y, Low HQ, Erwin T, Yang NH, Li JY, Zhao X, Jiao YL, Mao LG, Yin G, Jiang ZX, Wang XD, Yu JP, Hu ZH, Gong CH, Liu YQ, Liu RY, Wang DM, Wei D, Liu JX, Cao WK, Cao HZ, Li YP, Yan WG, Wei SY, Wang KJ, Hibberd ML, Yang S, Zhang XJ, Liu JJ (2009) Genomewide association study of leprosy. N Engl J Med 361:2609–2618
Author information
Authors and Affiliations
Corresponding author
Additional information
C. C. Cardoso and A. C. Pereira contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cardoso, C.C., Pereira, A.C., Brito-de-Souza, V.N. et al. IFNG +874 T>A single nucleotide polymorphism is associated with leprosy among Brazilians. Hum Genet 128, 481–490 (2010). https://doi.org/10.1007/s00439-010-0872-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-010-0872-x