Abstract
Background
Forkhead box C1 (FOXC1), a member of the Forkhead box (Fox) transcription factor family, plays an essential role in lymphatic vessel formation, angiogenesis and metastasis. Observational studies examining the relationship between the protein biomarker FOXC1 and breast cancer prognosis have reported conflicting findings. This systematic review and meta-analysis evaluates the prognostic value of the FOXC1 expression in association with patient survival in breast cancer and other types of cancers in order to identify the overall prognostic effectiveness of FOXC1.
Methods
This study followed the guidelines established in the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA). We conducted a broad search on the online bibliographic databases EMBASE, PubMed, Science Direct and Scopus, limiting search to publications from 2010 to 2018. The prognostic value was demonstrated by a random effects model meta-analysis using the hazard ratio (HR) with 95% confidence interval (CI) for overall survival (OS) in various cancer patients. The heterogeneity was measured by the I2 statistic. Publication bias and quality assessment for the selected articles was performed. Subgroup analysis was conducted based on the data available from the selected articles.
Results
A total of 16 studies met the predefined selection criteria established for our systematic review and meta-analysis, with multiple studies using diverse methodologies and reported on differing clinical outcomes, falling under a common banner of FOXC1 expression and survival in cancer. Overall, we observed a statistically non-significant association between FOXC1 protein expression and patients survival (HR: 1.186 and 95% CI 1.122–1.255, p = 0.000, I2 = 88.83%).
Conclusion
In summary, FOXC1 protein expression indicated poor survival outcome in various carcinomas, especially in patients with breast cancer, suggesting it as a possible biomarker for the prognosis in multiple carcinomas. Further clinical evaluation and large-scale cohort studies are required to accurately identify its possible clinical utility.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Our systematic and meta-analysis of Forkhead box C1 (FOXC1) indicates that there is a prognostic effect of said biomarker in different cancers. |
Based on our results, FOXC1 expression is positively correlated with poor patient survival. |
This review highlights the importance of FOXC1 among the FOX family proteins as a potential therapeutic biomarker. |
1 Introduction
Forkhead box (Fox) proteins comprise a family of evolutionarily conserved transcriptional regulators that play important roles such as development, differentiation and invasion in both healthy biological function and cancer development [1]. Early studies revealed that these proteins are overexpressed across different cancers and diseases. Forkhead box C1 (FOXC1) was initially identified as an essential transcription factor that controls the development of structures derived from the neural crest and development of the eye and meninges [2, 3].
Furthermore, recent evidence has shown that FOXC1 is overexpressed and correlated with metastasis and poor prognosis in several cancer types, including hepatocellular carcinoma [3], pancreatic ductal adenocarcinoma [2], lymphoma, lung cancer, oral cancer, melanoma, cervical cancer, breast cancer [4] and gastric cancer [4]. Hepatocellular carcinoma cells express a high level of FOXC1, which is associated with poor patient overall survival (OS) and high recurrence rates [3]. Therefore, FOXC1 has a substantial influence on aggressive metastatic cancer phenotypes. A recent study demonstrated that FOXC1 has emerged as a possible master regulator and marker for breast cancer, which is known to have a propensity for spreading to the lung and brain [5].
An American Cancer Society report estimated that approximately 252,710 new cases of invasive breast cancer and 40,610 breast cancer deaths are expected to have occurred among US women in 2017 [6]. Numerous studies reported that FOXC1 had predictive value for the different type of cancers, including in breast cancer prognosis. Breast cancer is a highly heterogeneous disease with distinct clinical and molecular features. Breast cancer microarray datasets showed that FOXC1 is highly expressed in basal-like breast cancer (BLBC), which is associated with worse survival, and these data are consistent with the results from a retrospective immunohistochemistry study of archived breast tumour tissue [7, 8].
Previous studies showed that FOXC1 expression is positively associated with brain metastasis and shorter brain metastasis-free survival in breast cancer. In addition, FOXC1 expression positively correlates with breast cancer lung metastasis [9]. Many observational studies have separately reported the relationship between FOX protein expression and cancer patients’ clinical outcomes, including patient survival. These studies showed inconclusive and conflicting findings. In this study, we conducted a meta-analysis to summarise all available evidence from pooled studies on the association between the expression of FOXC1and cancer patients’ survival to ameliorate this issue of contradicting studies and provide an idea of the overall prognostic effectiveness of FOXC1 across all published studies via pooling of individual study results.
2 Materials and Methods
2.1 Search Strategy
Our study was conducted following of the checklist of items established by the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement. A database search of EMBASE, PubMed, Science Direct and Scopus was carried out in October 2017 using the MeSH (Medical Subjective Headings) terms ‘FOXC1’ AND ‘Cancer’ and other appropriate search terms (Table 1). We initially screened the titles and abstracts of the articles, with two reviewers independently identifying relevant articles captured by the online search. Non-English articles and duplicates were removed. We also manually screened the reference lists of retrieved articles to identify other potentially relevant studies. Any discrepancies were resolved by common consensus.
2.2 Study Selection
Studies were retrieved from the online databases and collated into an EndNote file (Clarivate Analytics, Boston, MA, USA), with duplicates being removed. Titles and abstracts were screened carefully and cross-checked with the inclusion criteria. Articles were selected only when they met the inclusion criteria. The full texts of selected citations were retrieved and assessed by two independent reviewers, with consultation of a third reviewer in the case of disagreements.
2.3 Inclusion and Exclusion Criteria
The full-text articles were further evaluated using predefined selection criteria. The inclusion criteria were as follows:
-
1.
Studies that demonstrate the expression pattern of FOXC1 in the patient samples using protein or gene detection in a different type of cancer.
-
2.
Studies that reveal the association between the FOXC1 expression level and the prognosis of the patient with breast cancer.
-
3.
Studies that describe the association between FOXC1 protein levels and OS, disease-free survival (DFS) or clinicopathological features.
-
4.
Studies that displayed the Kaplan–Meier (KM) curve through the hazard ratio (HR) and in which 95% confidence interval (CI) values were available.
The exclusion criteria were as follows:
-
1.
Studies without full text and for which data could not be extracted from the abstract.
-
2.
Studies that explored FOXC1 expression either in vivo or in vitro.
-
3.
Letters to the editor, case studies, reviews, meta-analyses and conference proceedings.
-
4.
Studies published with improper information and no KM curve.
-
5.
Studies that merged the FOXC1 protein with any other genes and nomenclature.
-
6.
Studies in which the HR and 95% CI values could not be retrieved from the KM curve were excluded to produce a better systematic review and meta-analysis viewpoint.
2.4 Quality Assessment
The methodological quality was assessed using a quality assessment template based on the US National Heart, Lung and Blood Institute (NHLBI) Quality Assessment Tool for Quality Assessment of Systematic Reviews and Meta-Analyses [10]. These guidelines were adopted from previously published articles [11, 12]. This template has 14 elements which discuss the standard of the selected studies and helps to rate them on a scale of good, satisfactory or bad.
2.5 Data Extraction and Data Items
Data were extracted independently by two investigators. To standardise the data extraction process, a predefined Microsoft Excel® 2010 version (Microsoft Corp., Redmond, WA, USA) spreadsheet was prepared based on previous studies focusing on similar topics and the PRISMA guidelines [13]. The following data were extracted:
-
1.
Title, first author, year of publication, country of study.
-
2.
Study design, type of cancer, study population and FOXC1 expression using protein or gene detection.
-
3.
Clinicopathological features of cancer patients, FOXC1 (positive and negative patient), gender, age, tumour size, lymph node, the outcome of the analysis, survival data (including HR, relative risk, DFS and OS including HRs and 95% CIs).
-
4.
Representativeness of the exposed cohort.
-
5.
Experimental, statistical and analytical methods used.
2.6 Meta-Analysis and Assessment of Heterogeneity
The association between the prognostic value of FOXC1 expression levels and breast cancer patient survival was evaluated by HR (95% CI) values pooled across all included studies. Results were plotted on forest plots generated using CMA (Comprehensive Meta-Analysis) software (version 3.3.070; Biostat, Englewood, NJ, USA). A random-effects model was used to compare the odds ratios (ORs) between the individual studies depending on between-study heterogeneity [14]. The numerical value of the I2 statistic was used to categorise between-study heterogeneity as follows: 0–40% was non-heterogeneous, 30–60% was moderate, 50–90% was substantial and 75–100% was considerable heterogeneity [15]. The Higgins statistic (I2) was used as a gauge of inconsistency in study findings or outcomes and indicated the amount of overlap between the CI and the outcomes of the individual studies [16]. The Q value revealed the observed variability within and between vaccine trials [17]. A p value of < 0.01 was considered statistically significant for the Q test. Both I2 and the Q value ignore the threshold effect [18], and hence the tau squared (τ2) test was assessed, which helps to estimate the variation in test accuracy from the observed studies [19]. The z test was also included in the meta-analysis to indicate the number of standard deviations from the study mean that each study may deviate [14, 15]. The subgroup analysis was conducted as an additional parameter that is based on the heterogeneity of relative contributions of one or more key variables on the time period, any tumour stage or any other demographic factors [20].
2.7 Publication Bias
To assess any systematic review and meta-analysis, estimation of publication bias is mandatory to estimate the effect publication bias has on the results of the study. Publication bias was estimated visually using the symmetry of funnel plots (constructed using log [OR] and standard error), using Egger’s and Begg’s graphical bias indicator test [11, 15, 21].
3 Results
3.1 Study Selection
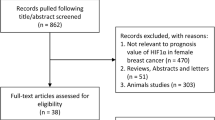
A literature search related to FOXC1 in cancer patients’ clinicopathological characteristics yielded 831 articles (Fig. 1). After removing duplicates (182 articles), a total of 649 studies were included for further evaluation. Among these, 302 articles that were not related to cancer were excluded. Letters to the editor, conference abstracts and non-English articles were also screened out. Also, 282 articles were removed as they were not related to FOXC1 protein, have performed only in vitro or in vivo studies, did not discuss survival outcome or contained incorrect information. A few articles that discussed microRNA (miRNA) significance and did not discuss FOXC1 prognosis were also removed. A total of 65 articles were included in a full-text search and eligibility screening. On full-text screening, a further 49 articles were removed due to insufficient data, inability to calculate HR and 95% CI values, improper patient data, absence of KM curve/HR values, low sample size or analysis of FOXC1 expression in conjunction with other genetic biomarkers. Sixteen articles on FOXC1 protein that discussed the prognosis of multiple carcinomas were included in the systematic review and meta-analysis [7, 22,23,24,25,26,27,28,29,30,31,32,33,34,35,36]. A complete manual search of all of the collected articles was performed, along with the previously published meta-analysis [37] and review articles [38,39,40] also being considered as data sources for relevant papers.
3.2 Study Characteristics
The main study characteristics of the included studies are summarised in Table 2. In brief, among the 16 included studies, 11 are from China, four are from the USA and one is from Korea. Nine types of cancer (breast cancer, cervical cancer, tongue cancer, pancreatic ductal adenocarcinoma, oesophageal squamous cell carcinoma, adenoid cystic carcinoma, non-small cell lung cancer, hepatocellular carcinoma and gastric cancer) were assesed in the included studies. A total of, 9891 patient samples across all included studies were assessed in this analysis. Reverse transcription polymerase chain reaction (RT-PCR), western blotting, immunohistochemistry, nCounter® expression assay (NanoString, Seattle, WA, USA) and microarray were the predominant methods used for detecting FOXC1 protein expression in the studies. Among the included studies in this analysis, 11 discussed age and seven discussed gender ratio as risk factors for survival. Only six studies mentioned follow-up details, with the follow-up times ranging from 32 months to 10 years.
3.3 Meta-Analysis and Survival Outcome
The meta-analysis was conducted using CMA software (version 3.3.070). Pooled HRs and 95% CIs were used to construct forest plots to assess the prognostic impact of FOXC1 expression in cancer survival. The OS HR values were extracted from 14 studies [7, 22,23,24,25,26,27,28,29,30,31,32,33,34,35,36] and pooled for meta-analysis (Fig. 2). The results showed a non-significant correlation between FOXC1 expression and cancer survival (HR: 1.186, 95% CI 1.122–1.255) at a p value of 0.000 and Z value of 5.967; however, the pooled effect size metric demonstrated that the likelihood of death ratio is increased by 18.6% in multiple carcinoma patients throughout the survival analysis. The observed Q value is 141.855 with I2 heterogeneity of 66.606%, indicating a moderate to high degree of heterogeneity.
Forest plot showing the meta-analysis of the association between Forkhead box C1 (FOXC1) expression and survival in different cancers. The pooled hazard ratio (HR) values were calculated using CMA (Comprehensive Meta-Analysis) statistical software (version 3.3070; Biostat, Englewood, NJ, USA). The black diamond represents the combined effect estimate of survival of patients with multiple carcinomas randomly assigned to FOXC1 protein expression evaluation. The red square with the line indicates the effect size and 95% confidence interval of the FOXC1 protein of the included studies. A risk ratio > 1 suggests no difference in risk of FOXC1 protein in multiple carcinomas whereas a risk ratio < 1 suggests a reduced risk of patients’ survival. ‘Favours survival’ refers to better survival and ‘favours death’ indicates worse survival
3.4 Forkhead Box C1 (FOXC1) Expression Within the Subgroup and Variation Among Multiple Carcinomas
Among the 20 cohorts included in the subgroups, ten different cancers have been discussed: breast cancer (n = 9), cervical cancer (n = 2), oesophageal squamous cell carcinoma (n = 1), tongue cancer (n = 1), pancreatic ductal adenocarcinoma (n = 1), adenoid cystic carcinoma (n = 1), non-small cell lung cancer (n = 3), hepatocellular carcinoma (n = 1) and gastric cancer (n = 1). The mixed effects of individual cancers has been analysed and the HR (95% CI) values are as follows: breast cancer 2.538 (1.814–3.549), cervical cancer 1.572 (0.916–2.697), oesophageal squamous cell carcinoma 1.278 (0.982–1.663), tongue cancer 1.830 (1.082–3.095), pancreatic ductal adenocarcinoma 1.328 (0.972–1.815), adenoid cystic carcinoma 1.970 (1.188–3.266), non-small cell lung cancer 1.189 (1.117–1.266), hepatocellular carcinoma 0.587 (0.453–0.760) and gastric cancer 0.273 (0.144–0.519). The Q value expressed 41.93 degrees of freedom (df), with the p value < 0.01 indicating a significant result (Fig. 3).
3.5 Publication Bias and Sensitivity Analysis
3.5.1 Funnel Plot
The funnel plot (Fig. 4) exhibited slight asymmetry, indicating that publication bias is a likely possibility.
Funnel plot of studies correlating overall patient survival with Forkhead box C1 (FOXC1) protein expression. Each dot represents an individual study. The funnel plot measures the study size and precision on the vertical axis and effect size on the horizontal axis and the plot shape describes the asymmetry. Smaller studies appear at the bottom and larger studies appear on the right side of the plot
3.5.2 Fail-Safe N
The classic fail-safe N and Orwin fail-safe N help to adjust for the missing and small studies in publication bias. This meta-analysis included data from 16 studies in multiple carcinomas, which yielded a Z value of 10.546 and a corresponding two-tailed p value of 0.0000. The fail-safe N was 560. This means that we would need to locate and include 560 ‘null’ studies for the combined two-tailed p value to exceed 0.050, which means the missing studies would be nullified for every observed study. As in the case of the classic fail-safe N, the Orwin fail-safe N endorses the missing studies and shifts the effect size towards null.
3.5.3 Begg and Mazumdar Rank Correlation Test
The Begg and Mazumdar rank correlation test suggests computing the rank order correlation (Kendall’s tau b). In this case, Kendall’s tau b (corrected for ties, if any) was 0.3526, with a one-tailed p value (recommended) of 0.01486 or a two-tailed p value of 0.0297, which is based on the continuity-corrected normal approximation.
3.5.4 Egger’s Test of the Intercept
In this case the intercept (B0) was 1.758 (95% CI – 0.00608 to 3.5774, with t = 2.0306, df = 18). The one-tailed p value (recommended) was 0.02866, and the two-tailed p value was 0.05733.
3.5.5 Duval and Tweedie’s Trim and Fill
Publication bias analysis indicated that six studies were missing. The imputation process was carried out and the funnel plot’s regression line was adjusted to better capture the missing studies, as seen in Fig. 5. The imputed point estimate after Trim and Fill was found to be 1.2498 (1.1852–1.3077). When applying the random effects model, the point estimate was found to be 1.2808 (1.072–1.5300).
Funnel plot with observed and imputed studies. Large studies appear toward the top of the graph and tend to cluster near the mean effect size. Smaller studies appear toward the bottom of the graph and (since there is more sampling variation in effect size estimates in the smaller studies) will be dispersed across a range of values
3.5.6 Subgroup Analysis on Tumour Size in Multiple Carcinomas
A subgroup analysis on tumour size from the nine studies [7, 24, 29,30,31, 33,34,35,36] was performed and is reported in a forest plot to study the prognostic effect on age (Fig. 6). The HR (95% CI) value is 1.345 (1.048–1.725), with a p value of 0.020 and Z value of 2.330. The Q value was 47.220 with an I2 heterogeneity of 83.058%.
3.5.7 Quality Assessment
Table 3 describes the quality of the selected studies, which is assessed using an appropriate tool. All of the studies included in the study have scored good quality—an important strength of the study. The primary quality score is provided based on the extraction of HR and 95% CI values, which were crucial for this study.
4 Discussion
FOXC1 is a master regulator of gene expression that plays an essential role in embryonic development, consistent with the fact that FOXC1 mutations are associated with developmental anomalies [41, 42]. More recently, however, studies have linked FOXC1 activity to the aggressive phenotype in cancer cells. FOXC1 enhances cell invasion, proliferation, metastasis, epithelial mesenchymal transition, and migration in BLBC [4]. We have selected recent studies reported between 2010 and 2018 for systematic meta-analysis. The results presented by this systematic review and meta-analysis, regarding the prognostic utility of FOXC1 in cancer, conform to prior studies, wherein the expression of FOXC1 was found to positively correlate with metastasis to the brain and lung in breast cancer [9]. However, unlike previous studies, this study expands the scope of utility of FOXC1 as a prognostic marker to all types of cancer. The pooled results, across all published studies in this field, indicate that increased FOXC1 expression is indicative of poor patient clinical outcomes and, subsequently, OS, across all patients, regardless of cancer type. A limitation of this study, resulting from a lack of sufficiently high-quality clinical studies published in this field, is the low power of analysis of the meta-analysis. A future updated systematic review and meta-analysis will be considered when further relevant literature is published. This study aimed to assist the clinical decision-making process as a prognostic factor that helps medical professionals determine not just survival but predict clinical outcomes, such as metastasis in cancer patients. The results obtained should aid physicians’ and patients’ ability to make informed decisions and could result in a better quality of life for cancer patients.
Fox proteins are highly conserved among the Fox gene family; however they serve different functions in cancer and other diseases. Previously, Xiao et al. [43] studied the prognostic value of FOXP1 in multiple carcinomas from 22 studies. They reported that the FOXP1 protein was associated with favourable prognosis in lymphoma patients (HR: 0.38, 95% CI 0.30–0.48, p < 0.001) with decreased expression. Moreover, it was associated with worse prognosis in breast cancer patients (HR: 1.93, 95% CI 1.33–2.80, p = 0.001). The current study has reported that the FOXC1 protein is associated with a worse prognosis in breast cancer patients through subgroup analysis (HR: 2.538, 95% CI 1.814–3.549) which is increased by a 1.53% death ratio.
4.1 Main Findings
This study was performed to gain insights into the prognostic significance of FOXC1 in various cancers. As recently published studies provide up-to-date data on molecular markers such as FOXC1, the time period of 2010–2018 was chosen. To our knowledge, this is the first systematic review and meta-analysis study to conduct a thorough analysis of the prognostic effect of FOXC1 in several cancer types. From the pool of studies screened, only 16 studies qualified for the systematic review and meta-analysis. One of the major limitations is the low number of studies performed in with FOXC1 as the biomarker.
4.2 Strengths
The studies included in our systematic review and meta-analysis are selected from globally published studies, and we adhered to the standard PRISMA guidelines. Being on FOXC1 as a prognostic marker in cancer, this study’s novelty in the sphere of systematic reviews and meta-analyses is its greatest strength.
4.3 Limitations
This systematic review and meta-analysis has several shortcomings. First, there is a scarcity of high-quality clinical research regarding biomarkers such as FOXC1 in cancers. Second, few studies have detected the association between FOXC1 protein levels and OS, DFS or clinicopathological features. Third, a publication bias existed that could not be avoided in the observational studies [44, 45]. A degree of publication bias also exists between study heterogeneity, being moderately high. Fourth, because of the lack of a unified survival endpoint [OS/DFS/event-free survival (EFS)], more subgroup analysis on individual survival endpoints would be helpful to more accurately predict the biomarker for cancers. Fifth, HR and 95% CI values were not available in many studies, and hence focusing on either analysis will increase the strength of the analysis. Therefore, more clinical studies are needed to elaborate and verify the results obtained in this study.
5 Conclusions
The systematic review and meta-analysis presented here suggests that FOXC1 is associated with patient prognosis in various cancers. The results regarding the prognosis of gastric cancer and hepatocellular carcinoma were associated with improved prognosis whereas all other cancers associated FOXC1 expression with worse prognosis, especially in breast cancer. FOXC1 could, therefore, be suggested as a promising biomarker for cancer prognosis pending further evaluation and large-scale cohort studies to provide robust clinical evidence.
References
Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7(11):847–59.
Kidson SH, Kume T, Deng K, Winfrey V, Hogan BL. The forkhead/winged-helix gene, Mf1, is necessary for the normal development of the cornea and formation of the anterior chamber in the mouse eye. Dev Biol. 1999;211(2):306–22.
Mears AJ, Jordan T, Mirzayans F, Dubois S, Kume T, Parlee M, et al. Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld-Rieger anomaly. Am J Hum Genet. 1998;63(5):1316–28.
Han B, Bhowmick N, Qu Y, Chung S, Giuliano AE, Cui X. FOXC1: an emerging marker and therapeutic target for cancer. Oncogene. 2017;36(28):3957–63.
Ray PS, Wang J, Qu Y, Sim M-S, Shamonki J, Bagaria SP, et al. FOXC1 is a potential prognostic biomarker with functional significance in basal-like breast cancer. Cancer Res. 2010;70(10):3870–6.
DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A. Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin. 2017;67(6):439–48.
Jensen TW, Ray T, Wang J, Li X, Naritoku WY, Han B, et al. Diagnosis of basal-like breast cancer using a FOXC1-based assay. J Natl Cancer Inst. 2015;107(8):djv148.
Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. J Proc Natal Acad Sci. 2010;107(35):15449–54.
Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17(24):2815–34.
National Institute of Health National Heart, Lung and Blood Institute. Quality Assessment Tool for Quality Assessment of Systematic Reviews and Meta-Analyses. 2015. https://www.nhlbi.nih.gov/health-topics/study-quality-assessment-tools. Accessed 11 Dec 2018.
Sabarimurugan S, Royam MM, Das A, Das S, Gothandam K, Jayaraj R, et al. Systematic review and meta-analysis of the prognostic significance of miRNAs in melanoma patients. Mol Diagn Ther. 2018;22(6):653–69.
Kumarasamy C, Devi A, Jayaraj R. Prognostic value of microRNAs in head and neck cancers: a systematic review and meta-analysis protocol. Syst Rev. 2018;7(1):150.
Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: the PRISMA statement. J Ann Intern Med. 2009;151(4):264–9.
Hooijmans CR, IntHout J, Ritskes-Hoitinga M, Rovers MM. Meta-analyses of animal studies: an introduction of a valuable instrument to further improve healthcare. ILAR J. 2014;55(3):418–26.
Deeks JJ, Higgins JP, Altman DG. Analysing data and undertaking meta-analyses. In: Higgings JPT, Green S, editors. Cochrane handbook for systematic reviews of interventions: cochrane book series. Chichester: Wiley; 2008. p. 243–96.
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–60.
Delgado Rodríguez M, Massons JMD. Revisión sistemática de estudios. Metaanálisis. Barcelona: Signo. 2005.
Jayaraj R, Kumarasamy C, Madhav MR, Pandey V, Sabarimurugan S, Ramesh N, et al. Comment on “Systematic review and meta-analysis of diagnostic accuracy of miRNAs in patients with pancreatic cancer”. Dis Mark. 2018;2018:6904569.
Jayaraj R, Kumarasamy C. Systematic review and meta-analysis of cancer studies evaluating diagnostic test accuracy and prognostic values: approaches to improve clinical interpretation of results. Cancer Manag Res. 2018;10:4669–70.
Madhav MR, Nayagam SG, Biyani K, Pandey V, Kamal DG, Sabarimurugan S, et al. Epidemiologic analysis of breast cancer incidence, prevalence, and mortality in India: protocol for a systematic review and meta-analyses. Med (Baltim). 2018;97(52):e13680.
Madurantakam RM, Kumarasamy C, Baxi S, Gupta A, Ramesh N, Kodiveri MG, et al. Current evidence on miRNAs as potential theranostic markers for detecting chemoresistance in colorectal cancer: a systematic review and meta-analysis of preclinical and clinical studies. J Mol Diagn Ther. 2019;23(1):65–82.
Cao S, Wang Z, Gao X, He W, Cai Y, Chen H, et al. FOXC1 induces cancer stem cell-like properties through upregulation of beta-catenin in NSCLC. J Exp Clin Cancer Res. 2018;37(1):220.
Han B, Qu Y, Jin Y, Yu Y, Deng N, Wawrowsky K, et al. FOXC1 activates smoothened-independent hedgehog signaling in basal-like breast cancer. Cell Rep. 2015;13(5):1046–58.
Huang L, Huang Z, Fan Y, He L, Ye M, Shi K, et al. FOXC1 promotes proliferation and epithelial-mesenchymal transition in cervical carcinoma through the PI3K-AKT signal pathway. Am J Transl Res. 2017;9(3):1297–307.
Huang Y, Huang H, Li M, Zhang X, Liu Y, Wang Y. MicroRNA-374c-5p regulates the invasion and migration of cervical cancer by acting on the Foxc1/snail pathway. Biomed Pharmacother. 2017;94:1038–47.
Kim J-Y, Jung HH, Ahn S, Bae S, Lee SK, Kim SW, et al. The relationship between nuclear factor (NF)-κB family gene expression and prognosis in triple-negative breast cancer (TNBC) patients receiving adjuvant doxorubicin treatment. Sci Rep. 2016;6:31804.
Lin Z, Sun L, Chen W, Liu B, Wang Y, Fan S, et al. miR-639 regulates transforming growth factor beta-induced epithelial–mesenchymal transition in human tongue cancer cells by targeting FOXC 1. Cancer Sci. 2014;105(10):1288–98.
Pan F, Yao J, Chen Y, Zhou C, Geng P, Mao H, et al. A novel long non-coding RNA FOXCUT and mRNA FOXC1 pair promote progression and predict poor prognosis in esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7(6):2838–49.
Ray PS, Bagaria SP, Wang J, Shamonki JM, Ye X, Sim MS, et al. Basal-like breast cancer defined by FOXC1 expression offers superior prognostic value: a retrospective immunohistochemical study. Ann Surg Oncol. 2011;18(13):3839–47.
Sizemore ST, Keri RA. The forkhead box transcription factor FOXC1 promotes breast cancer invasion by inducing matrix metalloprotease 7 (MMP7) expression. J Biol Chem. 2012;287(29):24631–40.
Wang L, Gu F, Liu C-Y, Wang R-J, Li J, Xu J-Y. High level of FOXC1 expression is associated with poor prognosis in pancreatic ductal adenocarcinoma. Tumor Biol. 2013;34(2):853–8.
Wang W-W, Chen B, Lei C-B, Liu G-X, Wang Y-G, Yi C, et al. miR-582-5p inhibits invasion and migration of salivary adenoid cystic carcinoma cells by targeting FOXC1. Jpn J Clin Oncol. 2017;47(8):690–8.
Wei L-X, Zhou R-S, Xu H-F, Wang J-Y, Yuan M-H. High expression of FOXC1 is associated with poor clinical outcome in non-small cell lung cancer patients. Tumor Biol. 2013;34(2):941–6.
Xia L, Huang W, Tian D, Zhu H, Qi X, Chen Z, et al. Overexpression of forkhead box C1 promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma. Hepatology. 2013;57(2):610–24.
Xu Y, Shao QS, Yao HB, Jin Y, Ma YY, Jia LH. Overexpression of FOXC1 correlates with poor prognosis in gastric cancer patients. Histopathology. 2014;64(7):963–70.
Xu Y, Yao R, Li J, Zhou Y, Mao F, Pan B, et al. FOXC1 overexpression is a marker of poor response to anthracycline-based adjuvant chemotherapy in sporadic triple-negative breast cancer. Cancer Chemother Pharmacol. 2017;79(6):1205–13.
Kume T, Shackour T. Meta-analysis of the likelihood of FOXC1 expression in early-and late-stage tumors. Oncotarget. 2018;9(93):36625–30.
Bach D-H, Long N, Luu TT, Anh N, Kwon S, Lee S. The dominant role of Forkhead box proteins in cancer. Int J Mol Sci. 2018;19(10):3279.
Elian FA, Yan E, Walter MA. FOXC1, the new player in the cancer sandbox. Oncotarget. 2018;9(8):8165–78.
Yang Z, Jiang S, Cheng Y, Li T, Hu W, Ma Z, et al. FOXC1 in cancer development and therapy: deciphering its emerging and divergent roles. Ther Adv Med Oncol. 2017;9(12):797–816.
Berry FB, Saleem RA, Walter MA. FOXC1 transcriptional regulation is mediated by N-and C-terminal activation domains and contains a phosphorylated transcriptional inhibitory domain. J Biol Chem. 2002;277(12):10292–7.
Nishimura DY, Searby CC, Alward WL, Walton D, Craig JE, Mackey DA, et al. A spectrum of FOXC1 mutations suggests gene dosage as a mechanism for developmental defects of the anterior chamber of the eye. Am J Hum Genet. 2001;68(2):364–72.
Xiao J, He B, Zou Y, Chen X, Lu X, Xie M, et al. Prognostic value of decreased FOXP1 protein expression in various tumors: a systematic review and meta-analysis. Sci Rep. 2016;6:30437.
Sabarimurugan S, Kumarasamy C, Baxi S, Devi A, Jayaraj R. Systematic review and meta-analysis of prognostic microRNA biomarkers for survival outcome in nasopharyngeal carcinoma. PLoS One. 2019;14(2):e0209760.
Jayaraj R, Kumarasamy C, Ramalingam S, Devi A. Systematic review and meta-analysis of risk-reductive dental strategies for medication related osteonecrosis of the jaw among cancer patients: approaches and strategies. Oral Oncol. 2018;86:312–3.
Acknowledgements
We would like to acknowledge the Meta-Analysis Concepts and Applications Workshop Manual by Michael Borenstein for its guidelines on reporting meta-analysis, subgroup analysis and publication bias (www.meta-analysis-workshops.com).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Nadana Sabapathi, Shanthi Sabarimurugan, Madurantakam Royam Madhav, Chellan Kumarasamy, Xingzhi Xu, Gaixia Xu, and Rama Jayaraj declare that they have no conflicts of interest related to this systematic review.
Funding
This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
Ethics approval and consent to participate
Ethical approval is not a requirement because all data in this review were retrieved from published studies. Since there is no specific direct patient involvement, ethical committee approval is not required.
Author contributions
RJ, XX and GX conceived this study and provided supervision and mentorship to NS. RJ and NS led the development of the study design, wrote the first draft, and coordinated and integrated comments from co-authors XX, GX, SS, MRM and CK. The editing of the final draft were done by SS, MRM and CK. RJ provided methodological guidance on the overall development of the protocol. All authors read, refined and approved the final version of the manuscript.
Rights and permissions
About this article
Cite this article
Sabapathi, N., Sabarimurugan, S., Madurantakam Royam, M. et al. Prognostic Significance of FOXC1 in Various Cancers: A Systematic Review and Meta-Analysis. Mol Diagn Ther 23, 695–706 (2019). https://doi.org/10.1007/s40291-019-00416-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-019-00416-y