Abstract
Citrus tristeza virus (CTV), an important graft transmissible pathogen infecting citrus (Citrus spp.) is prevalent in different citrus growing areas of the country. Sikkim is one of important citrus producing states in the Northeast region of India where Khasi mandarin (C. reticulata) is commercially cultivated. Apart from this, there is limited plantation of Assam lemon (C. limon), Rangpur lime (C. limonia) and Pomelo (C. grandis) in this region. We performed molecular characterization based on coat protein sequence analysis of CTV isolates from Sikkim state of India. Thirty six field samples suspected with Tristeza disease were tested by reverse transcription-polymerase chain reaction (RT-PCR) using CP specific primers (CN150/CN151). Out of 36, 11 samples were found CTV positive. Eight of the 36 samples showing prominent field symptoms were bio-indexed on Mexican lime (C. aurantifolia) indicator plant for CTV. Five of them showed CTV specific vein clearing and vein flecking symptoms. CTV incidence of 30.55% was recorded in the Sikkim based on combined results of ELISA and PCR. In phylogenetic analysis, all CTV isolates were grouped into eight clades, which represented as a genogroup, Gr-I to Gr-VIII. The Sikkim isolates used in the present study were segregated into two genogroup, namely Gr-I and Gr-VI. Sikkim CTV isolates SK13, SK20, SK18, SK6, SK11, SK14, SK12 and SK19 showed 98–100 per cent nucleotide identity among them and clustered with previously reported Indian isolate K10 and Florida severe isolate T3. Other isolates viz. SK5, SK7 and SK22 shared 97–99% nucleotide identity within themselves and clustered with Indian isolate K5. These results suggest the presence of genetic diversity among CTV isolates and at least two strains of CTV in Sikkim.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Northeast hills of India are among one of the natural habitats of many citrus species. Availability of favorable climatic conditions probably ensued the occurrence of numerous wild/semi-wild cultivars of citrus in this region. Sikkim is one of the most important citrus producing states in the Northeastern region of India. Mandarin (Citrus reticulata Blanco), locally known as Khasi mandarin is the predominant cultivar in Sikkim (Kishore et al. 2010). In addition to Khasi mandarin, other cultivars of citrus are also grown viz. Rangpur lime, lemon and Pomelo. Citrus tristeza virus is widely prevalent and one of the major contributors to citrus decline in India (Vasudeva and Capoor 1958; Ghosh et al. 2009; Kishore et al. 2010; Ahlawat 2012; Biswas et al. 2012).
CTV belongs to the genus Closterovirus, family Closteroviridae and is one of the major destructive pathogen of citrus farming worldwide (Bar-Joseph et al. 1981; Herron et al. 2006). Virons of CTV are long flexuous filamentous (2000 nm × 11 nm) and phloem limited in the host plant (Bar-Joseph et al. 1989; Flores et al. 2013). The virions contain positive-sense single stranded monopartite RNA genome, approximately 20 kb in size, having 12 open reading frames (ORFs) which potentially encode 19 proteins (Bar-Joseph et al. 1989; Karasev 2000; Barzegar et al. 2010; Albiach-Marti 2012). CTV is one of the largest known viruses with numerous biological strains present worldwide (Brlansky et al. 2003). It infects most citrus species, cultivars and hybrids (Albiach-Marti 2012). CTV is transmitted vegetatively via infected budwood, and through aphid species Toxoptera citricida, Aphis gossypii and Toxoptera aurantii in a semi persistent manner. Among different aphid species, T. citricida, commonly known as brown citrus aphid, is the most efficient vector of CTV transmission (Marroquin et al. 2004).
CTV causes a wide variety of symptoms in different citrus species depending on the virus strain, rootstock scion combination and prevailing temperature in the environment (Ghosh et al. 2008). Different strains of CTV produce symptoms of seedling yellowing and stem pitting (Ahlawat 2012). Field symptoms include stem pitting, vein clearing, vein flecking, general stunting, slow and quick decline (Ghosh et al. 2008). Different techniques have been used for detection of CTV (Ahlawat 2012). The most common method is biological indexing where a suspected scion is graft inoculated onto an indicator plant, Mexican lime (C. aurantifolia) Leaves of graft inoculated plants show characteristic vein clearing symptoms within few weeks post-inoculation (Ahlawat 2012). CTV is also detected by enzyme linked immunosorbent assay (ELISA), dot Immunobinding assay (DIBA), electron microscopy, Immuno electron microscopy (IEM) with polyclonal or monoclonal antibodies (Ahlawat 2012). Reverse transcription-polymerase chain reaction (RT-PCR) and real time RT-PCR are the most trustworthy, convenient and sensitive techniques for CTV detection (Ruiz–Ruiz et al. 2007). One-step RT-PCR (Hung et al. 2003), two-step RT-PCR (Metha et al. 1997; Ananthakrishnan et al. 2010) and multiplex RT-PCR (Roy et al. 2005), immunocapture (IC) RT-PCR have also been used for the detection of CTV (Saponari et al. 2008). Recently, LAMP-based CTV detection has been reported (Warghane et al. 2017a). The present attempt was made to study the genetic diversity and characterization of Sikkim CTV isolates using biological indexing, RT-PCR, nucleotide sequencing of coat protein gene and phylogenetic analysis.
Materials and methods
Sample collection, CTV incidence and diagnosis
Survey was conducted during the years 2013–2016. Total 36 samples were collected randomly representing different cultivars from different districts in Sikkim (Fig. 1). Leaves, young twigs, and bud sticks showing CTV symptoms, were collected and brought to the Plant Virology Laboratory, ICAR-Central Citrus Research Institute, Nagpur, Maharashtra, India. Direct antigen coated enzyme linked immunosorbent assay (DAC-ELISA) (Bar-Joseph et al. 1989) was used to detect CTV (Biswas 2008; Tarafdar et al. 2013). The virus titer in the infected samples was measured using optical density (OD) values at 405 nm in ELISA reader (Bio Rad Model 680, USA). Overall OD values of 3–4 folds compared to healthy control were considered as positive.
Biological indexing
Biological indexing of eight delegate samples showing prominent symptoms for CTV among 36 was done by graft inoculation on each of eight Mexican limes (C. aurantifolia) indicator seedlings. Grafted seedlings were maintained in polyhouse at 30 °C during day and 25 °C at night.
Total RNA isolation
Five to six symptomatic leaf of each sample was used to isolate total gRNA separately. Leaves were washed with tap water, followed by blotting with tissue paper and wiping with 70% ethanol. Leaf midribs were ground in liquid nitrogen and approximately 100 mg of grounded tissue was used for extraction of total genomic RNA. Total RNA was isolated using RNeasy plant mini kit (Qiagen, GmbH, and D-40724 Hilden, Germany) according to manufacturer’s instructions.
Reverse transcription- polymerase chain reaction (RT-PCR)
Total RNA was used to perform RT-PCR using CTV-CP specific primers CN150 (forward; 5′ATA TAT TTA CTC TAG ATC TAC CAT GGA CGA CGA AAC AAA3′) and CN151 (reverse; 5′GAA TCG GAA CGC GAA TTC TCA ACG TGT GTT AAA TTT CC3′) to amplify coat protein (P25) gene (Karasev et al. 1995; Ghosh et al. 2009). The RT-PCR was carried out in two steps. In first step, cDNA was synthesized in a 15 µl reaction volume containing 1.5 µl 1 × first strand buffer, 15.6U of RNAsin (Promega), 0.5 mM dNTPs and 0.4 µM reverse primer, 120U of M-MLV reverse transcriptase (Promega) and 4–5 µg of total RNA. The reaction was carried out in T100™ thermal cycler (Bio-Rad, USA) at 42 °C for 50 min followed by a denaturation at 72 °C for 10 min. An aliquot (1.75 µl/15 µl) of cDNA was used for PCR amplification in a 25 µl reaction volume containing 1 × PCR buffer, 1.5 mM MgCl2, 0.2 mM dNTPs mix, 0.2 µM of forward and reverse primer, and 1.25 U of GoTaq flexi DNA polymerase (Promega, Madison, USA). The thermal profile for the amplification was as followed: one cycle of 2 min at 94 °C followed by 35 cycles of 20 s at 94 °C, 20 s at 61 °C, 1 min at 72 °C and a final extension of 10 min at 72 °C. The resulting RT-PCR products were separated by electrophoresis in a 1% agarose gel and visualized by ethidium bromide staining in an UV GelDocumantation system (Bio-Rad, USA).
Sequencing of PCR amplicons
The PCR amplified amplicons of coat protein gene for all eleven CTV positive samples were eluted from an agarose gel by using GenElute™ Gel extraction kit (SIGMA-ALDRICH Co.St. Louis, USA) as per manufacturer’s instructions. We did not find any amplification for remaining 25 samples. The eluted samples were sequenced at Sanger sequencing facility from Eurofins Genomics India Pvt. Ltd., Bangalore using BigDye V3.1 technology and Genetic Analyzer (ABI3730XL, ABI, USA).
Sequence and phylogenetic analysis
Sequence similarities were found out using the BLAST algorithm of NCBI and visual FASTA result tool of European Molecular Biology laboratory-European Bioinformatics Institute (EMBL-EBI). The Phylogenetic tree was constructed by Maximum Likelihood method using MEGA6. The CP gene sequences of CTV isolates viz. T36 (Florida severe U16304), T30 (AF260651), VT (U56902), B165 (EU076703), HA16-5 (GQ454870), and NZRB-G90 (FJ525432) representing six recognized CTV genotypes and additional sequences (Table S1) were used from the previous studies (Roy and Brlansky 2010; Melzer et al. 2010; Biswas et al. 2012; Tarafdar et al. 2013).
Recombination analysis
The recombination detection program RDP4 was used for analysis of recombination events (Martin et al. 2015). Default settings were used with a standard Bonferroni-corrected highest acceptable p value cut-off of 0.05. The recombination event detected by two or more algorithms in CP gene of CTV was considered as evidence of putative recombination (Table S1).
Results
CTV detection and estimation of disease incidence
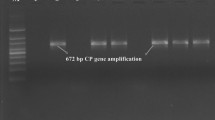
Overall, 30.55% of samples showed positive in ELISA reaction among the collected samples from various locations of Sikkim (Table 1). Out of 36 samples tested by RT-PCR using coat protein gene specific primer pairs (CN150/CN151), eleven samples viz. SK5, SK6, SK7, SK11, SK12, SK13, SK14, SK18, SK19, SK20, and SK22 were found positive for CTV showing ~ 672 bp amplicons (Fig. 2) while no amplification was observed in the remaining 25 samples (Table 1).
Biological indexing
About two and half months after graft inoculation, five samples (SK5, SK6, SK13, SK20 and SK22) out of 8 grafts inoculated samples showed characteristic symptoms of tristeza disease. Symptoms in Mexican lime indicator seedlings include vein clearing, leaf cupping, and leaf curling. Three samples viz. SK8, SK21 and SK25 did not show any CTV symptoms in the indicator seedlings and found negative for CTV by RT-PCR as well as ELISA.
Sequence analysis
The pair wise sequence analysis of the coat protein gene showed that the Sikkim isolates used in present study shared 91–100% nt identity among themselves (Table S1). These isolates showed 92–96% nt identity with the previously characterized Sikkim CTV isolates RL1 (MF078629), RS4 (MF078627) and RR5 (MF078628). Primarily, our overall sequence analysis conclude that Indian CTV isolates from North East region viz. Meghalaya, Assam, Manipur, West Bengal and Sikkim showed 89–100% nt identity among themselves (Table S1, Fig. 4).
Phylogenetic analysis
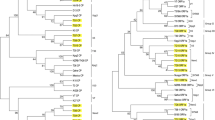
Overall phylogenetic analysis constructed a tree depicting eight clades, which were represented as geno-groups, Gr-I to Gr-VIII and the Sikkim isolates were placed under Gr-I and Gr-VI (Fig. 3). Eight isolates SK13, SK20, SK18, SK6, SK11, SK14, SK12 and SK19 were similar (98–100%) to each other and grouped with previously reported Indian isolates K10 and Florida severe isolate T3 in Gr-I by showing identify of 98–99%. The isolates SK7, SK5 and SK22 shared 97–99 per cent nt identity among them and grouped with previously reported Indian isolate K5 in Gr-VI by showing identify of 97–99%. Based on CP gene analysis, NE Indian CTV isolates were placed in five geno-groups (Gr-I, Gr-III, Gr-V–Gr-VII) while remaining Indian CTV isolates distributed into seven geno-groups (Gr-I, Gr-III–Gr-VIII) and worldwide CTV isolates into eight geno-groups (Gr-I to Gr-VIII) (Table S1, Fig. 3).
Phylogenetic analysis among CTV isolates based on sequence of the coat protein gene. The tree was generated using the Neighbour-Joining (NJ) method in MEGA 6. Significance of the nodes was estimated at 1000 bootstrap replications and the bootstra p values were shown next to the branches. The sequences generated in the present study are represented by bold fonts. The CTV genogroups with representative genotypes (isolates) generated in this study are demarcated with third bracket and arrow
Recombination
The CP gene sequence recombination analysis of eleven Sikkim CTV isolates revealed, recombination events only observed in six isolates; SK6, SK11, SK13, SK14, SK18 and SK20. The recombination events were detected at position from 438 to 672 nt in CP gene by a maximum probability (p = 2.945 × 10−1) compared to the Florida severe CTV isolate T36 as major and previously reported Indian isolates Kpg2 as minor donors. However, out of seven algorithms employed only one algorithm (Sis can) detected these recombination events. Therefore, the observed recombinations were not considered as true events. To confirm the true recombination event, further studies is required with additional CTV isolate from the Sikkim.
Discussion
Sikkim is an important citrus producing area in India. Citrus plants in many orchards show sparse foliage, vein clearing, vein corking and leaf yellowing symptoms that are indicative of tristeza disease contributing widespread decline of citrus production in the state. Kishore and colleagues studied the incidence of CTV in Sikkim Himalayan region by serological technique (DAS-ELISA) (Kishore et al. 2010) and have observed CTV incidence up to 46.32% in Khasi mandarin. The disease prevalence varied depending on the age of the plant and sampling locations.
ELISA is a robust, effective and reliable detection method of CTV and can be used for numerous samples at a time (Palchoudhury et al. 2017). In the present study, DAC-ELISA was successfully used for CTV detection. Current advancements in the field of molecular biology have provided a new paradigm for classification, identification and characterization of the plant viruses. Molecular target such as conserved nucleotide sequence of CP and other viral genes have been very useful in differentiating viral strains and genotypes (Gupta et al. 2017). It is also helpful to determine relationship of the genus, species and viral strains. Different citrus pathogens such as Candidatus Liberibacter spp., citrus mosaic badna virus, Indian citrus ringspot virus and citrus viroids have been successfully detected and characterized based on the genome sequences (Ahlawat 2012; Ghosh et al. 2015; Warghane et al. 2017b). In the present study, CP gene of CTV was amplified using primers CN150/CN151 from samples collected from different districts of Sikkim, India. These primers amplified a product of ~ 672 bp (Ghosh et al. 2008, 2009; Morales et al. 2013). Successful amplification was observed in 11 samples out of 36 tested from citrus cultivars viz. Khasi mandarin, Kali Jumbheri and Bimbra. However, Pomelo, Assam lemon and Rangpur lime samples showed no amplification of the CP gene and were presumed CTV negative.
The isolates from Sikkim showed 95–100% identity with isolates from India and other geographic regions. For example, isolates SK6, SK11, SK12, SK13, SK14, SK18 and SK20 showed 98% to 100% identity with isolate from Darjeeling (GQ475549), West Bengal. On the other hand, the isolates SK5, SK7 and SK22 showed 99 per cent identity to CTV isolates reported from Assam. Using the CP gene sequence, the genetic diversity of CTV isolates from different parts of India has been studied (Biswas et al. 2012). These authors characterized the CTV isolates in seven groups and showed that the Indian isolates tested represented all seven groups. The phylogenetic analysis revealed that Sikkim CTV isolates used in present study grouped into Gr-I (SK11, SK12, SK13, SK14, SK16, SK18, SK19) and Gr-VI (SK5, SK7, SK22) (Fig. 2). The previously characterized Sikkim CTV isolates viz., RS4, RR5 and RL1 (Palchoudhury et al. 2017) were closely related with Indian CTV isolate Kpg3 and Hawaii isolate HA16-5 and placed in Gr-VII. The recombination analysis revealed that, only one algorithm detected recombination events in six present isolates and recently it is also reported that recombination events were not detected in Sikkim CTV isolates (Palchoudhury et al. 2017). As it is detected by only one algorithm, it should not be considered as true recombination event (Table 2). Thus, our results suggest that CP gene of Sikkim CTV isolates are less recombination prone and presumably are more conserved in nature (Fig. 4).
Colour-coded pair wise nucleotide identity (%) matrix of CTV isolates based on CP gene. Each colour cell represents a percent identity score between two CTV isolates (one indicated horizontally to the left and the other vertically at the bottom). A coloured key indicates the correspondence between pair wise identities and colours displayed in the matrix. The sequences generated in the present study are represented by bold font
In summary, this study has established that the CTV is prevalent in the state of Sikkim and CTV genogroup are not restricted to citrus species and location. Further, the RT-PCR based detection using coat protein gene specific primers could be used for routine diagnosis of the disease.
References
Ahlawat YS (2012) Virus diseases of citrus and management. Studium Press, New Delhi, pp 25–40
Albiach-Marti MR (2012) Molecular virology and pathogenicity of Citrus tristeza virus. In: Maria Ga (ed) InTech. https://doi.org/10.5772/27052
Ananthakrishnan G, Venkataprasanna T, Roy A, Brlansky RH (2010) Characterization of the mixture of genotypes of a Citrus tristeza virus isolate by reverse transcription-quantitative real-time PCR. J Virol Methods 164(1–2):75–82
Bar-Joseph M, Roistacher CN, Garnsey SM, Gumpf DJ (1981) A review on tristeza, an ongoing threat to citriculture. Proc Int Soc Citric 1:419–423
Bar-Joseph M, Marcus R, Lee RF (1989) The continuous challenge of Citrus tristeza virus control. Annu Rev Phytopathol 27:291–316
Barzegar A, Heshmat R, Haleh HS (2010) Comparison of the minor coat protein gene sequences of aphid transmissible and non transmissible isolates of Citrus tristeza virus. J Gen Plant Pathol 76:143–151. https://doi.org/10.1007/s10327-009-0216-7
Biswas KK (2008) Molecular diagnosis of Citrus tristeza virus in Mandarin (Citrus reticulata) orchards of Hills of West Bengal. Indian J Virol 19(1):26–31
Biswas KK, Tarafdar A, Diwedi S, Lee RF (2012) Distribution of genetic diversity and recombination analysis of Citrus tristeza virus of India. Virus Genes 45:139–148
Brlansky RH, Damsteegt VD, Howd DS, Roy A (2003) Molecular analyses of Citrus tristeza virus sub isolates separated by aphid transmission. Plant Dis 87:397–401
Flores R, Ruiz-Ruiz S, Soler N, Sanchez-Navarro J, Fagoaga C, Lopez C, Navarro L, Moreno P, Pena L (2013) Citrus tristeza virus p23: a unique protein mediating key virus–host interactions. Front Microbiol 4:98
Ghosh DK, Aglave B, Baranwal VK (2008) Simultaneous detection of one RNA and one DNA virus from naturally infected citrus plants using duplex PCR technique. Curr Sci 94(10):1314–1318
Ghosh DK, Aglave B, Roy A, Ahlawat YS (2009) Molecular cloning, sequencing and Phylogenetic analysis of coat protein gene of a biologically distinct Citrus tristeza virus isolate occurring in central India. J Plant Biochem Biotechnol 18(1):105–108
Ghosh DK, Bhose S, Motghare M, Warghane A, Mukherjee V, Ladaniya MS, Gowda S (2015) Genetic diversity of the indian populations of ‘Candidatus Liberibacter asiaticus’ Based on the tandem repeat variability in a genomic locus. Phytopathology 105:1043–1048
Gupta N, Jain RK, Rao GP, Baranwal VK (2017) Molecular characterization and phylogenetic analysis of coat protein gene of Leek yellow stripe virus infecting garlic in India. Indian Phytopathol 70(1):114–121. https://doi.org/10.24838/ip.2017.v70.i1.48991
Herron CM, Mirkov TE, Graca JV, Lee RF (2006) Citrus tristeza virus transmission by the Toxoptera citricida vector: in vitro acquisition and transmission and infectivity immunoneutralization experiments. J Virol Methods 134:205–211
Hung TH, Wu ML, Su HJ (2003) A rapid method based on the one step reverse transcriptase-polymerase chain reaction (RT-PCR) technique for detection of different strains of Citrus tristeza virus. J Phytopathol 148(7–8):469–475. https://doi.org/10.1046/j.1439-0434.2000.00539
Karasev AV (2000) Genetic diversity and evolution of closteroviruses. Annu Rev Phytopathol 38:293–324
Karasev AN, Boyko VP, Gowda S, Nikolae OV, Hilf ME, Koonin EV, Niblett CL, Cline K, Gumpf DJ, Lee RF, Garnsey SM, Lewandowski DJ, Dawson WO (1995) Complete sequence of Citrus tristeza virus genome. Virology 208:511–520
Kishore K, Rahman H, Kalita H, Pandey B, Monika N (2010) Prevalence of Citrus tristeza virus in Mandarin of Sikkim Himalayan Region. Indian J Virol 21:140–143
Marroquin C, Olmos A, Gorris MT, Bertolini E, Martinez MC, Carbonell EA, Mendoza AH, Cambra M (2004) Estimation of the number of aphids carrying Citrus tristeza virus that visit adult citrus trees. Virus Res 100:101–108
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1(1):003. https://doi.org/10.1093/ve/vev003
Melzer M, Borth W, Sether DM, Ferreira F, Gonsalves D, Hu SJ (2010) Genetic diversity and evidence for recent modular recombination in Hawaiian Citrus tristeza virus. Virus Genes 40:111–118
Metha P, Brlansky RH, Gowda S, Yokomi RK (1997) Reverse-transcription polymerase chain reaction detection of Citrus tristeza virus in aphids. Plant Dis 81(9):1066–1069. https://doi.org/10.1094/PDIS.1997.81.9.1066
Morales J, Orlando A, Tamayo P, Penaranda J (2013) Characterizations of Citrus tristeza virus from Colombia. Rev Prot Veg 28(1):45–53
Palchoudhury S, Ghimiray P, Biswas MK, Biswas KK (2017) Citrus tristeza virus variants and their distribution in mandarin orchards in Northeastern Himalayan hill region of India. Int J Curr Microbiol App Sci 6(6):1680–1690. https://doi.org/10.20546/ijcmas.2017.606.196
Roy A, Brlansky RH (2010) Genome analysis of an orange stem pitting Citrus tristeza virus isolate reveals a novel recombinant genotype. Virus Res 151:118–130
Roy A, Fayad A, Barthe G, Brlansky RH (2005) A multiplex polymerase chain reaction method for reliable, sensitive and simultaneous detection of multiple viruses in citrus trees. J Virol Methods 129:47–55
Ruiz-Ruiz S, Morenoa P, Guerri J, Ambros S (2007) A real-time RT-PCR assay for detection and absolute quantitation of Citrus tristeza virus in different plant tissues. J Virol Methods 145:96–105
Saponari M, Manjunath K, Yokomia RK (2008) Quantitative detection of Citrus tristeza virus in citrus and aphids by real-time reverse transcription-PCR (TaqMan®). J Virol Methods 147:43–53
Tarafdar A, Godara S, Dwivedi S, Jayakumar BK, Biswas KK (2013) Characterization of Citrus tristeza virus and determination of genetic variability in North-east and South India. Indian Phytopathol 66(3):302–307
Vasudeva RS, Capoor SP (1958) Citrus decline in Bombay state. Bulletin of Plant Protection F.A.O, Rome
Warghane A, Misra P, Bhose S, Biswas KK, Sharma AK, Reddy MK, Ghosh DK (2017a) Development of reverse transcription-loop mediated isothermal amplification (RT-LAMP) assay for rapid detection of Citrus tristeza virus. J Virol Methods 250:6–10
Warghane A, Misra P, Ghosh DK, Shukla PK, Ghosh DK (2017b) Diversity and characterization of Citrus tristeza virus and Candidatus Liberibacter asiaticus associated with citrus decline in major citrus growing areas of India. Indian Phytopathol 70(3):359–367
Acknowledgements
This research was funded by ICAR-CRP, Gov. of India F. No.16-11/PP/ICAR-CRP/17-18/06. Authors thank Dr. Sidderame Gowda, CREC, UF, USA for reviewing and editing this MS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Warghane, A., Kokane, A., Kokane, S. et al. Molecular detection and coat protein gene based characterization of Citrus tristeza virus prevalent in Sikkim state of India. Indian Phytopathology 73, 135–143 (2020). https://doi.org/10.1007/s42360-019-00180-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42360-019-00180-3