Abstract
Background
Melanoma is the most aggressive and deadly form of skin cancer. The molecular variability involving microRNA (miRNA) expression plays a significant role in melanogenesis, which leads to poor prognostic effects in melanoma. Since there is a scarcity of comprehensive data on the prognostic role of miRNAs in melanoma patients, this study focuses on filling this knowledge gap through a systematic review and meta-analysis.
Methods
The included studies were extracted from several bibliographic databases between 2012 and 2018 using multiple keywords according to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines. The hazard ratios (HRs) and 95% confidence intervals (CIs) for different survival endpoints were compared to the high and low expression levels of miRNAs. The mean effect size of HR values was estimated using a random-effects model of meta-analysis. Inverted funnel plot symmetry was used to assess publication bias. Subgroup analysis was carried out individually for multiple miRNAs across different studies.
Results
A total of 24 studies across eight countries were included, of which 16 studies were eligible for meta-analysis. Twenty-five miRNA expression levels were studied from 2669 melanoma patients to estimate the association between the prognostic role of miRNAs and survival outcome in these 16 studies. The overall pooled effect size (HR) for up- and downregulated miRNAs was 1.043 (95% CI 0.921–1.181; p = 0.506), indicating that the miRNA expression increased the likelihood of death in melanoma patients by 4.3%. Subgroup analysis for miRNA10b, miRNA16 and miRNA21 showed a poor prognosis. The quality assessment revealed that 16 studies were good quality and eight studies were of fair quality.
Conclusion
This is one of the first pooled meta-analysis studies on the role of miRNAs in the prognosis of melanoma. Our findings are inconclusive but suggest that miRNA expression could predict poor survival in melanoma patients. Therefore, miRNA expression could act as promising prognostic marker for melanoma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
This meta-analysis reports the prognostic significance of different microRNAs (miRNAs) in melanoma patients. |
The effect size of estimated miRNA expressions in melanoma patients was associated with poor survival. |
Both upregulated and downregulated miRNA expression and their significance in survival were observed. |
Subgroup analysis for miRNA10b, miRNA16 and miRNA21 was studied and revealed an association with poor prognosis in melanoma. |
1 Introduction
Melanoma is one of the most malignant skin tumours and is found to occur predominantly in western countries. The incidence of melanoma has rapidly increased in recent years, with the incidence rate amongst white men and women in the USA currently being 1 in 45 and 1 in 58, respectively [1]. World Health Organization (WHO) statistics report 132,000 melanoma skin cancers every year globally. It is believed that with the depletion of ozone layer, the atmosphere lost its protective filters and allowed more UV rays onto the Earth’s surface, which could be the primary reason for the development of melanoma and non-melanoma skin cancers [2]. Moreover, it is estimated that an additional 10% decrease in the ozone layer would result in an additional 4500 melanoma skin cancers [3].
Ulcerated melanoma is often observed in tumour stages 0–IV and leads to a worse prognosis because a tumour has invaded the surrounding tissues. The prognosis for melanoma patients remains favourable when detected in the early stages, but it is associated with poor survival during the advanced stages (metastasis). Many recent studies have reported the importance of microRNAs (miRNAs) in cancers, including melanoma [4,5,6]. miRNA plays a pivotal role by contributing to the effect on cellular dysfunction, apoptosis, invasion, migration and metastasis of melanoma cells. miRNA regulates cell cycle and proliferation in melanoma, and a few miRNAs may be involved in cell migration, invasion and survival [7]. Recent studies have highlighted the significant role of miRNAs in the development and prognosis of melanoma [8, 9]. The current evidence demonstrates that there are 63 differentially expressed miRNAs in melanoma patients. A previous study has investigated the expression profiles of miRNAs by meta-analysis and predicted targets that are responsible for several essential genes involved in the metastasis of melanoma [10].
1.1 Rationale
1.1.1 Importance of the Issue
There are significant challenges in the disease management of melanoma patients, particularly in early prediction of disease progression and the clinical outcomes for those patients who have responded to adjuvant therapies [11]. Therefore, predicting patient outcomes in the early stages of the disease using a molecular biomarker could be helpful to improve disease management [12, 13]. Since miRNAs have been reported to play a significant role in melanoma prognosis, they could be considered as a prognostic biomarker for melanoma. A previous study demonstrated the critical role of miRNAs in primary melanoma and the development of distant metastatic melanoma [14]. In addition, miRNA dysregulation could be manifested due to the penetration of UV rays from sunlight. Hence, we anticipate that miRNA expression has a significant molecular role in the cancer prognosis of melanoma patients.
Although the miRNA expression profile in melanoma has been studied by a previously published meta-analysis study and other individual studies, there is a scarcity of studies that demonstrate the association between miRNA expression and melanoma patients’ survival. Therefore, a quantitative synthesis or meta-analysis is needed to estimate the association between the prognostic role of miRNAs and survival outcomes in melanoma patients. This analysis provides a more precise estimate of the prognostic effect.
1.1.2 How Will Our Study Address this Issue?
Li et al. [10] carried out a study using systematic review and meta-analysis on expression profiles of miRNA in melanoma patients, but no research group has reported the prognostic effects of miRNAs in melanoma patients quantitatively using a large number of patient samples. The clinical utility of these miRNAs has also not been established yet. Therefore, this systematic review and meta-analysis were carried out to summarise the studies that have been published globally. This approach helps us to predict specific miRNAs as early prognostic biomarkers which will aid in clinical outcomes of individualised treatment of melanoma patients. This comprehensive analysis will help to determine the possible prognostic trends of individual miRNAs and the mean effect size of survival outcomes in melanoma patients using evidence from published studies.
1.1.3 How Will it Help?
The results of this study will help to predict the impact of miRNA expression on the prognosis of melanoma patients. It will also help us in using biomarkers to predict survival risks, and aid in carrying out further research focusing on the molecular role of miRNAs in predicting pathological progression including distant metastasis. Moreover, these findings will help clinicians’ decision making during treatment, and aid timely relief from post-treatment effects. The intent of the study is to provide more knowledge on effect size estimates of hazard ratios (HRs) and 95% confidence intervals (CIs) for probable survival outcomes in melanoma patients during the course of the disease and motivate researchers to carry out large-scale prospective studies on melanoma prognosis. The findings from this study may also assist further prognostic research for skincare and translational capacity in dermatology practices.
2 Methods
2.1 Search Strategy and Study Selection
The systematic review and meta-analysis on the role of miRNAs in melanoma were carried out according to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines [15]. The study search was carried out in bibliographic databases including EMBASE, Google Scholar, MEDLINE, PubMed, Science Direct, Scopus and Web of science to identify relevant studies published between January 2011 and April 2018. A few studies were also obtained from the reference lists of these selected articles. Two authors (SS and MRM) performed the article search independently, and the corresponding author (RJ) performed a final review of the entire search and extracted articles.
2.2 Selection Criteria
Studies included in our systematic review and meta-analysis were selected using the following inclusion and exclusion criteria.
Inclusion criteria:
-
1.
Studies that discussed the role of miRNAs and the survival outcome of melanoma patients.
-
2.
Studies that reported HR and 95% CI values.
-
3.
Articles published between 2012 and 2018.
-
4.
Studies that interpreted survival analysis (clinical endpoint) with univariate or multivariate analysis.
-
5.
Studies that were in accordance with the PRISMA guidelines for systematic reviews and meta-analysis.
-
6.
Studies with Kaplan–Meier (KM) curves were also selected if it was possible to retrieve HR and 95% CI values from the KM curves.
Exclusion criteria:
-
1.
Conference abstracts, reviews and letters to the editor were not within the scope of our analysis.
-
2.
Studies reporting in vitro, in silico and other non-human (animal) miRNA expression.
-
3.
Unpublished articles/theses and incomplete studies.
-
4.
Studies that discussed tumour histology, tumour differentiation and other malignant potential specifically if they failed to discuss the survival outcomes of miRNA in melanoma.
-
5.
Articles that reported prognosis results for a sample size less than ten.
2.3 Type of Participants
This current study involves clinical data from the patient cohorts in the included studies that were identified through the online search. There were no limitations of gender, age, tumour stage, lymph nodes or any treatment parameters. Similarly, data on tissue, serum, skin lesions and plasma samples from the included studies were included in this study to investigate the expression of miRNAs in melanoma patients. The follow-up period was extracted from the included studies.
2.4 Data Extraction and Management
Data were extracted from the included studies independently; any discrepancies were discussed amongst all authors, and the final decision was taken through a team decision. Using Microsoft Excel® (Microsoft Corp., Redmond, WA, USA), data items were extracted after a manual check by the review team for evaluation of study quality and data synthesis. Data items such as author names, publication year, gender, study period, diagnostic methods, follow-up period, sunlight exposure, ulceration, TNM (tumour node metastasis) stages, endpoint determination, survival status and miRNA expressions were retrieved for further analysis. Endnote (Clarivate Analytics, Philadelphia, PA, USA) was used as a bibliographic reference manager to avoid duplication of the included studies.
2.5 Quality Assessment and Risk of Bias of Individual Studies
Two reviewers performed quality assessment of the included studies independently using the quality assessment tool from the National Heart, Lung, and Blood Institute (NHLBI): Quality Assessment of Systematic Reviews and Meta-Analyses and Quality Assessment Tool for observational Cohort and Cross-Sectional Studies [16]. Before attempting to extract the data items from the individual studies, a quality assessment was performed. This assessment tool was applied to all the selected full-text articles, and they were then categorised into good, fair or poor. Fourteen elements of quality assessment, including patient population and selection, study comparability, follow-up and outcome of interest, are addressed by this tool [16].
The risk of bias is inversely correlated with the quality of the study. While a high risk of bias translates to a rating of poor quality, a low risk of bias translates to a rating of good quality. The name and version of the assessment tool used, the risk of bias graph/summary, incomplete outcome data and other potential sources of bias were also assessed. Data were summarised using descriptive statistics (frequency, percentage).
2.6 Assessment of Heterogeneity and Statistics
Comprehensive Meta-Analysis (CMA) software (version 3.3.070; Biostat, Englewood, NJ, USA) was used to perform the meta-analysis to generate forest plots using HRs and associated 95% CIs of survival endpoints (overall survival [OS], disease-free survival [DFS], recurrence-free survival [RFS], metastasis-free survival [MFS], progression-free survival [PFS] and post-recurrence survival [PRS]) from the selected studies. The pooled effect size estimate of the HR was estimated using random-effects model meta-analysis. The effect size of the HR on melanoma patients indicates the important aspects of the prognostic impact of miRNAs. The mean effect size of the HR is a crucial tool in reporting and interpreting the effectiveness of miRNAs on melanoma patients’ prognosis, generally referring to the comparative and collective effect size estimates of the different studies. The mean effect estimate of the HR is more frequently used in meta-analysis than its statistical significance or the sample size of the individual studies [17, 18]. Heterogeneity was assessed using the Higgins I2 statistic and Cochran’s Q-test [19]. Cochran’s Q-test was used secondary to the I2 statistic given its low power of heterogeneity detection. In case of any discrepancy, the I2 statistic was given precedence [20]. The forest plots were analysed to elucidate the outcomes of patient survival in melanoma. Subgroup analysis was carried out for the repeated miRNAs with the same methods for cumulative meta-analysis.
2.7 Publication Bias
Publication bias was estimated to assess any bias in the included studies. Funnel plots, Orwin and classic fail-safe N tests [21], Begg and Mazumdar Rank correlation tests [22], the Harbord-Egger’s test of the intercept [23] and Duval and Tweedie’s trim and fill method [24] calculations were used.
2.7.1 Funnel Plots
Funnel plots for each, study effect size about their sample size were constructed with scattered plots, where a lack of skew or asymmetry indicated an absence of publication bias.
2.7.2 Classic Fail-Safe and Orwin Fail-Safe N Tests
Both the Orwin fail-safe N test and classic fail-safe N test were applied to determine studies that are missing from the systematic review and meta-analysis, even with the advent of complete online bibliographic searching. These tests help to enumerate the studies in a meta-analysis which transfers the effect size towards null and also makes it possible to interpret whether it is true or false [21].
2.7.3 Begg and Mazumdar Rank Correlation Test
The Begg and Mazumdar rank correlation test uses the correlation between the ranks of effect sizes and the ranks of their variances. Inverse correlation may occur between study size and effect size due to the sizeable prognostic effects in melanoma patients. Therefore, we estimated the rank order correlation (Kendall’s tau-b) between the prognostic effect and the standard error (which is driven primarily by sample size). Positive values indicate a trend towards a higher level of test accuracy in studies with smaller sample sizes [22].
2.7.4 Egger’s Test of the Intercept
The Egger regression gives the degree of funnel plot asymmetry as measured by the intercept from a regression of standard normal which deviates against precision [23]. This test was performed to determine whether there is an association between test accuracy estimates (estimate divided by its standard error) and their precision (reciprocal of the standard error of the estimate).
2.7.5 Duval and Tweedie’s Trim and Fill Method
A rank-based method was used to estimate the number of missing studies from the meta-analysis due to suppression of the most extreme results on one side of the funnel plot and to produce an adjusted estimate of test accuracy by inputting suspected missing studies. It initially trims the asymmetric studies from the right-hand side to locate the unbiased effect (in an iterative procedure), and then fills the plot by re-inserting the trimmed studies on the right and their inputted counterparts to the left of the mean effect [24].
3 Results
3.1 Study Selection and Data Extraction
In total, 32,375 studies were identified by searching the bibliographic databases EMBASE, Google Scholar, MEDLINE, PubMed, Science Direct, Scopus and Web of Science with the help of search strings or keywords (Table 1). Of these, 31,543 studies were excluded because they were either irrelevant topics or inappropriate studies. Finally, 620 articles were screened for further evaluation after removing 212 duplicates. A third reviewer was constantly monitoring all steps of the review process to ensure the complete study search and selection of most relevant studies for inclusion. After removing the duplicates and abstract screening, 498 articles were excluded as the articles were reviews, letters to the editor and case studies. This exclusion step resulted in 122 studies after careful manual screening. Double verification of the reference lists of the existing narrative reviews and meta-analysis revealed no further relevant missed studies [10]. Moreover, further manual screening for full-text studies, any non-human studies and availability of miRNA expression details resulted in the removal of 73 studies. Finally, 49 studies were eligible for inclusion in our systematic review. Of these, 25 studies were excluded because melanoma patients’ survival, HR values or KM curves were not provided. Also, some studies failed to mention the association between miRNA expressions and survival outcomes. Therefore, 24 studies were included in the systematic review. Furthermore, eight studies were not included in the meta-analysis due to non-availability of HR values [25,26,27,28] or difficulty in estimating the HR values [29,30,31,32]. Therefore, 16 studies were found eligible for meta-analysis. The complete study selection is shown in Fig. 1.
3.2 Study Characteristics
The 24 studies were from Australia, China, France, Germany, Italy, the UK and the USA and were published from 2012 to 2018. The total population included in this study was 3531 subjects from 24 studies. The gender details were available in 14 of the included 24 studies in the systematic analysis. In the 14 studies, 1135 males and 992 females were reported (Table 2) [9, 33,34,35,36,37,38,39,40,41,42,43,44,45,46,47]. Our quantitative synthesis demonstrated that 180 patients were exposed to sunlight in four studies [25, 33, 37, 38]. Our pooled studies reported the miRNA expression levels and prognostic significance of 627 miRNAs. The miRNA expression was detected from various patients’ samples, such as peripheral blood, skin lesions, tissue, serum and plasma. The miRNAs were mostly quantified by reverse transcription polymerase chain reaction (RT-PCR), but the TaqMan® assay (ThermoFisher Scientific, Waltham, MA, USA) was used in a few studies. In total, 16 studies were examined for ulceration in melanoma patients: 616 of 2258 melanoma patients were positive for ulceration. Seven studies did not mention the follow-up period. A few studies did not provide HR values, and hence we extracted the HR values from their KM curves. The main study characteristics are illustrated in Table 2.
3.3 Meta-analysis
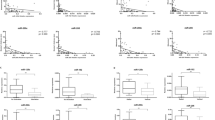
The prognostic significance of 25 miRNAs was studied in 2669 melanoma patients from 16 included studies (Fig. 2). Among these, 13 miRNAs were downregulated and 12 miRNAs were upregulated. The overall effect size estimate of the pooled HR for (up- and downregulated) miRNA expressions was 1.043 (95% CI 0.921–1.181), indicating that the miRNAs increased the likelihood of death of melanoma patients by 4.3%. The studies considered in this analysis were sampled from a universe of possible studies defined by a set of inclusion and exclusion criteria as outlined in Sect. 2.
Meta-analysis of miRNAs in melanoma patients. Pooled HRs and relative weight values for melanoma patients’ survival data were calculated and analysed using Comprehensive Meta-Analysis (CMA) software (version 3.3.070). Impact of HR value on melanoma patients: raw HR data with 95% CIs from individual studies are graphically represented in the plot. A meta-analysis statistically combines HRs from all studies and gives a more precise estimate of the effect. In the forest plot, the black diamond graphically represents the pooled effect estimate of survival for melanoma patients randomly assigned to miRNA evaluation. The red box and line indicate the effect size of miRNA expression in the included studies with a 95% CI. An HR of 1 suggests no difference in risk of melanoma patients’ survival (line of no effect or difference). An HR > 1 indicates an increased risk of patients’ survival whereas a HR < 1 suggests a reduced risk of patients’ survival. The estimated effect of miRNA expression in our meta-analysis is the relative risk of dying from melanoma. In our study, the miRNAs increased the likelihood of melanoma patients’ death by 4.3%. Upregulated miRNAs showed a significant relationship with patients’ survival. Impact of relative weight on melanoma patients: the size of the red box in the forest plot refers to the relative weight of the study. The bigger the box, the greater the weight of the study. CI confidence interval, HR hazard ratio, miRNA microRNA
3.3.1 Does MicroRNA (miRNA) Expression Affect Melanoma Patients’ Survival?
Of the 13 downregulated miRNAs, five miRNAs were associated with good patient survival (miRNA15b, miRNA33a, miRNA205, miRNA610 and miRNA4706), whereas the remaining eight miRNAs were associated with poor survival of melanoma patients (miRNA16, miRNA23a, miRNA203, miRNA206, miRNA513b, miRNA514a, miRNA4487 and miRNA4731). Similarly, the 12 upregulated miRNAs also displayed a mixed outcome (both good and poor) on melanoma patients’ survival. Of these upregulated miRNAs, miR200b was associated with good survival whereas the remaining miRNAs displayed poor survival (miRNA10b, miRNA21, miRNA142-3p, miRNA142-5p, miRNA150, miRNA199a-5p, miRNA210, miRNA301a, miRNA424, miRNA513c and miRNA579-3p).
The Z-value for the null hypothesis test (the mean risk ratio of which is 1.0) is 0.665, with a corresponding p-value of 0.091. Therefore, we can accept the null hypothesis that the risk of death is not the same in both upregulated and downregulated miRNA groups, and also conclude that the risk of death is higher in upregulated expression groups.
3.3.2 How Much Does the Effect Size of Melanoma Patient Survival Vary Across Studies?
The Q-value is 188.45 with 25 degrees of freedom (df) and a p-value of 0.427. Since the observed variance falls within the range that can be attributed to sampling error, we cannot reject the null that the actual effect size is the same in all studies. While the data do not prove that the effect size varies across populations, we can assume that it does. Moreover, we can proceed to estimate the extent of the variation. The I2 statistic tells us what proportion of the observed variance reflects differences in exact effect sizes rather than sampling error. Here, I2 is 85.143%. T2 (T = tau) is the variance of accurate effect sizes (in log units). Here, T2 is 0.054. T is the standard deviation of true effects (in log units). Here, T (tau) is 0.232.
3.3.3 Does the Effect Size Vary by Subgroup of Melanoma Patient Survival?
While the mean effect size across all studies is modest (HR 1.043), it is possible that the mean HR varies by subgroup. We used subgroup analysis to compare the effect size in studies that employed a high expression and low expression of miRNAs.
The mean risk ratios in the two groups were found to be 1.581 and 0.900, respectively. The mean effect size of the HR value of upregulated miRNAs in melanoma patient survival was 1.581 (95% CI 1.241–2.015; p- = 0.000), indicating that the miRNAs increased the likelihood of death of melanoma patients by 58%. The HR and CI values of downregulated miRNAs in melanoma patients’ survival was 0.900 (95% CI 0.779–1.039; p- = 0.151), denoting that miRNAs increased the likelihood of survival in melanoma patients by 10%.
The Q-value for the differences was 15.378 with 25 df and p = 0.000. Therefore, there was no evidence that the HR varied with melanoma patients’ survival.
3.4 miRNA Subgroup Analysis
In total, three miRNA subgroup analyses were performed in this study: miRNA10b, miRNA16 and miRNA21 as two different research groups studied these miRNAs.
3.4.1 miRNA10b
Two studies have investigated the survival outcome of miRNA10b in melanoma patients [33, 43]. The elevated expression level of miRNA10b displayed a mean HR value of 1.737 (95% CI 1.336–2.258; p = 0.000). The Z-value was 4.119 (Fig. 3).
3.4.2 miRNA16
Two studies investigated the prognostic significance of miRNA16 in melanoma patients [37, 44]. Both studies reported that downregulation of miRNA16 significantly indicated poor survival analysis in the subgroup meta-analysis. The mean effect size of the HR was 0.463 (95% CI 0.635–3.372; p = 0.372). The Z-value was 0.893 (Fig. 4).
3.4.3 miRNA21
Two studies reported on the prognostic significance of miRNA21 [40, 42]. The observed random effect size of the HR was 2.546 (95% CI 1.35–4.77; p = 0.004). The Z-value was 2.909 with I2 = 0.000 (Fig. 5).
3.5 Publication Bias and Sensitivity Analysis
We used many modules to study the sensitivity and bias of this meta-analysis, as discussed in the following sections.
3.5.1 Funnel Plot
Figure 6 shows the funnel plots, which were slightly asymmetric (existence of publication bias) across survival outcome studies. Funnel plots in Figs. 6 and 7 have been constructed using CMA software (version 3.3.070, USA). The funnel plots in Figs. 6 and 7 represent the possible bias between the included studies. Ideally, studies that have no publication bias would display a set of points (each representing an individual study included in the metaanalysis) that are symmetrically present across the central line of the funnel plot. In this study, each point represents an individual cohort or study that was included in the meta-analysis for the generation of the forest plot. Every single study/cohort included in the forest plot is represented as a point on the funnel plot. The publication bias is assessed, and the funnel plot is constructed simultaneously alongside the forest plot, with the aid of log HR values (HR values used in the meta-analysis) and the standard error.
3.5.2 Classic Fail-Safe and Orwin Fail-Safe N Tests
This meta-analysis included data from 29 melanoma studies, which yielded a Z-value of 3.18371 and a corresponding two-tailed p-value of 0.00145. The p-value for the observed studies was 48.00. The HR in the observed studies was found to be 0.98668, which did not fall between the mean HR in the missing studies. Therefore, we could not calculate the Orwin fail-safe N.
3.5.3 Begg and Mazumdar Rank Correlation Test
The corrected Kendall’s tau values were 0.20443 and 1.55692. The one-tailed and two-tailed p-values were 0.05974 and 0.11949, respectively.
3.5.4 Egger’s Test of the Intercept
In this study the intercept (B0) was found to be 1.35186 (95% CI 0.0524–2.6513, with t = 2.13456, df = 27. The one-tailed p-value (recommended) was 0.02101 and the two-tailed p-value was 0.04203.
3.5.5 Duval and Tweedie’s Trim and Fill
The funnel plot for the trimmed and imputed study is displayed in Fig. 7. Funnel plots in Figs. 6 and 7 have been constructed using CMA software (version 3.3.070, USA). The funnel plots in Figs. 6 and 7 represent the possible bias between the included studies. Ideally, studies that have no publication bias would display a set of points (each representing an individual study included in the metaanalysis) that are symmetrically present across the central line of the funnel plot. In this study, each point represents an individual cohort or study that was included in the meta-analysis for the generation of the forest plot. Every single study/cohort included in the forest plot is represented as a point on the funnel plot. The publication bias is assessed, and the funnel plot is constructed simultaneously alongside the forest plot, with the aid of log HR values (HR values used in the meta-analysis) and the standard error. Under the fixed-effect model, the hazard point estimate (95% CI) for the combined studies was 0.98668 (0.95457–1.01987). Using the Trim and Fill method, the imputed point estimate (95% CI) was 0.97364 (0.94213–1.00621). Under the random-effects model, the point estimate (95% CI) for the combined studies was 1.13569 (1.01135–1.27532), indicating that the miRNAs increased the likelihood of death of melanoma patients by 13%. Using the Trim and Fill method, the imputed point estimate of the HR value (95% CI) was 1.00008 (0.88714–1.12741).
3.6 Quality Assessment
Quality assessment of the included studies was based on the National Institutes of Health (NIH-NHLBI) Quality Assessment Tool for Observational Cohort and Cross-Sectional Studies (Table 3). The majority of the studies (15/22) had good quality scores, indicating the good methodological quality of included studies. Furthermore, seven studies had satisfactory scores. Though the scores differ between good and satisfactory based on each criterion, the cardinal score (good—16 studies, satisfactory—eight studies) was designated based on the results of HR and 95% CI values.
4 Discussion
We conducted a comprehensive systematic review to explore the association between miRNA expressions and melanoma patients’ prognosis through systematic review and meta-analysis. Previous studies have reported the vital role of miRNAs in melanocytes invasion and cell destruction and the involvement of various expressions of miRNAs in primary and metastatic melanoma [7]. Hence, miRNAs could be considered a promising biomarker. Indeed, biomarkers are increasingly used to diagnose patients (diagnostic biomarkers), predict survival (prognostic biomarkers) and measure the therapeutic response (theragnostic biomarkers) [48].
However, some queries remain unanswered. Does miRNA expressions affect melanoma patients’ survival? How much does the effect size of melanoma patient survival vary across studies? Does the effect size vary by subgroup of melanoma patient survival? With this systematic review and meta-analysis, we aimed to specifically answer these three questions. This meta-analysis demonstrated that the likelihood of death in both the up- and downregulated miRNA expressions were not same, and that the survival risk is higher in upregulated miRNA expression groups. To our knowledge, this is the first meta-analysis involving such a wide range of miRNAs to describe their role in melanoma prognosis.
A previous meta-analysis study has looked at miRNA expression profiles and biological functions in the metastasis of melanoma [10]. The results herein demonstrate the prognostic effects of 25 miRNAs (13 were downregulated and 12 were upregulated) and studied their survival outcomes. The findings in this current study summarised these 25 upregulated and downregulated miRNAs and their association with melanoma prognosis, the effect size of the HR values of 2669 patients and different modules of publication bias of the included studies. If we implement these miRNAs in routine dermatology care, it should yield valuable prognostic information.
A previous study showed systematic cross-validation of miRNA prognostic signatures from publicly available metastatic melanoma and summarised four studies using meta-signatures [11]. Our findings from 24 published studies focused on both primary and metastatic melanoma. In contrast to the previous study, we extracted HR values from the published studies and not from the genomic data. Also, the meta-analysis of our study was supported by the evaluation of different modules of publication bias, including funnel plots, the Trim and Fill method, and Egger’s and Begg’s tests. However, it is important to note that prognostication in melanoma requires standardisation of these specific miRNAs. Therefore, prospective large-scale observational clinical studies are warranted to assess the reproducibility and inter-observation.
Song et al. [49] performed a meta-analysis of ten upregulated and 14 downregulated miRNA expressions in prostate cancer. Their findings showed that five miRNAs (miRNA21, miRNA30c, miRNA129, miRNA145 and let 7c) could predict poor RFS of prostate cancer, and miRNA375 exhibited poor OS. Of the 13 downregulated miRNAs in our current study, five miRNAs were associated with good survival, whereas the remaining eight miRNAs potentially indicated poor survival. The 12 upregulated miRNAs displayed both poor and good survival in melanoma patients. These findings demonstrate that upregulated miRNAs show markedly worse survival prognosis (58%). These findings from 24 studies with 2823 patients support the inclusion of miRNAs into melanoma pathological progression as markers of more aggressive disease.
miRNA21 is the most predominant miRNA signature that is strongly involved in poor prognosis in multiple cancers [50,51,52]. It has been reported to be a tumour promoter in many cancers, including colorectal cancer [6], breast cancer [53] and pancreatic ductal adenocarcinoma [54]. A study explained the prognostic effects of miRNA21 in 2258 oesophagal cancer patients from 25 studies, indicating miRNA21 overexpression as a significant predictor of poor OS in oesophagal cancer [55]. Similarly, Xie and Wu [56] also revealed the prognostic importance of miRNA21 in 777 oral cancer patients from nine eligible studies and reported miRNA21 overexpression leading to poor prognosis in oral cancer patients. Our findings on miRNA21 were consistent with those of these studies, with our results demonstrating that the overexpression of miRNA21 was associated with poor survival in 322 melanoma patients. Since previous studies had reported the prognostic significance of miRNA, we performed a subgroup analysis of miRNA21 [39, 57]. Results for both of these studies indicate upregulated expression. Jamali et al. [57] also investigated the prognostic implications of miRNAs in head and neck squamous cell carcinoma (HNSCC). The findings showed that significantly elevated expressions of miRNA21 were associated with poor survival in patients with HNSCC [57]. Along with miRNA21, they also reported the prognostic significance of miRNA203 and miRNA205. The authors interpreted that the downregulation of miRNA203 and miRNA205 were associated with a poor prognosis. Similarly, in this current study, the downregulation of miRNA203 was associated with a poor prognosis but not miRNA205, which on the contrary displayed good prognosis. Furthermore, miRNA205 estimation from a different set of samples was associated with favourable prognosis in melanoma patients [39].
Huang et al. [58] studied the miRNA prognosis in various cancers (lung cancer, breast cancer, glioma, colorectal cancer and gastric carcinoma). The meta-analysis revealed that upregulation of miRNA10b could confer an unfavourable factor for OS (HR = 1.853, 95% CI 1.521–2.258; p < 0.01) but not DFS (HR = 1.309, 95% CI 0.699–2.453; p = 0.4) or RFS (HR = 2.692, 95% CIs 0.877–8.265; p = 0.084). It should be noted that the current study measured only DFS. Also, Wang et al. [53] undertook a meta-analysis of the role of miRNA10b in the prognosis of breast cancer and deduced that high miRNA10b expression in patients with breast cancer was significantly associated with poor DFS. Two previous studies have reported the prognostic significance of miRNA16 in melanoma patients [41, 50], both of which have demonstrated the lower expression levels of miRNA16 in melanoma patients. Hence, the survival outcome could not be determined by the endpoint results alone. This subgroup meta-analysis provides further evidence on standardisation of the clinical utility of miRNAs.
A network of miRNAs are capable of controlling key signalling pathways responsible for growth and development of resistance to targeted drug therapies [59]. Immunotherapy plays a crucial role in melanoma which is implied on CTLA4 (cytotoxic T lymphocyte-associated protein 4) and PD1/PD-L1 (programmed cell death-1/programmed death-ligand 1) [60]. Targeted therapies with MAPK (mitogen-activated protein kinase) pathway kinase inhibitors such as BRAF and NRAS mutations are among the major oncogenic drivers of melanoma proliferation and survival [61]. da Silveira Nogueira Lima et al. [62] conducted a meta-analysis on BRAF- or MAPK kinase (MEK)-targeted therapies for advanced melanoma from electronic databases. BRAFi–MEKi (BRAF inhibitor–MEK inhibitor) combination (HR = 0.50, 95% confidence interval 0.34–0.74) and the PD-1 inhibitor (PD-1i) (HR = 0.52, 95% confidence interval 0.36–0.75) have shown a better prognostic effect in melanoma patients. The current meta-analysis results on miRNA prognosis have been associated with worse prognostic effect in melanoma patients, as discussed previously, since the treatment alone does not produce a better indication of the melanoma patients’ risk and recurrence. The other molecular pathways and genetic factors may play a crucial role in the developmental of melanoma progress. In this study, miRNA has shown an association with melanoma patients’ survival. Hence, this information will help future researchers to focus more on miRNA signatures and prognosticators that are considered to add more value to the management of melanoma patients.
4.1 Strength of the Current Study
This meta-analysis has several strengths. The studies chosen for the systematic review and meta-analysis are recently published studies across the globe. The studies chosen in this analysis have various survival endpoints (OS, DFS, RFS, MFS, PFS and PRS) and this enables analysis of all of the included melanoma survival data. The sample size in the selected studies was more than 50, which is a reasonable sample size (subjective). In addition, the inclusion of 2669 melanoma patients provides statistical strengths for robust meta-analyses and subgroup analyses. The systematic review will help us to understand the various modality and demographic characteristics of each study. It also has three subgroup analyses. Both the upregulated and downregulated miRNA expression was studied individually, and hence it should be easy to compare the death risk of melanoma patients with the mean effect size of the plot. In addition, the studies included in the analysis meet the requirements of the PRISMA guidelines. Finally, the quality assessment tool was analysed and displayed reasonable methodological quality for the majority of the studies.
4.2 Limitations of the Meta-analysis
There are a few limitations of this study. This meta-analysis is based on previously published studies, not on individual melanoma patient data. Publication bias exists and could not be avoided in the observational studies. The diverse source of the samples in the included studies and different follow-up periods may result in publication bias. However, the imputed and observed point estimates using the Trim and Fill method indicates robustness. Since a few studies did not provide HR values, we calculated the HR and 95% CI values from the KM curve, and hence the marginal error in the publication bias may exist. Another limitation arises due to non-inclusion of four studies in the quantitative analysis, though the HR and 95% CI values are provided. This problem occurs because the miRNA expressions seem imprecise, requiring further validation. Consequently, data on local, regional and distance recurrence values were not available for all included studies. Since there was non-availability of prognostic results for sunlight exposure and ulceration and other histological sites/TNM staging, we could not perform additional subgroup analysis or meta-regression. In addition, miRNA expression was detected via various platforms such as microarray profiling, TaqMan® assay, quantitative RT-PCR (qRT-PCR) and Fluidigm Real-Time PCR is a digital PCR (BiomarkTM system, USA), which may also influence the variation in results, and hence publication bias may exist.
5 Conclusion
Our study suggests that miRNA expression is associated with poor survival prognosis in melanoma patients. However, following total meta-analysis, the results indicated a worse prognosis, making our findings inconclusive. Our study suggests that current and future researchers should focus more on prospective large-scale cohort studies to validate the prognostic significance of miRNAs. Further clinical investigation with a large-scale sample size will help determine a more accurate impact on the significance of miRNA in melanoma survival outcomes and its use as a biomarker to predict diagnosis and appropriate therapeutic measures in melanoma patients.
References
Howell PM Jr, Li X, Riker AI, Xi Y. MicroRNA in melanoma. Ochsner J. 2010;10(2):83–92.
Lucas R, Norval M, Neale R, Young A, De Gruijl F, Takizawa Y, et al. The consequences for human health of stratospheric ozone depletion in association with other environmental factors. Photochem Photobiol Sci. 2015;14(1):53–87.
World Health Organization. Skin cancers. http://www.who.int/uv/health/uv_health2/en/index1.html. Accessed 18 Aug 2018.
Dar AA, Majid S, de Semir D, Nosrati M, Bezrookove V, Kashani-Sabet M. miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J Biol Chem. 2011;286(19):16606–14.
Mueller DW, Rehli M, Bosserhoff AK. miRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. J Investig Dermatol. 2009;129(7):1740–51.
Peng Q, Zhang X, Min M, Zou L, Shen P, Zhu Y. The clinical role of microRNA-21 as a promising biomarker in the diagnosis and prognosis of colorectal cancer: a systematic review and meta-analysis. Oncotarget. 2017;8(27):44893.
Deng Z, Hao J, Lei D, He Y, Lu L, He L. Pivotal MicroRNAs in melanoma: a mini-review. Mol Diagn Ther. 2016;20(5):449–55.
Saldanha G, Potter L, Lee YS, Watson S, Shendge P, Pringle JH. MicroRNA-21 expression and its pathogenetic significance in cutaneous melanoma. Melanoma Res. 2016;26(1):21–8.
Wang K, Zhang Z-W. Expression of miR-203 is decreased and associated with the prognosis of melanoma patients. Int J Clin Exp Pathol. 2015;8(10):13249.
Li J-Y, Zheng L-L, Wang T-T, Hu M. Functional annotation of metastasis-associated microRNAs of melanoma: a meta-analysis of expression profiles. Chin Med J (Engl). 2016;129(20):2484–90.
Jayawardana K, Schramm S-J, Tembe V, Mueller S, Thompson JF, Scolyer RA, et al. Identification, review, and systematic cross-validation of microRNA prognostic signatures in metastatic melanoma. J Investig Dermatol. 2016;136(1):245–54.
Aris M, Barrio MM. Combining immunotherapy with oncogene-targeted therapy: a new road for melanoma treatment. Front Immunol. 2015;6:46.
American Cancer Society. Cancer facts & figures 2012. American cancer society (ACS). Atlanta: American Cancer Society; 2012. p. 2012.
Hanniford D, Zhong J, Koetz L, Gaziel-Sovran A, Lackaye DJ, Shang S, et al. A miRNA-based signature detected in primary melanoma tissue predicts development of brain metastasis. Clin Cancer Res. 2015;21(21):4903–12.
Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097.
US Department of Health & Human Services. Study quality assessment tools. https://www.nhlbi.nih.gov/health-topics/study-quality-assessment-tools.
Borenstein M, Hedges LV, Higgins JP, Rothstein HR. Effect sizes based on means. In: Introduction to meta-analysis. 1st edn. Hoboken: John Wiley; 2009. p. 21–32.
Coe R. It’s the effect size, stupid: what effect size is and why it is important. Annual Conference of the British Educational Research Association; 12–14 Sep 2002; Exeter.
Huedo-Medina TB, Sánchez-Meca J, Marín-Martínez F, Botella J. Assessing heterogeneity in meta-analysis: Q statistic or I 2 index? Psychol Methods. 2006;11(2):193.
Thorlund K, Imberger G, Johnston BC, Walsh M, Awad T, Thabane L, et al. Evolution of heterogeneity (I 2) estimates and their 95% confidence intervals in large meta-analyses. PLoS One. 2012;7(7):e39471.
Rosenthal R. The file drawer problem and tolerance for null results. Psychol Bull. 1979;86(3):638–41.
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;1088–101.
Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–34.
Duval S, Tweedie R. Trim and fill: a simple funnel-plot–based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56(2):455–63.
Li P, He Q-Y, Luo C-Q, Qian L-Y. Circulating miR-221 expression level and prognosis of cutaneous malignant melanoma. Med Sci Monit. 2014;20:2472–7.
Li N. Low expression of Mir-137 predicts poor prognosis in cutaneous melanoma patients. Med Sci Monit. 2016;22:140–4.
Lin N, Zhou Y, Lian X, Tu Y. Expression of microRNA-106b and its clinical significance in cutaneous melanoma. Genet Mol Res. 2015;14(4):16379–85.
Nguyen T, Kuo C, Nicholl MB, Sim M-S, Turner RR, Morton DL, et al. Downregulation of microRNA-29c is associated with hypermethylation of tumor-related genes and disease outcome in cutaneous melanoma. Epigenetics. 2011;6(3):388–94.
Armand-Labit V, Meyer N, Casanova A, Bonnabau H, Platzer V, Tournier E, et al. Identification of a circulating microRNA profile as a biomarker of metastatic cutaneous melanoma. Acta Derm Venereol. 2016;96(1):29–34.
Fleming NH, Zhong J, da Silva IP, Vega-Saenz de Miera E, Brady B, Han SW, et al. Serum-based miRNAs in the prediction and detection of recurrence in melanoma patients. Cancer. 2015;121(1):51–9.
Luan W, Qian Y, Ni X, Bu X, Xia Y, Wang J, et al. miR-204-5p acts as a tumor suppressor by targeting matrix metalloproteinases-9 and B-cell lymphoma-2 in malignant melanoma. OncoTargets Ther. 2017;10:1237–46.
Pencheva N, Tran H, Buss C, Huh D, Drobnjak M, Busam K, et al. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. 2012;151(5):1068–82.
Bai M, Zhang H, Si L, Yu N, Zeng A, Zhao R. Upregulation of serum miR-10b is associated with poor prognosis in patients with melanoma. J Cancer. 2017;8(13):2487–91.
Cui L, Li Y, Lv X, Li J, Wang X, Lei Z, et al. Expression of MicroRNA-301a and its functional roles in malignant melanoma. Cell Physiol Biochem. 2016;40(1–2):230–44.
Fattore L, Mancini R, Acunzo M, Romano G, Laganà A, Pisanu ME, et al. miR-579-3p controls melanoma progression and resistance to target therapy. Proc Natl Acad Sci USA. 2016;113(34):E5005–13.
Friedman EB, Shang S, de Miera EV, Fog JU, Teilum MW, Ma MW, et al. Serum microRNAs as biomarkers for recurrence in melanoma. J Transl Med. 2012;10(1):155.
Guo S, Guo W, Li S, Dai W, Zhang N, Zhao T, et al. Serum miR-16: a potential biomarker for predicting melanoma prognosis. J Investig Dermatol. 2016;136(5):985–93.
Guo W, Wang H, Yang Y, Guo S, Zhang W, Liu Y, et al. Down-regulated miR-23a contributes to the metastasis of cutaneous melanoma by promoting autophagy. Theranostics. 2017;7(8):2231–49.
Hanna JA, Hahn L, Agarwal S, Rimm DL. In situ measurement of miR-205 in malignant melanoma tissue supports its role as a tumor suppressor microRNA. Lab Investig. 2012;92(10):1390–7.
Jiang L, Lv X, Li J, Li J, Li X, Li W, et al. The status of microRNA-21 expression and its clinical significance in human cutaneous malignant melanoma. Acta Histochem. 2012;114(6):582–8.
Ono S, Oyama T, Lam S, Chong K, Foshag LJ, Hoon DS. A direct plasma assay of circulating microRNA-210 of hypoxia can identify early systemic metastasis recurrence in melanoma patients. Oncotarget. 2015;6(9):7053–64.
Saldanha G, Potter L, Shendge P, Osborne J, Nicholson S, Yii N, et al. Plasma microRNA-21 is associated with tumor burden in cutaneous melanoma. J Investig Dermatol. 2013;133(5):1381–4.
Saldanha G, Elshaw S, Sachs P, Alharbi H, Shah P, Jothi A, et al. microRNA-10b is a prognostic biomarker for melanoma. Mod Pathol. 2016;29:112–21.
Stark MS, Klein K, Weide B, Haydu LE, Pflugfelder A, Tang YH, et al. The prognostic and predictive value of melanoma-related microRNAs using tissue and serum: a microRNA expression analysis. EBioMedicine. 2015;2(7):671–80.
Tembe V, Schramm SJ, Stark MS, Patrick E, Jayaswal V, Tang YH, et al. MicroRNA and mRNA expression profiling in metastatic melanoma reveal associations with BRAF mutation and patient prognosis. Pigment Cell Melanoma Res. 2015;28(3):254–66.
Tian R, Liu T, Qiao L, Gao M, Li J. Decreased serum microRNA-206 level predicts unfavorable prognosis in patients with melanoma. Int J Clin Exp Pathol. 2015;8(3):3097–103.
Zhang G, Ai D, Yang X, Ji S, Wang Z, Feng S. MicroRNA-610 inhibits tumor growth of melanoma by targeting LRP6. Oncotarget. 2017;8(57):97361–70.
Taylor MA. Circulating microRNAs as biomarkers and mediators of cell–cell communication in cancer. Biomedicines. 2015;3(4):270–81.
Song CJ, Chen H, Chen LZ, Ru GM, Guo JJ, Ding QN. The potential of microRNAs as human prostate cancer biomarkers: a meta-analysis of related studies. J Cell Biochem. 2018;119(3):2763–86.
Lubov J, Maschietto M, Ibrahim I, Mlynarek A, Hier M, Kowalski LP, et al. Meta-analysis of microRNAs expression in head and neck cancer: uncovering association with outcome and mechanisms. Oncotarget. 2017;8(33):55511–24.
Yang J, Han S, Huang W, Chen T, Liu Y, Pan S, et al. A meta-analysis of microRNA expression in liver cancer. PLoS One. 2014;9(12):e114533.
Zhou X, Wang X, Huang Z, Wang J, Zhu W, Shu Y, et al. Prognostic value of miR-21 in various cancers: an updating meta-analysis. PLoS One. 2014;9(7):e102413.
Wang Y, Zhang Y, Pan C, Ma F, Zhang S. Prediction of poor prognosis in breast cancer patients based on microRNA-21 expression: a meta-analysis. PLoS One. 2015;10(2):e0118647.
Frampton AE, Krell J, Jamieson NB, Gall TM, Giovannetti E, Funel N, et al. microRNAs with prognostic significance in pancreatic ductal adenocarcinoma: a meta-analysis. Eur J Cancer. 2015;51(11):1389–404.
Fu W, Pang L, Chen Y, Yang L, Zhu J, Wei Y. The microRNAs as prognostic biomarkers for survival in esophageal cancer: a meta-analysis. Sci World J. 2014;2014:523979.
Xie Y, Wu J. MicroRNA-21 as prognostic molecular signatures in oral cancer: a meta-analysis. Int J Clin Exp Med. 2017;10(7):9848–56.
Jamali Z, Aminabadi NA, Attaran R, Pournagiazar F, Oskouei SG, Ahmadpour F. MicroRNAs as prognostic molecular signatures in human head and neck squamous cell carcinoma: a systematic review and meta-analysis. Oral Oncol. 2015;51(4):321–31.
Huang Q, Song Q, Zhong W, Chen Y, Liang L. MicroRNA-10b and the clinical outcomes of various cancers: a systematic review and meta-analysis. Clin Chim Acta. 2017;474:14–22.
Fattore L, Costantin S, Malpicci D, Ruggiero CF, Ascierto PA, Croce CM, et al. MicroRNAs in melanoma development and resistance to target therapy. Oncotarget. 2017;8(13):22262–78.
Ascierto PA, Marincola FM. 2015: the year of anti-PD-1/PD-L1s against melanoma and beyond. EBioMedicine. 2015;2:92–3.
Menzies AM, Long GV. Systemic treatment for BRAF mutant melanoma: where do we go next? Lancet Oncol. 2014;15:e371–81.
da Silveira Nogueira Lima JP, Georgieva M, Haaland B, de Lima Lopes GD. A systematic review and network meta-analysis of immunotherapy and targeted therapy for advanced melanoma. Cancer Med. 2017;6(6):1143–53.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
SS, MRM, AD, SD, GKM and RJ declare that they have no conflict of interest.
Funding
This research received no specific Grant from any funding agency in the public, commercial or not-for-profit sectors.
Ethics Approval and Consent to Participate
Ethical approval is not a requirement because all data in this review were retrieved from published studies. Since there is no any specific direct patient involvement, human research ethical committee approval is not required.
Rights and permissions
About this article
Cite this article
Sabarimurugan, S., Madurantakam Royam, M., Das, A. et al. Systematic Review and Meta-analysis of the Prognostic Significance of miRNAs in Melanoma Patients. Mol Diagn Ther 22, 653–669 (2018). https://doi.org/10.1007/s40291-018-0357-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-018-0357-5