Abstract
Myxofibrosarcoma (MFS) is an aggressive sarcoma with a highly complex karyotype. Complete resection is the only curative treatment for MFS because it is refractory to chemotherapy. To improve clinical outcomes, it is critical to develop novel treatments for MFS. Although patient-derived cell lines play a key role in cancer research, only 12 MFS cell lines have been reported to date, and considering the diversity of the disease, more cell lines need to be established. Hence, in the present study, we established a novel MFS cell line, NCC-MFS4-C1, using a surgically resected tumor tissue from a patient with MFS. NCC-MFS4-C1 cells exhibited copy number alterations similar to those of the original tumors and showed constant proliferation, spheroid formation, and aggressive invasion. By screening a drug library, we found that actinomycin D, bortezomib, docetaxel, eribulin, and romidepsin significantly reduced the proliferation of NCC-MFS4-C1 cells. Therefore, the NCC-MFS4-C1 cell line may be a useful resource for researching MFS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Myxofibrosarcoma (MFS) is a malignant fibroblastic neoplasm histologically characterized by variably myxoid stroma and pleomorphism [1]. Highly complex karyotypes and genomic profiles have been observed [2], and no specific genetic aberrations have been reported for MFS; however, a previous genomic study revealed frequent amplification in the region of chromosome 5p [3]. MFS often occurs in the lower extremities and dermal or subcutaneous tissues [4]. MFS represents approximately 5% of all soft tissue sarcoma diagnoses and is one of the most common sarcomas diagnosed in elderly patients. However, the absolute number of patients with MFS is extremely small [5, 6]. Surgical resection is the optimal treatment; however, complete resection is often difficult to achieve because of the strong infiltrative nature of MFS [7, 8]. Incomplete resection leads to local recurrence at high rates of 30–40% [5, 6, 9, 10], and the distant metastasis rate ranges from 20 to 35%. Consequently, an unfavorable clinical outcome with a 5-year mortality rate of 30–35% has been reported for MFS cases [4, 5, 9,10,11]. Although systemic chemotherapy is essential to improve clinical outcomes, the benefits of systemic treatments have not been established, and previous studies have reported that conventional chemotherapy does not improve the clinical outcomes in patients with distant metastases [5, 12, 13]. Therefore, a novel treatment has long been desired.
Patient-derived cell lines are important resources in cancer research. At the early passages, they hold the characteristics of the original tumors [14,15,16]; genomic profiling and determination of drug sensitivity study have been performed using patient-derived cell lines [17,18,19,20]. However, these large-scale studies did not include sarcoma cell lines. For example, only 28 sarcoma cell lines, which account for merely 2.2% of all 1300 cell lines investigated, were included in a recent large-scale study [21], suggesting that because of the paucity of adequate cell lines, the latest technologies have not benefited research on sarcomas [22]. Regarding MFS, according to the cell line database, Cellosaurus [23], only 12 MFS cell lines, including the cell lines established in our laboratory, have been reported worldwide [24,25,26,27,28,29,30,31,32]. These findings indicate that it is necessary to generate novel MFS cell lines to improve clinical outcomes.
Here, we established a novel MFS cell line, NCC-MFS4-C1, using a surgically resected tumor tissue from a patient with MFS. Furthermore, we determined the characteristics of NCC-MFS4-C1 cells and demonstrated their utility in a drug screening study.
Materials and methods
Patient history
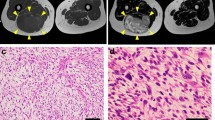
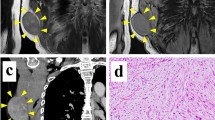
The patient donor was a 65-year-old female diagnosed with myxofibrosarcoma. The patient visited another hospital with the major complaint of a mass in her left lower leg. Following resection, the tumor was pathologically diagnosed as MFS, and the resection margin was positive. The patient was then referred to the National Cancer Center Hospital (Tokyo, Japan) for further treatment. The patient underwent an additional wide resection, but 9 months after the surgery, metastasis was detected in the left inguinal area of the thigh. Thus, a revision surgery was performed. However, 9 months after reoperation, magnetic resonance imaging detected tumor recurrence in the deep thigh (Fig. 1A, B). Therefore, the patient underwent another reoperation. Pathological observations revealed that the tumor exhibited pleomorphic spindle cell proliferation in a focally myxoid background. No specific lines of differentiation were observed (Fig. 1C, D). A portion of the resected tumor excised during the second operation was used to generate the cell line in this study. The ethical committee of the National Cancer Center Hospital approved this study using clinical materials, and the patient donor provided written informed consent.
Clinical and pathological data. Magnetic resonance imaging revealed a deep-seated multilocular tumor in the left thigh showing A low intensity on a T1-weighted image and B iso to high intensity on a T2-weighted image. C, D Hematoxylin and eosin staining showing pleomorphic spindle cell proliferation in a focally myxoid background
Histological analysis
Four-micrometer-thick sections were prepared for histological examination using paraffin-embedded tumor tissues. The sectioned tissues were subjected to deparaffinization and stained with hematoxylin and eosin (H&E).
Cell culture
The surgically resected tumor specimen was used to generate a cell line as described in our previous study [32]. Dulbecco’s modified Eagle medium/nutrient mixture F-12 (Gibco, Grand Island, NY, USA) supplemented with 10% heat-inactivated fetal bovine serum (Gibco), 100 μg/mL penicillin, and 100 μg/mL streptomycin (Nacalai Tesque, Kyoto, Japan) was used to maintain the cells at 37 °C in a humidified atmosphere containing 5% carbon dioxide. We confirmed that the cells proliferated for more than 8 months under tissue culture conditions and were passaged more than 30 times.
Authentication and quality control of the established cell line
We achieved authentication of the established cell line by examining short tandem repeats (STRs) using the GenePrint 10 System (Promega, Madison, WI, USA) according to our previous study [32]. We analyzed the STR pattern of the established cell line using GeneMapper software (Thermo Fisher Scientific), and matched it to those of published cell lines using a function of Cellosaurus with a standard match threshold of 80% [33]. For quality control, DNA fragments of mycoplasma in the tissue culture medium were examined as previously described [32].
Genetic analysis
We conducted single nucleotide polymorphism (SNP) array genotyping using the Infinium OmniExpressExome-8 version 1.4 BeadChip (Illumina, San Diego, CA, USA), according to our previous study [24]. We analyzed the SNP array data using R version 4.0.3 (R Foundation for Statistical Computing, http://www.R-project.org), and DNAcopy package version 1.64.0 (Bioconductor, https://bioconductor.org/). We defined chromosome regions with copy numbers > 3 and < 1 as amplifications and deletions, respectively. We annotated genes with copy number alterations (CNAs) using the biomaRt package, version 2.46.0 (Bioconductor), and “Cancer Gene Census” in the Catalogue of Somatic Mutations in Cancer database (GRCh 37 version 91).
Cell proliferation assay
We assessed cell proliferation according to our previous report [32]. In brief, we seeded the cells at a density of 1 × 105 cells/well onto 12-well culture plates on day 0. The number of cells was counted at multiple time points, and the doubling time was calculated based on the growth curve. We repeated all experiments three times.
Spheroid formation assay
We confirmed the ability of spheroid formation according to a previous report [32]. To monitor the morphology of the tumor cells in the spheroids, we prepared sectioned spheroids. Briefly, we prepared spherical colonies for paraffin sections using iPGell (Genostaff, Tokyo, Japan), according to the manufacturer’s instructions. The cell blocks were fixed with 10% formalin neutral buffer solution and the fixed cells were embedded in paraffin. Then, 4-µm-thick paraffin sections were created and stained with H&E for microscopic observations.
Invasion assay using real-time cell analyzer
The invasion potential of the cells was evaluated using a real-time cell analyzer (xCELLigence, Agilent, Santa Clara, CA, USA), which allows the continuous monitoring of cell behavior in a level-free manner, according to our previous study [32]. We used MG63 osteosarcoma cells (JCRB; Ibaraki Osaka, Japan) as controls because they exhibit consistent growth and possess a representative histological subtype of a sarcoma [34]. The process of invasion was monitored every 15 min for 72 h, and the results were plotted as a function of time after seeding.
Tumorigenicity assessment
The animal experiment in this study was performed in compliance with the guidelines of the Institute for Laboratory Animal Research, National Cancer Center Research Institute. The female Balb/c nude mouse was purchased from Charles River Japan Inc. (Kanagawa, Japan); housed in specific pathogen-free conditions, with food and water available ad libitum and held in a 12-h light and dark cycle. The cells were resuspended in PBS (−) at a concentration of 1 × 106 cells/mL, and 0.1 mL of the cell suspension was injected subcutaneously in nude mouse on the back in four sites. In the days post tumor inoculation, tumor diameters were measured with a digital caliper weekly. The tumor volumes were calculated using the following formula: volume = (length × width2)/2.
Screening for the antiproliferative effects of anticancer agents
We assessed the anti-proliferative effects of 214 anticancer agents according to our previous study [32]. The anticancer agents used in this study are listed in Supplementary Table 1. We evaluated the anti-proliferative effects of the selected drugs in a dose–response manner as reported in our previous study [32]. We determined the IC50 value, which is defined as the sample concentration required to inhibit cell growth by 50% in comparison with the growth of the control cells, from the dose–response curves.
Results
Authentication of the established cell line
We authenticated the NCC-MFS4-C1 cell line by analyzing the STR status of ten microsatellites and found that STR allele patterns of the cells were almost identical to those of the corresponding original tumor tissue (Table 1, Supplementary Fig. 1). In addition, we confirmed that the STR patterns of NCC-MFS4-C1 cells did not match those of any other cell line in the public cell bank, Cellosaurus, and concluded that NCC-MFS4-C1 was a novel MSF cell line. Additionally, we detected no mycoplasma DNA fragments in the tissue culture medium of NCC-MFS4-C1 cells (data not shown), and thus, we concluded that the established cells were free from mycoplasma contamination.
Characterization of the cell line
To examine genomic aberrations in the NCC-MFS4-C1 cell line, we performed SNP array analysis. SNP array analysis revealed CNAs in NCC-MFS4-C1 cells that corresponded to the original tumor. Partial allelic amplification was detected in chromosome 14q, while deletions were detected in chromosomes 1p, 5q, 6q, 7q, 8p, 9p, 10q, 12q, and 14q. The deletion in the chromosome 9p region included the tumor suppressor genes, CDKN2A and CDKN2B (Fig. 2, Supplementary Table 2).
When NCC-MFS4-C1 cells grew under two-dimensional culture conditions, they contained spindle cells (Fig. 3A, B). Moreover, we found that NCC-MFS4-C1 cells formed spheroids when seeded onto a low-attachment microplate. Microscopic observations of H&E-stained spheroid sections revealed dense proliferation of pleomorphic atypical oval cells (Fig. 3C). When the cells were seeded under monolayer culture conditions, they exhibited consistent proliferation (Fig. 3D); based on the growth curve, it took approximately 64 h for the population to double (Fig. 3D). In the invasion assay, we found that NCC-MFS4-C1 cells exhibited more aggressive invasion than MG63 cells (Fig. 3E), and the invasion of NCC-MFS4-C1 cells depended on the time after seeding (Fig. 3E).
Characterization of NCC-MFS4-C1 cells. A, B NCC-MFS4-C1 cells showing spindle cell morphology under monolayer culture conditions. C NCC-MFS4-C1 cells formed spheroids and exhibited dense proliferation of pleomorphic atypical oval cells. D Growth curve of NCC-MFS4-C1 cells. The Y-axis indicates the relative proliferation of NCC-MFS4-C1 cells, and the X-axis represents the hours after seeding. E Real-time cell analyzer invasion assay revealed that NCC-MFS4-C1 cells possessed invasion ability, which was more prominent than MG63 osteosarcoma cells
The cells were inoculated to the nude mice, and caused tumorigenesis under the described condition (Fig. 4A). The tumor consisted of proliferation of atypical cells (Fig. 4B, C) observed their growth. The tumor size consistently increased after transplantation during the observation period (Fig. 4D).
Sensitivity to anticancer agents
We treated NCC-MFS4-C1 cells with 214 anticancer agents at a fixed concentration of 10 µM and evaluated their effects on cell proliferation (Supplementary Table 3). We found that 19 agents showed notable inhibitory effects on the growth of NCC-MFS4-C1 cells. Thus, we calculated the IC50 values of these 19 drugs as well as five additional drugs that are frequently used in standard chemotherapy for soft tissue sarcomas. The IC50 values of the five most effective agents (actinomycin D, bortezomib, docetaxel, eribulin, and romidepsin) are listed in Table 2, and those of other drugs were in Supplementary Table 4. The growth curves used to calculate the IC50 values of the five drugs showing the most prominent anti-proliferative effects on the cells are shown in Fig. 5.
Discussion
MFS is an aggressive sarcoma, and because of its unfavorable clinical features and a paucity of effective anticancer drugs, the prognosis of patients with MFS remains poor. Since MFS is a genetically complex disease and its unique genetic aberrations have not been reported, screening of anticancer drugs using a patient-derived cell line may be a potential approach. Although large-scale drug sensitivity analyses, consisting of genomic biomarker investigations, have been performed using a considerable number of cell lines [17,18,19,20], only a few sarcoma cell lines were included in those studies. For example, only 12 MFS cell lines have been reported to date, indicating that MFS research did not benefit from recent drug screening studies. Hence, we established a novel MFS cell line, NCC-MFS4-C1, which can be used for drug screening.
To date, a few genetic studies have been performed for the CNAs of MFS, and the common genetic aberrations, which can be used as biomarkers or therapeutic targets, have not yet been detected. Although frequent amplification of the chromosome 5p region was reported, the reported CNAs are not specific to MFS [3]. In the NCC-MFS4-C1 cell line, deletions in chromosome 9p were observed. The CNAs observed in NCC-MFS4-C1 cells differed from those of the previously reported MFS cell lines (NCC-MFS1-C1 [30], NCC-MFS2-C1 [31], and NCC-MFS3-C1 [32]). These observations may be consistent with the highly complex genomic features of MFS and indicate that more MFS cell lines should be established to explore the genetic complexity of the disease.
NCC-MFS4-C1 cells exhibited spindle cell sarcoma morphology and demonstrated constant growth and spheroid formation. In addition, NCC-MFS4-C1 formed tumors and exhibited constant proliferation, when they were inoculated in the nude mice. Notably, NCC-MFS4-C1 cells allowed the screening of anti-proliferative effects of 214 anticancer agents. Since these characteristics are suitable for in vitro studies, NCC-MFS4-C1 cells will be useful for the study of MFS.
The five anticancer agents with the lowest IC50 values included actinomycin D, bortezomib, docetaxel, eribulin, and romidepsin. The MFS cell lines established in our previous studies, such as NCC-MFS1-C1 [30], NCC-MFS2-C1 [31], and NCC-MFS3-C1 [32] cells, also showed high sensitivity to bortezomib and romidepsin. Bortezomib is a proteasome inhibitor that was approved for the treatment of multiple myeloma and mantle cell lymphoma [35]. The efficacy of bortezomib for MFS has been examined in only one study [36]. In that study, amplification of the chromosome 5p region was observed in the MFS cell lines. Although the amplification of SKP2 on chromosome 5p has been associated with the antiproliferative effects of bortezomib on MFS, NCC-MFS1-C1, NCC-MFS2-C1, NCC-MFS3-C1, and NCC-MFS4-C1 cells did not exhibit such genomic aberrations. Although the amplification of chromosome 5p was not observed, the growth of these MFS cell lines was hindered by bortezomib. These observations may suggest the presence of other molecular mechanisms which affect the sensitivity to bortezomib. Romidepsin is a histone deacetylase (HDAC) inhibitor, and is usually used for the treatment of peripheral T-cell lymphoma [37]. Only one study reported that an HDAC inhibitor was effective for MFS [38]. The molecular mechanisms which provide a clue for the action of HDAC inhibitors on MFS remain unclear, and further studies are worth conducting.
In conclusion, we generated a novel MFS cell line, NCC-MFS4-C1, from surgically resected tumor tissue of a patient with MFS. NCC-MFS4-C1 cells exhibited constant proliferation, spheroid formation, and invasion. Additionally, we demonstrated the utility of NCC-MFS4-C1 cells in drug screening, revealing the antiproliferative effects of bortezomib and romidepsin on MFS (Fig. 5). As MFS is a diverse and rare disease, and the number of established MFS cell lines remains insufficient, we need to establish more MFS cell lines for further research.
References
Board WCoTE. The 2019 World Health Organization classification of tumours of the breast. Histopathology. 2020;77(2):181–185
Widemann BC, Italiano A. Biology and management of undifferentiated pleomorphic sarcoma, myxofibrosarcoma, and malignant peripheral nerve sheath tumors: state of the art and perspectives. J Clin Oncol. 2018;36:160–7.
Barretina J, Taylor BS, Banerji S, et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet. 2010;42:715–21.
Mentzel T, Calonje E, Wadden C, et al. Myxofibrosarcoma. Clinicopathologic analysis of 75 cases with emphasis on the lowgrade variant. Am J Surg Pathol. 1996;20:391–405.
Sanfilippo R, Miceli R, Grosso F, et al. Myxofibrosarcoma: prognostic factors and survival in a series of patients treated at a single institution. Ann Surg Oncol. 2011;18:720–5.
Roland CL, Wang WL, Lazar AJ, Torres KE. Myxofibrosarcoma. Surg Oncol Clin N Am. 2016;25:775–88.
Iwata S, Yonemoto T, Araki A, et al. Impact of infiltrative growth on the outcome of patients with undifferentiated pleomorphic sarcoma and myxofibrosarcoma. J Surg Oncol. 2014;110:707–11.
Waters B, Panicek DM, Lefkowitz RA, et al. Low-grade myxofibrosarcoma: CT and MRI patterns in recurrent disease. AJR Am J Roentgenol. 2007;188:W193–8.
Dewan V, Darbyshire A, Sumathi V, Jeys L, Grimer R. Prognostic and survival factors in myxofibrosarcomas. Sarcoma. 2012;2012:830879.
Lee AY, Agaram NP, Qin LX, et al. Optimal percent myxoid component to predict outcome in high-grade myxofibrosarcoma and undifferentiated pleomorphic sarcoma. Ann Surg Oncol. 2016;23:818–25.
Huang HY, Lal P, Qin J, Brennan MF, Antonescu CR. Low-grade myxofibrosarcoma: a clinicopathologic analysis of 49 cases treated at a single institution with simultaneous assessment of the efficacy of 3-tier and 4-tier grading systems. Hum Pathol. 2004;35:612–21.
Mutter RW, Singer S, Zhang Z, Brennan MF, Alektiar KM. The enigma of myxofibrosarcoma of the extremity. Cancer. 2012;118:518–27.
Look Hong NJ, Hornicek FJ, Raskin KA, et al. Prognostic factors and outcomes of patients with myxofibrosarcoma. Ann Surg Oncol. 2013;20:80–6.
Saito S, Morita K, Kohara A, et al. Use of BAC array CGH for evaluation of chromosomal stability of clinically used human mesenchymal stem cells and of cancer cell lines. Hum Cell. 2011;24:2–8.
Tseng YY, Boehm JS. From cell lines to living biosensors: new opportunities to prioritize cancer dependencies using ex vivo tumor cultures. Curr Opin Genet Dev. 2019;54:33–40.
Ben-David U, Beroukhim R, Golub TR. Genomic evolution of cancer models: perils and opportunities. Nat Rev Cancer. 2019;19:97–109.
Basu A, Bodycombe NE, Cheah JH, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154:1151–61.
Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7.
Yang W, Soares J, Greninger P, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–61.
Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–23.
Goodspeed A, Heiser LM, Gray JW, Costello JC. Tumor-derived cell lines as molecular models of cancer pharmacogenomics. Mol Cancer Res. 2016;14:3–13.
Hattori E, Oyama R, Kondo T. Systematic Review of the Current Status of Human Sarcoma Cell Lines. Cells. 2019;8:157.
Bairoch A. The Cellosaurus, a cell-line knowledge resource. J Biomol Tech. 2018;29:25–38.
Kawashima H, Ogose A, Gu W, et al. Establishment and characterization of a novel myxofibrosarcoma cell line. Cancer Genet Cytogenet. 2005;161:28–35.
Ariizumi T, Ogose A, Kawashima H, Hotta T, Umezu H, Endo N. Multinucleation followed by an acytokinetic cell division in myxofibrosarcoma with giant cell proliferation. J Exp Clin Cancer Res. 2009;28:44.
Moneo V, Serelde BG, Fominaya J, et al. Extreme sensitivity to Yondelis (Trabectedin, ET-743) in low passaged sarcoma cell lines correlates with mutated p53. J Cell Biochem. 2007;100:339–48.
Miserocchi G, De Vita A, Mercatali L, et al. Characterization and drug sensitivity of a new high-grade myxofibrosarcoma cell line. Cells. 2018;7.(11):186.
Lohberger B, Stuendl N, Wolf E, Liegl-Atzwanger B, Leithner A, Rinner B. The novel myxofibrosarcoma cell line MUG-Myx1 expresses a tumourigenic stem-like cell population with high aldehyde dehydrogenase 1 activity. BMC Cancer. 2013;13:563.
Lohberger B, Stuendl N, Leithner A, et al. Establishment of a novel cellular model for myxofibrosarcoma heterogeneity. Sci Rep. 2017;7:44700.
Kito F, Oyama R, Sakumoto M, et al. Establishment and characterization of a novel cell line, NCC-MFS1-C1, derived from a patient with myxofibrosarcoma. Hum Cell. 2019;32:214–22.
Noguchi R, Yoshimatsu Y, Ono T, et al. Establishment and characterization of NCC-MFS2-C1: a novel patient-derived cancer cell line of myxofibrosarcoma. Hum Cell. 2021;34:246–53.
Tsuchiya R, Yoshimatsu Y, Noguchi R, et al. Establishment and characterization of NCC-MFS3-C1: a novel patient-derived cell line of myxofibrosarcoma. Hum Cell. 2021;34(4):1266–1273.
Capes-Davis A, Reid YA, Kline MC, et al. Match criteria for human cell line authentication: where do we draw the line? Int J Cancer. 2013;132:2510–9.
Billiau A, Edy VG, Heremans H, et al. Human interferon: mass production in a newly established cell line, MG-63. Antimicrob Agents Chemother. 1977;12:11–5.
Gandolfi S, Laubach JP, Hideshima T, Chauhan D, Anderson KC, Richardson PG. The proteasome and proteasome inhibitors in multiple myeloma. Cancer Metastasis Rev. 2017;36:561–84.
Li CF, Wang JM, Kang HY, et al. Characterization of gene amplification-driven SKP2 overexpression in myxofibrosarcoma: potential implications in tumor progression and therapeutics. Clin Cancer Res. 2012;18:1598–610.
Barbarotta L, Hurley K. Romidepsin for the treatment of peripheral T-Cell lymphoma. J Adv Pract Oncol. 2015;6:22–36.
Kawarazaki A, Horinaka M, Yasuda S, Kawashima H, Numajiri T, Sakai T. The HDAC inhibitor OBP-801 suppresses the growth of myxofibrosarcoma cells. J BUON. 2020;25:464–71.
Acknowledgements
We thank Drs. F. Nakatani, E. Kobayashi, S. Fukushima, M. Nakagawa, T. Komatsubara, M. Saito, and C. Sato of the Department of Musculoskeletal Oncology, and Drs. M Arakaki, and Y Suemitsu of the Department of Diagnostic Pathology, National Cancer Center Hospital, for sampling tumor tissue specimens from surgically resected materials. We also appreciate the technical assistance provided by Ms. Y. Kuwata (Division of Rare Cancer Research), and the technical support provided by Ms. Y. Shiotani, Mr. N. Uchiya, and Dr. T. Imai (Central Animal Division, National Cancer Center Research Institute). We would like to thank Editage (www.editage.jp) for help with English language editing and constructive comments on the manuscript. This research was technically assisted by the Fundamental Innovative Oncology Core at the National Cancer Center.
Funding
This research was supported by the Japan Agency for Medical Research and Development (grant number 20ck0106537h0001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
The ethical committee of the National Cancer Center approved the use of clinical materials for this study (approval number 2004–050).
Informed consent
Written informed consent for publication was provided by the patient.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yoshimatsu, Y., Noguchi, R., Tsuchiya, R. et al. Establishment and characterization of NCC-MFS4-C1: a novel patient-derived cell line of myxofibrosarcoma. Human Cell 34, 1911–1918 (2021). https://doi.org/10.1007/s13577-021-00589-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-021-00589-x