Abstract
Myxofibrosarcoma (MFS), an aggressive soft tissue sarcoma, presents a significant challenge because of its high recurrence rate, distal metastasis, and complex genetic background. Although surgical resection is the standard treatment for MFS, the outcomes are unsatisfactory and effective non-surgical treatment strategies, including drug therapy, are urgently warranted. MFS is a rare tumor that requires comprehensive preclinical research to develop promising drug therapies; however, only two MFS cell lines are publicly available worldwide. The present study reports two novel patient-derived MFS cell lines, NCC-MFS7-C1 and NCC-MFS8-C1. These cell lines have been extensively characterized for their genetic profile, proliferation, spheroid-forming capacity, and invasive behavior, confirming that they retain MFS hallmarks. Furthermore, we conducted comprehensive drug screening against these cell lines and six others previously established in our laboratory to identify potential therapeutic candidates for MFS. Among the screened agents, actinomycin D, bortezomib, and romidepsin demonstrated considerable antiproliferative effects that were superior to those of doxorubicin, a standard drug, highlighting their potential as novel drugs. In conclusion, NCC-MFS7-C1 and NCC-MFS8-C1 are valuable research resources that contribute to the understanding of the pathogenesis and development of novel therapies for MFS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Myxofibrosarcoma (MFS), a notably aggressive soft tissue sarcoma, is a malignant fibroblastic neoplasm characterized by a variable myxoid stroma and pleomorphic cells [1]. Tumors are most commonly located in the limbs, with a particular propensity for the lower extremities [2, 3]. MFS predominantly affects older adults, with a peak incidence in the sixth to eighth decades of life [4, 5]. Notably, surgical resection is the cornerstone of curative treatment for MFS [2, 6]. However, high rates of local recurrence (30–40%) and distant metastasis (15–35%) significantly affect the 5-year survival prospects, which hover around 60–70% [1, 2, 4, 7, 8]. These data highlight the need for comprehensive multidisciplinary treatment approaches that extend beyond surgery and include innovative drug therapies. Despite the clear necessity of such strategies, effective multidisciplinary methods, including established drug regimens, remain elusive.
The intricate genetic landscape of MFS underpins its aggressive clinical behavior and contributes to significant challenges in the development of effective therapeutic strategies. Key genetic alterations observed in MFS include mutations and copy number alterations in pivotal tumor suppressor genes and cell cycle regulators, notably TP53, RB1, CDKN2A, and CDKN2B [9, 10]. Moreover, frequent amplifications on chromosome 5p, which harbors oncogenes such as TRIO and RICTOR, have been implicated in driving the malignant potential of MFS [10]. These genetic aberrations not only confer a high degree of biological aggressiveness but also foster a heterogeneous tumor environment, complicating personalized treatment approaches.
Cell lines are indispensable in cancer research as they provide a foundational platform for elucidating disease mechanisms and evaluating therapeutic interventions [11, 12]. Their importance is amplified in the context of rare diseases such as MFS, where conducting extensive clinical trials is challenging owing to the limited patient population. This difficulty underscores the value of robust preclinical models, such as cell lines, for advancing our understanding and treatment of such diseases [13]. Despite the critical role of these models, the Cellosaurus database, a comprehensive resource for cell line information, lists only 16 MFS cell lines, with only two publicly available (Supplementary Table 1) [14]. This scarcity of MFS cell lines severely restricts research progress and the development of targeted therapies. Therefore, additional cell lines are strongly required to promote further research and improve the therapeutic outcomes of MFS.
This study introduced two novel MFS cell lines, NCC-MFS7-C1 and NCC-MFS8-C1, established from tumor samples resected from two different patients. We meticulously characterized the NCC-MFS7-C1 and NCC-MFS8-C1 cell lines and examined their genetic profiles, proliferation rates, spheroid formation capabilities, and invasive behaviors. Additionally, we expanded our investigation to include drug screening across these novel cell lines and six others previously established in our laboratory with the aim of identifying potential therapeutic indications for MFS [15,16,17,18,19,20].
Materials and methods
Patient data
Case 1
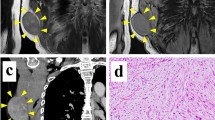
The donor patient was a 74-year-old female with myxofibrosarcoma. She visited a previous hospital because of a soft mass on her left back. Magnetic resonance imaging (MRI) revealed a soft tissue mass 9 × 8 cm in size bordering the left thoracic back region (Fig. 1A–C). The patient was referred to the National Cancer Center Hospital, Tokyo, Japan, for further treatment. The tumor was diagnosed as myxofibrosarcoma based on a needle biopsy. The patient underwent wide resection of the left thoracic back tumor, and tumor tissues obtained at the time of surgery were used to establish the cell line. Histologically, the tumor consisted of pleomorphic spindled cells with bizarre hyperchromatic nuclei in prominent myxoid stroma. High mitotic activity and abnormal mitosis were observed (Fig. 1D).
Clinical and pathological data of NCC-MFS7-C1. Magnetic resonance imaging demonstrated an enhanced mass of the left thoracic back (A: T2-weighted axial view, B: gadolinium-enhanced T1-weighted axial view, C: gadolinium-enhanced T1-weighted sagittal view). The histological image of the tumor showed morphology of myxofibrosarcoma (D: H&E)
Case 2
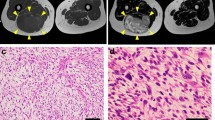
The donor patient was a 79-year-old female with a recurrent myxofibrosarcoma. She had undergone a wide resection of a right knee tumor four years ago at the National Cancer Center Hospital in Tokyo, Japan. The tumor was diagnosed as a myxofibrosarcoma. Four years after surgery, she developed a soft mass in her right leg. MRI revealed an enhanced mass in the right leg (Fig. 2A–C). The tumor was diagnosed as a recurrent myxofibrosarcoma based on a needle biopsy. The patient underwent wide resection of the right leg tumor, and tumor tissues obtained at the time of surgery was used to establish the cell line. Histologically, the tumor comprised of spindle-shaped cells with mild nuclear atypia in the myxoid stroma (Fig. 2D).
Clinical and pathological data of NCC-MFS8-C1. Magnetic resonance imaging demonstrated an enhanced mass of the right leg (A: T2-weighted axial view, B: gadolinium-enhanced T1-weighted axial view, C: gadolinium-enhanced T1-weighted sagittal view). The histological image of the tumor showed morphology of myxofibrosarcoma (D: H&E)
Histological analysis
Histology examination was performed on 4-μm-thick sections from a representative paraffinized tumor sample. The sections were deparaffinized and stained with hematoxylin and eosin (H&E).
Establishment of cell lines
Surgically excised MFS tumor tissues were sectioned into smaller fragments. These samples were then processed for digestion and homogenization in the gentleMACS™ Octo Dissociator with Heaters (Miltenyi Biotec B.V. & Co. KG, Bergisch Gladbach, Germany) utilizing the Tumor Cell Dissociation Kit human (Miltenyi Biotec) within the gentleMACS™ C Tube (Miltenyi Biotec) for 30 min at a temperature of 37 °C. Subsequently, the cells were cultivated on 60 mm collagen-type I-coated dishes (Sumitomo Bakelite Co. Ltd., Tokyo, Japan) in a DMEM/F-12 mixture (Thermo Fisher Scientific Inc., MA, USA) supplemented with GlutaMAX (Thermo Fisher Scientific Inc.), 5% inactivated fetal bovine serum, and supplements including Y-27632 (ROCK inhibitor; Selleck Chemicals, Houston, TX, USA), basic fibroblast growth factor, epidermal growth factor, insulin, hydrocortisone, penicillin, and streptomycin. This culture environment was kept at 37 °C with a 5% CO2 atmosphere, with periodic media changes and cell monitoring using microscopy. When the cells reached an appropriate density, they were washed, detached with Trypsin–EDTA (Nacalai Tesque Inc., Kyoto, Japan), and relocated to continue cultivation under the same optimal conditions.
Authentication of the cell lines
DNA was isolated from the tumor tissues and corresponding cell cultures for authentication and quality assessment. The extraction was facilitated using the Wizard® Genomic DNA Purification Kit (Promega Co., Madison, WI, USA) and the Qiagen DNeasy Blood and Tissue Kit (QIAGEN N.V., Hilden, Germany), while the DNA quantity was evaluated with a NanoDrop 8000 spectrophotometer (Thermo Fisher Scientific Inc.). The authenticity of the cell lines was verified through an analysis of short tandem repeats (STR) across 10 specific loci, employing the GenePrint 10 system (Promega Co.) in conjunction with the 3500xL Genetic Analyzer (Thermo Fisher Scientific Inc.).
Quality control of the cell lines
The presence of mycoplasma was investigated by isolating and fragmenting DNA from cells, using an e-Myco Mycoplasma PCR Detection Kit (Intron Biotechnology, Gyeonggi-do, Korea). Following polymerase chain reaction amplification, the DNA fragments were separated via 1.5% agarose gel electrophoresis and visualized by staining with SYBR Safe (Invitrogen). Images were assessed using an Amersham Imager 600 (GE Healthcare Biosciences, Little Chalfont, UK).
Genetic analysis
Single nucleotide polymorphism (SNP) array genotyping was performed using the Infinium OmniExpressExome-8 version 1.4 BeadChip (Illumina, San Diego, CA, USA), adhering to both the guidelines provided by the manufacturer and the methodologies outlined in previously published studies. Genomic DNA from both established cell lines and primary tumors was amplified before being subjected to hybridization on array slides using the iScan system (Illumina). The assessment of Log R ratios and B allele frequencies was carried out using Genome Studio 2011.1 and cnvPartition v3.2.0 software from Illumina, in conjunction the R software (version 4.0.3, provided by the R Foundation for Statistical Computing, http://www.R-project.org) and the DNAcopy package (version 1.64.0, from Bioconductor, http:// bioconductor.org). Regions within chromosomes displaying copy numbers exceeding three or falling below one were identified as amplifications or deletions, respectively. Genes revealing copy number alterations (CNAs) were annotated using the biomaRt package (version 2.46.0, Bioconductor) and cross-referenced with data from the “Cancer Gene Census” in the Catalog of Somatic Mutations In Cancer database (GRCh 37 version 91).
Cell proliferation assay
Cells were initially plated at a density of 2.5 × 104 cells/well in 24-well plates and monitored over 4 days. Cell proliferation was quantified at 24-h intervals using a Cell Counting Kit-8 (DOJINDO LABORATORIES, Kumamoto, Japan). Cells’ doubling time was calculated based on the growth curves. Each procedures was conducted in triplicate.
Spheroid formation assay
The methodology for assessing spheroid formation aligned with that used inn previous studies. Initially, cells were plated at a density of 1 × 105 cells/well in 96-well clear round bottom ultra-low attachment microplates (Corning Inc., Corning, NY, USA), and their ability to form spheroids was verified through microscopy (KEYENCE Co., Osaka, Japan). After a cultivation period of three days, the spheroids were extracted, encapsulated in iPGell (GenoStaff Co. Ltd., Tokyo, Japan), and fixed in a 10% neutral buffered formalin solution. For microscopic analysis, the encapsulated spheroids were embedded in paraffin, sectioned into slices of 4 μm thickness, stained with H&E, and examined under a microscope.
Invasion assay
The invasive potential of the established MFS cell lines was investigated using the real-time cell analyzer xCELLigence (Agilent Technologies Inc., Santa Clara, CA, USA). The upper chamber was prepared with a Matrigel coating (Corning Inc.) and seeded with 4.0 × 104 cells in a basal DMEM/F12 solution while the lower chamber was filled with a nutritive mix of DMEM/F12 with supplements. Electronic sensors, positioned underneath the membrane between the chambers, captured the impedance of migrating cells every 15 min and translated it into data over 72 h to quantify cell invasion. This method utilized the MG63 osteosarcoma cell line (Japanese Collection of Research Bioresources Cell Bank, Osaka, Japan) as a control.
Drug screening assay
The antitumor potential of 214 agents, including drugs approved by the Food and Drug Administration, was evaluated (Supplementary Table 2). Cells were seeded in 384-well plates at a density of 5.0 × 103 cells per well and incubated for 1 day before treatment. Following this period, each drug was applied to duplicate wells and incubated for an additional 3 days. Cell growth and viability were determined using the Cell Counting Kit-8 (DOUJINDO LABORATORIES) by comparing the outcomes with those of a dimethyl sulfoxide control. These results integrated with the findings of previous studies on NCC-MFS1-C1, NCC-MFS2-C1, NCC-MFS3-C1, NCC-MFS4-C1, NCC-MFS5-C1, and NCC-MFS6-C1. The analysis involved quantile normalization and unsupervised hierarchical clustering using R (version 4.0.3, limma package version 3.46.0, Bioconductor) and the gplots package (version 3.1.0, CRAN, https://cran.r-project.org).
To ascertain the half-maximal inhibitory concentration (IC50) values, 24 drugs showing promising results in preliminary screens were tested across a range of ten concentrations. IC50 values were calculated by analyzing these concentration-survival relationships via logistic regression, utilizing GraphPad Prism 9.11 (GraphPad Software, San Diego, CA, USA).
Results
Authentication and quality control of the established MFS cell lines
NCC-MFS7-C1 and NCC-MFS8-C1 cell lines originating from patients 1 and 2, respectively, were established and sustained for more than 20 passages over 3 months. Authentication of these cell lines was conducted by analyzing 10 microsatellite loci, which showed that the STR profiles were almost identical to those of primary tumor tissues (Table 1 and Supplementary Fig. 1). Additionally, no detection of mycoplasma-specific DNA was detected, confirming the absence of mycoplasma contamination.
Characterization of the established MFS cell lines
Analysis of SNP arrays for the established cell lines, NCC-MFS7-C1 and NCC-MFS8-C1, revealed specific chromosomal CNAs. In NCC-MFS7-C1, deletions were noted on chromosomes 5q, 6p, 16q, and 17p13.1, whereas amplifications were observed on chromosomes 7p, 7q, 17p11.2, 18q, and 19q. Conversely, NCC-MFS8-C1 exhibited deletions on chromosomes 1p, 3p, 6p, 9p, and 13q with no observed amplifications. Notably, deletions affected tumor suppressor genes such as TP53 on 17p13.1 in NCC-MFS7-C1 and both CDKN2A and CDKN2B on 9p21.3 in NCC-MFS8-C1 cells (Fig. 3, Table 2, Supplementary Table 3 and 4).
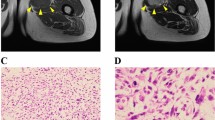
The NCC-MFS7-C1 and NCC-MFS8-C1 cell lines showed an elongated spindle or polygonal appearance (Fig. 4A, B) and formed spheroids when cultured on low-attachment plates (Fig. 4C, D). The spheroids contained oval and pleomorphic cells with atypical nuclei. Growth curve analysis showed that the population doubling times for NCC-MFS7-C1 and NCC-MFS8-C1 cells were 29 h and 68 h, respectively (Fig. 4E). Furthermore, both cell lines showed higher invasive capacity than did the MG63 osteosarcoma cell line (Fig. 4F).
Characterization of NCC-MFS7-C1 and NCC-MFS8-C1. Microscopic appearance of A NCC-MFS7-C1 (passage 25) and B NCC-MFS8-C1 (passage 25) cells in 2-dimensional culture conditions. Hematoxylin and eosin-stained spheroid sections of C NCC-MFS7-C1 (passage 26) and D NCC-MFS8-C1 (passage 26) cells were fabricated in 96-well low-attachment round-bottom plates. Growth curves of NCC-MFS7-C1 (passage 22) and NCC-MFS8-C1 (passage 22) cells (E). Each point represents the mean ± standard deviation (n = 3). The invasion capabilities of NCC-MFS7-C1 (passage 23) and NCC-MFS8-C1 (passage 23) cells were observed using a Real-Time Cell Analyzer (F). The MG63 osteosarcoma cell line was used as the control
Sensitivity of the antitumor agents to the established MFS cell lines
Extensive analysis was conducted to evaluate the effect of the 214 antitumor compounds on the proliferation of NCC-MFS7-C1 and NCC-MFS8-C1 cells (Supplementary Table 5). This included a comparison with data from six previously established MFS cell lines in our laboratory. Antitumor agents were categorized into effective (Cluster A) and ineffective (Cluster B) groups based on their efficacy (Fig. 5A and Supplementary Table 6). Cluster A had a higher proportion of cytotoxic drugs than did cluster B, including among all topoisomerase inhibitors were included in cluster A (Fig. 5B, C). The distribution of tyrosine kinase inhibitors across both clusters was similar; however, most anaplastic lymphoma kinase inhibitors were grouped into cluster A (Fig. 5D). Molecular targeted drugs were predominantly distributed in cluster A, with all histone deacetylase (HDAC) inhibitors belonging to cluster A (Fig. 5E).
Drug screening assay of myxofibrosarcoma cell lines. A The antitumor agents were categorized into two groups according to their antitumor effect: cluster A as the effective group and cluster B as the ineffective group. (B–E) The proportion of each agent type belonging to each cluster. The graphs were obtained after the normalization of the number of agents. Data regarding NCC-MFS1-C1, NCC-MFS2-C1, NCC-MFS3-C1, NCC-MFS4-C1, NCC-MFS5-C1, and NCC-MFS6-C1 were previously reported. Passage number of NCC-MFS7-C1 and NCC-MFS8-C1 were 23 and 22 respectively. MFS, myxofibrosarcoma
Further investigation into the screened antitumor agents involved calculating IC50 values for each, along with values for six other MFS cell lines previously identified (Supplementary Table 7). Notably, actinomycin D (an RNA polymerase and topoisomerase inhibitor) and bortezomib (a proteasome inhibitor) showed lower IC50 values than doxorubicin, the standard treatment agent for sarcomas, in most cell lines (7/8). Moreover, romidepsin (an HDAC inhibitor) showed significantly lower IC50 values than doxorubicin in all cell lines (8/8), indicating its potent antitumor activity. Table 3 and Fig. 6 provide a comprehensive view of these IC50 values and corresponding growth curves.
Cell viability of myxofibrosarcoma cell lines at different concentrations of antitumor agents. Antiproliferative effects of romidepsin (A), actinomycin D (B), bortezomib (C), and doxorubicin (D) on MFS cells are shown. Data regarding NCC-MFS1-C1, NCC-MFS2-C1, NCC-MFS3-C1, NCC-MFS4-C1, NCC-MFS5-C1, and NCC-MFS6-C1 were previously reported. Passage number of NCC-MFS7-C1 and NCC-MFS8-C1 were 26 and 24, respectively. Half-maximal inhibitory concentration (IC50) values are summarized in Table 3. MFS, myxofibrosarcoma
Discussion
The establishment of the NCC-MFS7-C1 and NCC-MFS8-C1 cell lines represents a significant advancement in MFS research. Given the highly malignant nature of MFS and the challenges in conducting clinical trials owing to its rarity, the preclinical study is crucial. However, the scarcity of available cell lines poses major challenges. These cell lines offer a valuable resource for deepening our understanding of MFS and exploring novel therapeutic strategies.
Genetic analysis of the newly established cell lines revealed significant alterations in key tumor suppressor genes, including TP53, CDKN2A, and CDKN2B. These findings align with those of previous studies highlighting the prevalence of such mutations in MFS, underscoring the relevance of these cell lines as models with representative genetic mutations in the disease [21,22,23]. However, the absence of CNA in genes such as RB1, ATRX, and HDLBP, which has been reported in other MFS studies, points to the genetic diversity inherent in MFS, further emphasizing the need for a broad range of patient-derived cancer models [9, 24].
Functionally, NCC-MFS7-C1 and NCC-MFS8-C1 cell lines exhibited characteristics typical of MFS, including the ability to consistently proliferate and invade. NCC-MFS7-C1, in particular, demonstrated higher proliferative and invasive capacities than did NCC-MFS8-C1. With an average doubling time of 66.1 h across previously established cell lines in our laboratory, NCC-MFS7-C1 emerged as a relatively fast-growing model, whereas NCC-MFS8-C1 presented a standard growth profile. Both cell lines showed spheroid formation capabilities, positioning them as promising tools for more advanced preclinical experiments.
A comprehensive evaluation of 214 antitumor agents against these cell lines revealed promising results. In particular, actinomycin D, bortezomib, and romidepsin showed superior efficacy than did doxorubicin, the standard drug frequently used to treat MFS and other sarcomas. Although actinomycin D is sometimes utilized in VAC (vincristine, actinomycin D, cyclophosphamide) regimens for sarcoma treatment, bortezomib and romidepsin are rarely employed in managing sarcomas [25, 26]. Initially, bortezomib was approved for multiple myeloma, and romidepsin for cutaneous T-cell lymphoma [27,28,29,30]. Despite a few reports suggesting the potential efficacy of bortezomib and HDAC inhibitors, OBP-801 (not romidepsin), in MFS models, these investigations are preclinical studies, confined to in vitro or in vivo data [31, 32]. Therefore, these agents are promising candidates for MFS treatment that deserve further validation in large-scale drug screening of cell lines and clinical trials.
Our study has three limitations. Although this study is the largest drug screening analysis conducted for MFS, the number of cell lines and drugs used in our study was insufficient, considering those for common cancers [11, 33]. This limitation will be addressed by establishing a more diverse collection of MFS cell lines and conducting larger-scale drug sensitivity assays. Furthermore, although the treatment for metastatic tumors is challenging in MFS, our study did not include cell lines from metastatic lesions. We will require the cell lines from the metastatic sites to develop the novel therapy for the patients with metastasis. In addition, the metastatic potentials of our cell lines were not examined in this study. However, the conventional experiments using animal models are quite artificial as the cells or tumors are inoculated ectopically in the animal body, and subcutaneously transplanted tumors rarely metastasize in the mouse, even when derived from a highly metastatic patient tumor [34]. To address this issue, we require a novel cancer model or a protocol to recapitulate the metastasis in patients with MFS.
In conclusion, this study introduced two novel fibrosarcoma (MFS) cell lines, NCC-MFS7-C1 and NCC-MFS8-C1. These cell lines are crucial for MFS research, as they will improve our understanding of the biological and genetic mechanisms of the disease and identify potential therapeutics. Sensitivity of various antitumor agents suggested that actinomycin D, bortezomib, and romidepsin are effective in treating MFS. These findings clearly indicates that the established cell lines will serve as valuable research models for future large-scale drug screening for MFS.
Data availability
Data will be available under reasonable conditions.
References
board WHOcte. Soft tissue and bone tumours. 5th ed. World Health Organization classification of tumours. World Health Organization International Agency for Research on Cancer; 2020.
Sanfilippo R, Miceli R, Grosso F, Fiore M, Puma E, Pennacchioli E, et al. Myxofibrosarcoma: prognostic factors and survival in a series of patients treated at a single institution. Ann Surg Oncol. 2011;18(3):720–5. https://doi.org/10.1245/s10434-010-1341-4.
Wakely PE Jr. Cytopathology of myxofibrosarcoma: a study of 66 cases and literature review. J Am Soc Cytopathol. 2021;10(3):300–9. https://doi.org/10.1016/j.jasc.2020.09.004.
Dewan V, Darbyshire A, Sumathi V, Jeys L, Grimer R. Prognostic and survival factors in myxofibrosarcomas. Sarcoma. 2012;2012: 830879. https://doi.org/10.1155/2012/830879.
Willems SM, Debiec-Rychter M, Szuhai K, Hogendoorn PC, Sciot R. Local recurrence of myxofibrosarcoma is associated with increase in tumour grade and cytogenetic aberrations, suggesting a multistep tumour progression model. Mod Pathol. 2006;19(3):407–16. https://doi.org/10.1038/modpathol.3800550.
Ghazala CG, Agni NR, Ragbir M, Dildey P, Lee D, Rankin KS, et al. Myxofibrosarcoma of the extremity and trunk: a multidisciplinary approach leads to good local rates of LOCAL control. Bone Joint J. 2016;98-b(12):1682–8. https://doi.org/10.1302/0301-620x.98b12.37568.
van der Horst CAJ, Bongers SLM, Versleijen-Jonkers YMH, Ho VKY, Braam PM, Flucke UE, et al. Overall survival of patients with Myxofibrosarcomas: An epidemiological study. Cancers. 2022;14(5):1102.
Teurneau H, Engellau J, Ghanei I, Vult von Steyern F, Styring E. High recurrence rate of myxofibrosarcoma: the effect of radiotherapy is not clear. Sarcoma. 2019;2019(1):8517371. https://doi.org/10.1155/2019/8517371.
Takeuchi Y, Yoshida K, Halik A, Kunitz A, Suzuki H, Kakiuchi N, et al. The landscape of genetic aberrations in myxofibrosarcoma. Int J Cancer. 2022;151(4):565–77. https://doi.org/10.1002/ijc.34051.
Heitzer E, Sunitsch S, Gilg MM, Lohberger B, Rinner B, Kashofer K, et al. Expanded molecular profiling of myxofibrosarcoma reveals potentially actionable targets. Mod Pathol. 2017;30(12):1698–709. https://doi.org/10.1038/modpathol.2017.94.
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–7. https://doi.org/10.1038/nature11003.
Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013. https://doi.org/10.1093/nar/gks1111.
Kondo T. Current status and future outlook for patient-derived cancer models from a rare cancer research perspective. Cancer Sci. 2021;112(3):953–61. https://doi.org/10.1111/cas.14669.
Bairoch A. The cellosaurus, a cell-line knowledge resource. J Biomol Tech. 2018;29(2):25–38. https://doi.org/10.7171/jbt.18-2902-002.
Kito F, Oyama R, Sakumoto M, Shiozawa K, Qiao Z, Toki S, et al. Establishment and characterization of a novel cell line, NCC-MFS1-C1, derived from a patient with myxofibrosarcoma. Hum Cell. 2019;32(2):214–22. https://doi.org/10.1007/s13577-018-00233-1.
Noguchi R, Yoshimatsu Y, Ono T, Sei A, Hirabayashi K, Ozawa I, et al. Establishment and characterization of NCC-MFS2-C1: a novel patient-derived cancer cell line of myxofibrosarcoma. Hum Cell. 2021;34(1):246–53. https://doi.org/10.1007/s13577-020-00420-z.
Tsuchiya R, Yoshimatsu Y, Noguchi R, Sin Y, Ono T, Sei A, et al. Establishment and characterization of NCC-MFS3-C1: a novel patient-derived cell line of myxofibrosarcoma. Hum Cell. 2021;34(4):1266–73. https://doi.org/10.1007/s13577-021-00548-6.
Yoshimatsu Y, Noguchi R, Tsuchiya R, Sin Y, Ono T, Sugaya J, et al. Establishment and characterization of NCC-MFS4-C1: a novel patient-derived cell line of myxofibrosarcoma. Hum Cell. 2021;34(6):1911–8. https://doi.org/10.1007/s13577-021-00589-x.
Tsuchiya R, Yoshimatsu Y, Noguchi R, Sin Y, Ono T, Akiyama T, et al. Establishment and characterization of NCC-MFS5-C1: A novel patient-derived cell line of myxofibrosarcoma. Cells. 2022;11(2):207. https://doi.org/10.3390/cells11020207.
Yoshimatsu Y, Noguchi R, Sin Y, Tsuchiya R, Ono T, Akiyama T, et al. Establishment and characterization of NCC-MFS6-C1: a novel patient-derived cell line of myxofibrosarcoma. Hum Cell. 2022;35(6):1993–2001. https://doi.org/10.1007/s13577-022-00749-7.
Ogura K, Hosoda F, Arai Y, Nakamura H, Hama N, Totoki Y, et al. Integrated genetic and epigenetic analysis of myxofibrosarcoma. Nat Commun. 2018;9(1):2765. https://doi.org/10.1038/s41467-018-03891-9.
Yamashita A, Suehara Y, Hayashi T, Takagi T, Kubota D, Sasa K, et al. Molecular and clinicopathological analysis revealed an immuno-checkpoint inhibitor as a potential therapeutic target in a subset of high-grade myxofibrosarcoma. Virchows Arch. 2022;481(4):1–17. https://doi.org/10.1007/s00428-022-03358-9.
Sambri A, De Paolis M, Spinnato P, Donati DM, Bianchi G. The biology of myxofibrosarcoma: state of the art and future perspectives. Oncol Res Treat. 2020;43(6):314–22. https://doi.org/10.1159/000507334.
Li GZ, Okada T, Kim YM, Agaram NP, Sanchez-Vega F, Shen Y, et al. Rb and p53-deficient myxofibrosarcoma and undifferentiated pleomorphic sarcoma require Skp2 for survival. Cancer Res. 2020;80(12):2461–71. https://doi.org/10.1158/0008-5472.Can-19-1269.
Nakano K, Ae K, Matsumoto S, Takahashi S. The VAC regimen for adult rhabdomyosarcoma: Differences between adolescent/young adult and older patients. Asia Pac J Clin Oncol. 2020;16(2):e47–52. https://doi.org/10.1111/ajco.13279.
Özkan A, Bayram İ, Sezgin G, Mirioğlu A, Küpeli S. Efficacy of replacing actinomycin-D with carboplatin in Ewing sarcoma consolidation treatment: Single-center experience. J Bone Oncol. 2022;35: 100435. https://doi.org/10.1016/j.jbo.2022.100435.
Sharma A, Preuss CV. Bortezomib. StatPearls. Treasure Island (FL): StatPearls Publishing Copyright © 2024, StatPearls Publishing LLC; 2024.
Cvek B. Proteasome inhibitors. Prog Mol Biol Transl Sci. 2012;109:161–226. https://doi.org/10.1016/b978-0-12-397863-9.00005-5.
Grant C, Rahman F, Piekarz R, Peer C, Frye R, Robey RW, et al. Romidepsin: a new therapy for cutaneous T-cell lymphoma and a potential therapy for solid tumors. Expert Rev Anticancer Ther. 2010;10(7):997–1008. https://doi.org/10.1586/era.10.88.
Shimony S, Horowitz N, Ribakovsky E, Rozovski U, Avigdor A, Zloto K, et al. Romidepsin treatment for relapsed or refractory peripheral and cutaneous T-cell lymphoma: Real-life data from a national multicenter observational study. Hematol Oncol. 2019;37(5):569–77. https://doi.org/10.1002/hon.2691.
Li CF, Wang JM, Kang HY, Huang CK, Wang JW, Fang FM, et al. Characterization of gene amplification-driven SKP2 overexpression in myxofibrosarcoma: potential implications in tumor progression and therapeutics. Clin Cancer Res. 2012;18(6):1598–610. https://doi.org/10.1158/1078-0432.Ccr-11-3077.
Kawarazaki A, Horinaka M, Yasuda S, Kawashima H, Numajiri T, Sakai T. The HDAC inhibitor OBP-801 suppresses the growth of myxofibrosarcoma cells. J buon. 2020;25(1):464–71.
Bashi AC, Coker EA, Bulusu KC, Jaaks P, Crafter C, Lightfoot H, et al. Large-scale pan-cancer cell line screening identifies actionable and effective drug combinations. Cancer Discov. 2024. https://doi.org/10.1158/2159-8290.Cd-23-0388.
Fidler IJ. Critical factors in the biology of human cancer metastasis: twenty-eighth G.H.A Clowes memorial award lecture. Cancer Res. 1990;50(19):6130–8.
Acknowledgements
We thank Drs. S. Fukushima, S. Osaki, K. Ogura (Department of Musculoskeletal Oncology and Rehabilitation Medicine, National Cancer Center Hospital) ; Dr. T. Funada, and Dr. Y. Kobayashi (Department of Diagnostic Pathology, National Cancer Center Hospital); and the National Cancer Center Hospital for sampling the tumor tissue specimens from the surgically resected materials. We appreciate the technical assistance provided by Mr. Y. Ohno (Division of Rare Cancer Research, National Cancer Center Research Institute) and Mrs. Y. Shiotani (Central Animal Division, National Cancer Center Research Institute). We would like to thank Editage (www.editage.jp) for providing English language editing services and for their constructive comments on this manuscript. The Fundamental Innovative Oncology Core of the National Cancer Center provided technical assistance for this study.
Funding
This study was supported by the Japan Agency for Medical Research and Development (grant number: 20ck0106537h).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
The Ethics Committee of the National Cancer Center approved the use of clinical materials for this study (approval number: 2004-050).
Informed consent
Written informed consent was provided by the patients.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Adachi, Y., Noguchi, R., Osaki, J. et al. Establishment and characterization of two novel patient-derived cell lines from myxofibrosarcoma: NCC-MFS7-C1 and NCC-MFS8-C1. Human Cell (2024). https://doi.org/10.1007/s13577-024-01124-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13577-024-01124-4