Abstract
Northeast (NE) India is known to produce different kinds of citrus species. Citrus tristeza virus (CTV), brown citrus aphid (Toxoptera citricidus) transmitted closterovirus, is a major problem to cause citrus decline in this region. CTV has destroyed more than one million of citrus trees in India. CTV contains flexuous filamentous particles (2000 × 11 nm) and positive sense ssRNA genome of about 19.3 kb having 12 ORFs encoding about 19 proteins. The survey was made in different citrus orchards of Assam to study the disease incidence and genetic diversity of CTV. Several citrus samples were collected from Citrus reticulata cvs Khasi, Kinnow and Nagpur mandarin; C. sinensis cvs Sweet orange and Valencia orange; C. limon cv. Assam lemon; C. jambhiri cv. Rough lemon, of three citrus Farms of Assam; HRS farm, AAU, Guwahati; CEC, Kamrup Rural; and URF, AAU, Jorhat. Different kinds of symptoms like decline, chlorosis, yellowing and poor growth with stunting of citrus trees were observed in these orchards. Based on Direct antigen coated-enzyme link immunosorbent assay (DAC-ELISA) and Polymerase chain reaction (PCR) it was found that incidence CTV ranged from 40.9 to 85.54% and the overall incidence of 65.7% was estimated. Twenty-one isolates of CTV, designated as CTV-Asm 1 to CTV-Asm 21 were collected from different locations and characterized based sequence analysis of 404 nt fragment of 5’ORF1a gene of CTV genome. The pair-wise sequence analysis showed 92–100 nt identity among the present CTV isolates. Phylogenetic tree analysis segregated the present isolates into two genogroups; of which 16 isolates clustered into one genogroup along with the decline CTV strain VT/Kpg3, and the remaining isolates into another genogroup along with Indian CTV isolate AR-1. Intra-farm genetic diversity among CTV isolates were observed in citrus orchards. The present study revealed that decline inducing CTV genotype VT/Kpg3 is prevalent in citrus growing areas of Assam of NE India.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Citrus tristeza virus (CTV), a phloem-limited, flexuous, filamentous longest known plant viruswith particle-size of 2000 × 11 nm, belonging to the genus Closterovirus (Family, Closteroviridae), contains positive sense single stranded RNA genome of 19.3 kb comprising 12 open reading frames (ORFs) potentially encoding at least 19 proteins (Karasev et al. 1995; Bar-Joseph et al. 2002; Biswas et al. 2019). CTV is one of the major causes of citrus decline in most of the citrus growing countries in the world and it infects nearly all the citrus species and their cultivars and relatives (Rocha-Pena et al. 1995; Lee and Bar-Joseph 2000). Till date, CTV has killed about 100 million citrus trees worldwide including about one million trees in India ((Bar-Joseph and Dawson 2008; Ahlawat 1997). Biologically, CTV differs in intensity of symptoms induced in different citrus species and its heir aphid transmissibility (Hilf et al. 2005).
CTV has diversity of phenotypes; decline, stem pitting, seedling yellows, vein clearing, vein flecking and vein corking (Lee and Bar-Joseph 2000; Dawson et al. 2013). Under field conditions, CTV infected citrus trees do not always produce visible symptoms. Thus, suitable detection techniques based on enzyme linked immune-sorbent assay (ELISA), polymerase chain reaction (PCR) and nucleotide sequence analysis of virus genes have been developed for quick diagnosis and estimation of genetic diversity of CTV (Clark and Bar-Joseph 1984; 1993; Rubio et al. 2001; Biswas 2008; Martin et al. 2009; Tarafdar et al. 2013; Biswas 2010; Biswas et al. 2012a; Palchoudhury et al. 2017).
CTV has exhibited a wide range of genetic diversity in the natural environment and existence of several genetically divergent CTV isolates have been reported from many citrus growing countries (Manjunath et al. 2000; Lee and Bar-Joseph 2000; Brlansky et al. 2003). Analysis of complete genome of several CTV isolates has determined and classified the CTV worldwide into seven genotypes, namely, VT, T36, T30, T3, B165, RB and HA16-5 (Roy and Brlansky 2010; Biswas et al. 2012a; Harper 2013). Extensive genetic diversity in CTV genotypes has been reported from time to time from citrus growing regions of India (Biswas 2010; Biswas et al. 2012a, b; Roy et al. 2005; Sharma et al. 2011; Singh et al. 2013; Palchoudhury et al. 2017). Recently, based on the analysis of CP gene and 5′ ORF1a gene fragments of about 105 CTV isolates collected from all the citrus growing geographical regions of India, occurrence of seven to nine CTV genotypes have been determined (Palchoudhury et al. 2017).
Tristeza was shown to be one of the most important diseases in citrus growing areas of India (Raychaudhury et al. 1977). It is a century-old problem and widely distributed in all the citrus growing regions of India infecting almost all the citrus species (Ahlawat 1997; Biswas 2008, 2010; Sharma et al. 2011; Biswas et al. 2014). In India, CTV causes leaf yellowing, growth cessation, stunting with poor fruit quality, and ultimately decline of the commercially grown citrus species cultivated in India (Chakroborty et al. 1992; Biswas 2008; Biswas et al. 2014). CTV is spread primarily through infected planting material and horizontally transmitted through several aphid species in a semi-persistent manner in India; of which brown citrus aphid (BrCA) (Toxoptera citiricidus) is the most efficient vector (Capoor and Rao 1967; Raychaudhury et al. 1977; Biswas 2008). A single aphid can transmit several CTV genotypes simultaneously in the plant (Biswas et al. 2004).
CTV exhibits a wide range of disease incidence of 26.3–60% in citrus growing region of India (Biswas et al. 2014); 26.3% in the Central, 47.1–56.0% in the Northeast (NE), 36–50% in the South and 16–60% in the North and Northwest India Biswas et al. 2014). Although NE India has ideal climatic conditions for citrus cultivation and produces several economically important citrus fruits with commercially important fruits Khasi mandarin (Citrus reticulata). However, citrus productivity in this region is far below and citrus industry is un-remunerative. One of the prime factors for this dwindling productivity of Khasi mandarin is citrus decline caused by CTV. Majority of the Khasi mandarin orchards are infected by CTV (Biswas et al. 2014; Tarafdar et al. 2013; Palchoudhury et al. 2017). As the cultivation of Khasi mandarin become non-profitable day by day, the farmers are compelled to abandon the citrus cultivation. Newly established citrus orchards also get affected by this disease very fast as infected planting materials are used and subsequently horizontally transmission by BrCA into new areas. Therefore, it is time to rejuvenate the mandarin orchards in NE India through intervention of biotechnological tools, and thereby to address the challenge being faced by citrus growers. Management is dependent upon prevention and reduction of virus inoculum in the field that can be achieved by use of disease-free or resistant citrus plantlets through a sound seed certification program. Under this context, efforts have been made to study the molecular epidemiology and determination of the status of CTV prevalent and identification of dominant strain in NE India, which in turn will help to rejuvenate Khasi mandarin industry in NE India.
Materials and methods
Survey, collection and maintenance of CTV isolates
Survey was made in three farms of Assam State: Horticulture Research Station (HRS), Kahikuchi, Guwahati; Center of Excellence for Citrus (CEC), Kamrup (Rural) and University Research Station (URS), Assam Agricultural University (AAU), Jorhat to study CTV incidence and collection of samples. CTV does not show visible and distinguishable symptoms in the infected citrus plants under field conditions. Thus, the citrus trees showing poor and stunted growth with decline syndrome were considered to be apparently CTV-infected trees. Twigs of these citrus trees from each of the orchard of the farms were collected and brought to laboratory for detection and molecular assays.
Detection of CTV by Direct antigen coated-enzyme linked immuno-sorbent assay
Direct antigen coated enzyme linked immuno-sorbent assay (DAC-ELISA) developed by Clark and Bar-Joseph (1984) and used earlier in India (Biswas 2008; Tarafdar et al. 2013) was followed to detect CTV in the infected samples using the antisera obtained from the Advanced Center for Plant Virology (ACPV), ICAR-Indian Agricultural Research Institute (IARI), New Delhi-110012. CTV is predominantly phloem-limited and thus, plant parts like barks, petioles, veins and veinlets containing phloem tissues were taken as source of virus extraction for DAC-ELISA. The virus titre in infected samples was measured using optical density (OD) values of the samples at 405 nm in ELISA reader.
Reverse transcriptase-PCR, cloning and sequencing of genomic regions of CTV isolate
Total plant RNA was isolated from tender bark tissues using SV total RNA isolation system (Promega, Madison, USA). The first strand cDNA was synthesized using M-MLV-Reverse transcriptase (Promega, Madison, USA) using the method standardized by Biswas (2010). The virus sequences were amplified by polymerase chain reaction (PCR) using the forward primer KLM488 F and reverse prime KLM 491R targeting ~ 404 nt fragments of 5′ ORF1a gene of CTV genome. The amplicons were purified using QIA quick PCR Purification Kit (Qiagen, Maryland), cloned into T&A cloning vector (RBC, UK) and grown in E. coli DH5α strain using standard method. The clones of viral DNA were sequenced outsourcing by vector derived M13 forward and M13 reverse primers in an automatic sequencer (ABI 3011, Chromous Biotech Pvt. Ltd., Bangalore, India). Two clones of each isolate were sequenced and consensus sequences were considered for further analysis.
Sequence analysis
The corresponding sequences of International and previously reported Indian CTV isolates were used for sequence comparison of the present CTV isolates. The multiple sequence alignments were carried out using the software Clustal W, version 1.6 (Thompson et al. 1997). Maximum likelihood phylogenetic tree were constructed using software MEGA 6.0 (Tamura et al. 2015). Sequence identity matrix was generated using Sequence demarcation tool (SDT) version 1.2 (Muhire et al. 2014). The putative recombination events were identified using recombination- detecting program (RDP4) version 4.55 implementing seven algorithms, RDP, GeneConv, Bootscan, MaxChi, Chimera, SiScan and 3SEQ (Martin et al. 2015) using default parameter values for the different detection programs. When the same recombination events were detected by more than two algorithms, they were considered to be evidence of putative recombination.
Results and discussion
Survey, symptomatology and collection of infected citrus samples from Assam
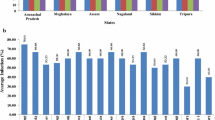
Three citrus farms of Assam, namely, HRS, Kahikuchi; CEC, Kamrup (Rural); and URF, Jorhat growing seven citrus cultivars, C. reticulata cvs Khasi Mandarin, Kinnow, Nagpur mandarin and Daisy Mandarin; C. sinensis cvs Sweet and Valencia orange; C. limon cv. Assam lemon; C. jambhiri cv. Rough lemon; were surveyed. Under field conditions, majorities of the citrus trees in the orchards of these farms showed poor growth with chlorosis of leaves. Decline symptoms along with stunted growth were commonly observed in the most of the farms surveyed (Fig. 1a). Citrus samples (twigs) from each farm were collected and brought to the laboratory for diagnosis and molecular assays. Earlier, a general decline symptoms along with chlorosis, poor and stunted growth of the mandarin tree in majority of the orchards of the NE India including Assam caused by CTV has also been reported (Tarafdar et al. 2013; Biswas et al. 2014; Palchoudhury et al. 2017). The BrCA (T. citricidus), the predominant insect vector of CTV was observed in plenty in citrus orchards of URF, AAU, Jorhat. In NE India prevalence of BrCA is very common (Biswas 2008; Tarafdar et al. 2013) and the horizontal spread through BrCA exposing multiple CTV infection to the citrus trees is the potential cause of higher incidence of CTV in NE India.
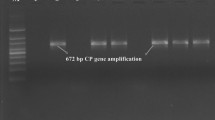
(A) Khasi Mandarin orchards in different farms of Assam; (a) Horticulture Research Station (HRS), Kahikuchi, Guwahati; (b) Center of Excellence for Citrus (CEC) Farm, Kamrup (Rural) district and (c) University Research Farm (URS), Assam Agricultural University (AAU), Jorhat. (B) Agarose gel electrophoresis showing PCR amplification of on 5′ORF 1a fragment (404nt) of Citrus tristeza virus (CTV) of Assam. Lanes M:1 kb ladder; +: positive control (CTV infected Kagzilime plant maintained in greenhouse), −: Healthy control (pooled samples of healthy Kagzilime, Sweet orange and Darjeeling mandarin plants maintained in greenhouse); (a) Lane 1–3: three CTV samples of CEC Farm, Kamrup (Rural) District; (B) Lane 4–6: three samples of HRS Farm, Assam Agricultural University (AAU), Kahikuchi, Guwahati; Lane 7–9: three samples of URS Farm, AAU, Jorhat
Detection of CTV and estimation of disease incidence
Altogether 70 samples from all the seven citrus cultivars under four Citrus species (Table 1) were collected from three citrus farms of Assam; HRS, Kahikuchi; CEC, Kamrup (Rural) and URF. DAC-ELISA results showed that majority of the citrus samples collected from these farms of Assam were infected by CTV showing OD values of 2–4 folds (Table.1). Khasi mandarin samples in URF, Jorhat showed threefold higher virus titre, whereas Khasi mandarin samples from HRS, Kahikuchiand CEC Farm, Kamrup (Rural) showed 1.5–2.1 fold higher virus titre compared to healthy control. Rough lemon samples of all the three farms showed maximum virus titre (OD value, fourfold). Nagpur mandarin, Kinnow mandarin, Daisy Mandarin, Sweet orange and Valencia sweet orange of CEC Kamrup (Rural)showed very lowtitre value (Table.1).
Based on DAC-ELISA, the incidence of CTV was estimated and the overall incidence was estimated to 65.7%; however, the higher CTV incidence up to 85.54% was found in URF, Jorhat followed by 71.4% in CEC Farm, Kamrup (Rural) and 40.9% in HRS, Kahikuchi. For confirmation of the infection by CTV, DAC-ELISA positive citrus samples were subjected to PCR test using universal primers KLM 488F and KLM 491R targeting the amplification 5′ ORF1a gene fragment (404 nt) of CTV (Fig. 1b). The ELISA positive samples showed PCR positive reaction showing the amplification of desired length of PCR amplicons in all the samples tested. The present finding showed that per cent tree infection by CTV in the three orchards surveyed was very high as 48.9–85.4%. It was found that CTV incidences in same citrus varieties or same citrus species in different locations were varied from one location to other location. This situation might be is occurred due to use of uncertified citrus planting materials or transmission efficiencies by brown citrus aphid. Although, occurrence of effective aphid vector in NE India is common (Biswas 2008; Tarafdar et al. 2013) but present of efficient aphid may vary in place to place. Moreover, differences of CTV incidence in same species in different locations occur due to randomly use of disease free or infected planting materials depending on the availability. Earlier, occurrence of the CTV disease was reported from Assam and other NE Indian staes through biological indexing (Raychaudhury and Sharma 1970; Bhagabati et al. 1989), DAC-ELISA and PCR (Biswas 2008; Borah et al. 2014; Kashyap et al. 2015; Biswas et al. 2014). CTV incidence of 48.2–60% (Biswas 2008; Kishore et al. 2010; Biswas et al. 2014; Palchoudhury et al. 2017) in the Darjeeling and Sikkim hills of NE India has been reported. Based on survey of three citrus species, Khasi mandarin, Assam lemon and Rough lemon, CTV incidence of 41.37–59.05% in Arunachal Pradesh, Meghalaya and Nagaland were reported earlier (Kashyap et al. 2015). Thus, previous (Biswas et al. 2014; Kishore et al. 2010; Palchoudhury et al. 2017) and the present studies show that incidence of CTV is very high in citrus growing areas of NE India.
Molecular cloning and sequencing of CTV genes
Citrus samples those were CTV positive through DAC-ELISA were considered to be different CTV isolates of CTV of different citrus farms of Assam. Altogether 21 CTV isolates randomly were designated as CTV-Asm1 to CTV-Asm 21 and they were taken for molecular characterization based on sequence analysis 5′ORF1a gene fragment of 404 nt length of CTV genome. The PCR products of desired length were obtained from all the present CTV isolates. Of these, the PCR products of 12 isolates, CTV-Asm 1, 3, 5,7, 9, 11, 13, 15,17,19, 20 and 21were cloned and remaining products were sent directly for sequencing by outsourcing (ABI 3011, Chromous Biotech, Bangalore). The clones of 12 CTV isolates were confirmed by standard colony PCR. Two positive clones from each CTV isolate were sent for sequencing by outsourcing. The consensus CTV sequences were identified, aligned and analyzed.
Sequence analysis of CTV isolates
In the present study, pair-wise sequence analysis using 5′ORF1a gene fragment of the present 21 isolates, the previously reported Indian and international CTV isolates showed overall 80–100% nt identities among them. A range of 78–99% nt identity were observed amongst the present Indian CTV isolates (Fig. 2), whereas 84–100% nt identity were observed amongst the present and previously reported Indian CTV isolates.
Colour-coded pair-wise percent nucleotide identity matrix of Citrus tristeza virus isolates based on sequences of 5′ORF1a gene fragment; Each colour cell represents a percent identity score between two CTV isolates (one indicated horizontally to the left and the other vertically at the bottom), A coloured key indicates the correspondence between pair-wise identities and colours displayed in the matrix
In the phylogenetic tree analysis, all the present 21 CTV isolates segregated into two genogroups (Fig. 3); of them, 16 isolates clustered into one genogroup along with recognized CTV genotype VT/Kpg3, and the remaining isolates, CTV-Asm 2, 4, 5,6 and 7 clustered into another genogroup along with previously recognized Indian CTV isolate AR-1. Overall, based on 5’ORF1a gene fragments, the Indian and International CTV isolates included in the present study segregated into 10 genogroups/genotypes. Thus, the present study showed that except one genogroup T36, CTV isolates from all the other nine CTV genogroups occur in India.
Phylogenetic relationships among Citrus tristeza virus isolates using maximum likelihood parameter (1000 bootstrap) based on sequences of 5′ORF1a gene fragment of CTV genome; The present isolates are highlighted in bold font and genogroups are marked in the right panel of the figure; VT, T36, T30, T3, T68-1, RB-G90 and HA16-5 are the representative isolates of seven International recognized genotypes; Others are Indian isolates characterized earlier
Earlier, Tarafdar et al. (2013) analyzed CTV isolates of Assam-Meghalaya based on 5′ORF1a gene and identified five genogroups; the Assam isolates clustered into four genogroups, VT/Kpg3 (severe), HA16-5 (recombinant), AG26 and AR-1; whereas, the Meghalaya isolates into two genogroups, VT/Kpg3 and T30 (mild). Palchoudhury et al. (2015) reported occurrence of two CTV genogroups VT/Kpg3 and AG 28/HA16-5 in the Khasi mandarin orchards of Manipur. Palchoudhury et al. (2017) reported occurrence of three CTV genogroups, VT/Kpg3, T3 and K10/B165 in the Darjeeling and Sikkim hills based on analysis of 5’ORF1a gene. Analyzing the 5’ORF1a gene of several CTV isolates, Biswas et al. (2012a, b) reported occurrence of overall eight CTV genotypes in India, where four genotypes, VT/Kpg3, K5, T30, and B165 occurred in the Darjeeling hills. The present study showed that majority of the CTV isolates of Assam are decline inducing CTV strain Kpg3/VT. The distinct CTV genotype AR-1 also occurs in this region,but its specific biological reaction is not known. The previous (Biswas et al. 2012a, b; Tarafdar et al. 2013) and the present study revealed that diversified CTV genotypes occur in citrus growing areas of NE India including Assam, Kpg3/VT strain is most prevalent in this region. Therefore, it is concluded that the decline inducing CTV strain Kpg3/VT is the major threat to the citrus industry in Assam and NE India.
Intra-farm genetic diversity among CTV isolates
In the present study, intra-farm genetic diversity among CTV isolates in the individual citrus farm of Assam was estimated. When the present six CTV isolates, CTV-Asm 1 to CTV-Asm 6, all of HRS farm, Kahikuchi were analyzed based on 5′ORF1a sequences, it was found that they clustered into two genotypes. The isolate CTV-Asm 1 and 3 were under Kpg3/VT genotype; whereas CTV-Asm 2, 4, 5 and 6 under AR-1 genotype indicating two different CTV genotypes are present in the HRS farm. Interestingly, the Khasi mandarin orchard in the same farm showed two different CTV genotypes Kpg3/VT, and AR1 (Table 1). Similarly, two CTV genotypes, Kpg3/VT and AR1 were also detected in citrus of URF, Jorhat. Intra-farm genetic diversity among CTV isolates in the individual citrus farm has also been reported earlier (Sharma et al. 2011; Biswas et al. 2012a, b). Occurrence of three distinct CTV genotypes in the experimental farm, ICAR-Indian Agricultural Research Institute (IARI), New Delhi has also been reported (Sharma et al. 2011). Biswas et al. (2012a, b) reported occurrence of two different CTV genotypes, Kpg3/VT genotype and B165 genotype at citrus farms of ICAR-IARI, Regional Station (RS), Kalimpong and Indian Institute of Horticulture Research, Bangalore; VT and T30 genotypes at citrus farm at ICAR-IARI-Regional Station, Pune. The previous (Sharma et al. 2011; Biswas et al. 2012a, b) and the present study revealed that intra-farm diversity among CTV genotypes are common in citrus growing individual farm in India.
Recombination analysis
To identify the recombination events, sequences of 5’ORF1a gene fragment of all the present CTV isolates were analyzed using recombination-detecting program, RDP4. The RDP4 detected CTV-Asm 18, 19 and 21 as recombinants with a breakpoint positioned at 337–88 nt supported by maximum probability, p = 1.882 × 10–08 detected by SiScan, and 7.208 × 10–01 by 3Seq algorithm (Table 2). CTV isolate T3 was major and CTV-Asm14 was minor parent for recombination of these CTV isolates. Evidence for recombination events in the origin of divergent CTV isolates has been documented earlier in India (Sharma et al. 2011; Biswas et al. 2012a, b; Tarafdar et al. 2013; Singh et al. 2013).
In conclusion, determination of genetic diversity, identification and distribution of variants in citrus growing regions of India are essential in understanding the molecular epidemiology of CTV. Additionally, the sequence analysis and establishment of phylogenetic relationship among large number of CTV isolates will lead to develop improved diagnostics and detection of specific CTV genogroup designing specific primers targeting conserve sequences of the particular genogroup, and design molecular-based management strategy using RNAi targeting conserved sequence of the virus. Present study also showed that most of the mandarin orchards in NE India suffer from citrus decline and disease incidence is as higher as 40.9–85.54%. Intra-farm genetic diversity of CTV are also common. The present study revealed that VT/Kpg3 which is decline inducing strain of CTV is dominant in citrus growing areas of Assam. As CTV disease spreads primarily through infected planting material and then secondarily by brown citrus aphid, supply of disease-free plantlets and keeping the orchards free from aphid with regular inspection are essential to maintain the citrus industry viable and profitable in NE India.
References
Ahlawat YS (1997) Viruses, greening bacterium and viroids associated with citrus (Citrus species) decline in India. Indian J Agric Sci 67:51–57
Bar-Joseph M, Dawson WO (2008) Citrus tristeza virus. Encyclopedia Virol 1:520–525
Bhagabati KN, Ahlawat YS, Chakroborty NK, Borthakur BC (1989) Distribution of greening, tristeza amd mosaic disease of citrus in North eastern states of India. Indian Phytopath 42:552–555
Biswas KK (2008) Molecular diagnosis of Citrus tristeza virus in mandarin (Citrus reticulata) orchards of hills of West Bengal. Indian J Virol 19:26–31
Biswas KK (2010) Molecular characterization of Citrus tristeza virus isolates from the Northeastern Himalayan region of India. Arch Virol 155:959–963
Biswas KK, Manjunath KL, Marais LJ, Lee RF (2004) Single aphids transmit multiple genotypes of Citrus tristeza virus, but often with changed population dynamics. Phytopath 94:S8
Biswas KK, Tarafdar A, Sharma SK (2012a) Complete genome of mandarin decline Citrus tristeza virus of Northeastern Himalayan hill region of India: comparative analyses determine recombinant. Arch Virol 157:579–583
Biswas KK, Tarafdar A, Diwedi S, Lee RF (2012b) Distribution, genetic diversity and recombination analysis of Citrus tristeza virus of India. Virus Genes 45:139–148
Biswas KK, Godara S, Nayak D (2014) Distribution of Citrus tristeza virus in the Darjeeling hills and their biological symptoms in mandarin orchards. Indian J Hort 71:408–411
Biswas KK, Palchoudhury S, Chakraborty P, Bhattacharyya UK, Ghosh DK, Debnath P, Ramadugu C, Keremane ML, Khetarpal RK, Lee RF (2019) Codon usage bias analysis of Citrus tristeza virus: higher codon adaptation to Citrus reticulata host. Viruses 11:331. https://doi.org/10.3390/v11040331
Borah M, Nath PD, Saikia AM (2014) Biological and serological techniques for detection of Citrus tristeza virus affecting Citrus species of Assam, India. Afr J Agric Res 804–3810
Brlansky RH, Damsteegt VD, Howd DS, Roy A (2003) Molecular analysis of Citrus tristeza virus sub isolate separated by aphid transmission. Plant Dis 87:397–401
Capoor SP, Rao DG (1967) Tristeza virus infection of Citrus in India. In: Proc. Inter. Symp. Subtropical and Tropical Hort. Horticulture Society of India, Bangalore, pp: 723–736.
Chakraborty NK, Ahlawat YS, Varma A, Chandra, KJ, Ramapandu S, Kapur SP (1992) Serological reactivity in Citrus tristeza virus strains in India. In: Proc. of the 12th Conference of the International Organization of Citrus Virologists, India, Riverside, International Organization of Citrus Virologists, University of California. pp108–112.
Clark MF, Bar-Joseph M (1984) Enzyme linked immune sorbent assay in plant virology. Methods Virol 7:51–85
Dawson WO, Garnsey SM, Tatineni S, Folimonova SY, Harper SJ, Gowda S (2013) Citrus tristeza virus-host interactions. Front Microbiol 4:1–10
Harper SJ (2013) Citrus tristeza virus: evolution of complex and varied genotypic groups. Front Microbiol 4:1–18
Hilf ME, Mavrodieva VA, Garnsey SM (2005) Genetic marker analysis of a global collection of isolates of Citrus tristeza virus: characterization and distribution of CTV genotypes and association with symptoms. Phytopath 95:909–917
Karasev AV, Boyko VP, Gowda S, Nikolaeva OV, Hilf ME, Koonin EV, Niblett CL, Cline K, Gumpf DJ, Lee RF, Garnsey SM, Lewandowski DJ, Dawson WO (1995) Complete sequence of the Citrus tristeza virus RNA genome. Virol 208:511–520
Kashyap A, Nath PD, Acharjee S, Biswas KK (2015) Prevalence of Citrus tristeza virus in North Eastern region of India and molecular characterization of its isolates Indian. J Hort 72(2):206–211
Kishore K, Rahman H, Kalita H, Pandey B, Minika N (2010) Prevalence of Citrus tristeza virus in mandarin of Sikkim Himalayan Region. Indian J Virol 21:140–143
Lee RF, Bar-Joseph M (2000) Tristeza. In: Timmer LW, Garnsey SM, Graham JH 2nd (eds) Compendium of Citrus diseases. American Phytopathological Society, St. Paul, pp 61–63
Manjunath KL, Lee RF, Niblett, CL (2000) Recent advances in the molecular biology of Citrus tristeza closterovirus. In: Proc of the 14th Conf of the Inter Organization of Citrus Virologists, Brazil (1998), Riverside, CA: Inter Organization of Citrus Virologists, University of California. pp:1–11.
Martin S, Sambade A, Rubio L, Vives MC, Moya P, Guerri J, Elena SF, Moreno P (2009) Contribution of recombination and selection to molecular evolution of Citrus tristeza virus. J Gen Virol 90:1527–1538
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1(1):vev003
Muhire BM, Varsani A, Martin DP (2014) SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 9:e108-277
Palchoudhury S, Bhattacharyya UK, Balram N, Chakroborty P, Biswas KK (2017) Molecular analysis of mild cross protecting strain of Citrus tristeza virus for management of disease.I n: Proc of Natl Sym “Diagnosis and Mgt of Plant Dis: Inegrated approaches and Recent Trend” organized by Ind Phytopath Soc at ICAR-Res Complex, Umiam, Meghalaya, Jan 9–11, 2017, pp-124
Raychaudhuri SP, Nariani TK, Ahlawat YS (1977) Dieback of Citrus in India. Proc Int Soc Citriculture 3:914–918
Rocha-Pena MA, Lee RF, Lastra R, Niblett CL, Ochoa-Corona FM, Garnsey SM, Yokomi RK (1995) Citrus tristeza virus and its aphid vector Toxoptera citricida: threats to citrus production in the Caribbean and Central and North America. Plant Dis 79:437–445
Roy A, Brlansky RH (2010) Genome analysis of an orange stem pitting Citrus tristeza virus isolate reveals a novel recombinant genotype. Virus Res 151:118–130
Roy A, Manjunath KL, Brlansky RH (2005) Assessment of sequence diversity in the 5-terminal region of Citrus tristeza virus from India. Virus Res 113:132–142
Rubio L, Ayllon MA, Kong P, Fernandez A, Polek M, Guerri J, Moreno P, Falk BW (2001) Genetic variation of isolates from California and Spain: evidence for mixed infections and recombination. J Virol 75:8054–8062
Sharma SK, Tarafdar A, Khatun D, Kumari S, Biswas KK (2011) Intra-farm diversity and evidence of genetic recombination of Citrus tristeza virus isolates in Delhi region of India. JPBB 21:38–43
Singh JK, Tarafdar A, Sharma SK, Biswas KK (2013) Evidence of recombinant Citrus tristeza virus isolate occurring in Acid Lime cv. Pant Lemon orchard in Uttarakhand Terai region of Northern Himalaya in India. Virus Dis 24:35–41
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2015) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tarafdar A, Godara S, Dwivedi S, Jayakumar BK, Biswas KK (2013) Characterization of Citrus tristeza virus and determination of genetic variability in North-east and South India. Indian Phytopath 66:302–307
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acid Res 24:4876–4882
Acknowledgements
The authors are thankful to DBT, Govt. of India (Code No. 24-33) and ICAR-IARI for financial support; Director, IARI; Head, Division of Plant Pathology; In-charge, ACPV, IARI, New Delhi for providing the laboratory facility. Authors are thankful to Swati Nayak for assistance in sequence data analysis and Mr. Vidyasagar Singh for his help in laboratory assisting.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Singh, S., Mulani, E., Bhattacharyya, U. et al. Decline inducing Citrus tristeza virus-VT/Kpg3 genotype occurs predominantly in citrus orchards of Northeast India. Indian Phytopathology 75, 853–861 (2022). https://doi.org/10.1007/s42360-022-00482-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42360-022-00482-z