Abstract
Starch, the primary determinant of wheat grain yield, contributes 65–75% grain dry weight and 80% of the endosperm dry weight. The starch biosynthetic pathway is regulated by several enzymes, of which the soluble starch synthase (SS) is the most crucial enzyme and is sensitive to environmental change. In the present study, we targeted identifying the unique haplotypes of SSI gene in 17 different wild and cultivated genotypes of wheat, selected for abiotic stress tolerance. Four unique SNPs corresponding to non-synonymous substitution were identified from wild progenitor species, Aegilops tauschii (accessions pau14102 and pau3747), Ae. speltoides (pau15081) and T. dicoccoides (pau7107), each present in exon-1. However, two unique SNPs corresponding to non-synonymous substitution were identified from cultivated wheats, Impala and C591, present in exon-2 and exon-4, respectively. These SNPs correspond to the N-terminal half of the GT5 domain in exon 1–6 and are expected as novel haplotypes contributing to better grain filling and starch deposition under changing environmental conditions. The positive effects of these haplotypes need further validation through molecular and phenotypic evaluation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bread wheat (Triticum aestivum L.) is the most important staple food worldwide contributing nearly 30% to global cereal production, with average productivity of 744.5 million tonnes (FAO 2020). Despite the global importance, wheat production is significantly lower than rice and maize (FAO 2020), and an increase by 70% from the present level is required to feed an additional 3 billion mouths by 2050 (Kaur et al. 2021). Maintaining crop productivity levels under changing climate, increasing vulnerability, and decreasing land is a major challenge in modern agriculture, and there is an immediate need of the hour to streamline suitable strategies to boost the yield under limited resources.
Grain weight is a major determinant of final wheat yield and starch accounts for 65–75% grain dry weight and 80% of the endosperm dry weight (Rahman et al. 2000). Photo-assimilates produced in the vegetative tissue of wheat plants were transported as sucrose (Tuncel and Okita 2013) and sink convert these photo-assimilates into starch, an important limiting factor to wheat grain yield (Zhenlin et al. 1999; Miralles and Slafer 2007). The starch biosynthetic pathway is regulated by a coordinated series of enzyme-catalyzed reactions, including granule bound starch synthase (GBSS), soluble starch synthase (SS), starch branching enzyme (SBE), starch debranching enzyme (SDE), isoamylases, pullulanase, starch phosphorylase (SP) and disproportionating enzyme (DPE) (Keeling and Myers 2010). Of these GBSS and SS catalyze the first unique step in starch synthesis (Jeon et al. 2010). SS is involved in the elongation of amylopectin polymers and is one of the most important enzymes with four isoforms (SSI, SSII, SSIII, and SSIV), (James et al. 2003). SSI isoform contributing 60–70% of the total SS activity is the major isoform responsible for synthesizing its shorter chains (dp 8–12) (Delvallé et al. 2005; Cao et al. 1999; Kumar et al. 2021). The enzyme is constitutively expressed at all stages of seed development (Park and Nishikawa 2012). It also participates in the regulation of other starch biosynthetic enzymes and (Crofts et al. 2017a, b) different SS isoforms play a synergistic role in the synthesis of amylopectin clusters, and a deficiency of an individual isoform have an impact on the grain development and starch accumulation (Fujita et al. 2007).
Reduction in starch quantity is one of the major factors for the reduction in yield and yield components of wheat in response to different biotic and abiotic stresses. Downregulation of starch deposition has been reported in the kernel as high temperature impairs the conversion of sucrose to starch (Keeling et al. 1994; MacLeod and Duffus 1988; Yamakawa and Hakata 2010; Phan et al. 2013; Sehgal et al. 2018; Yang et al. 2019). Among the enzymes involved in the wheat starch biosynthetic pathway, soluble starch synthase (SS) is the most labile to high temperature (Keeling et al. 1993; Hurkman et al. 2003; Kumar et al. 2021) and a decline in SS activity leads to reduced starch accumulation in wheat (Prakash et al. 2004). Terminal heat stress can reduce the yield of wheat crop by 11.1% by 2050 worldwide and negatively influence the heading and grain filling stages (Dubey et al. 2020). Heat stress pertaining to a 5 °C increase in temperature above 20 °C accelerates the rate of grain filling, whereas grain filling duration is reduced by 12 days in wheat (Yin et al. 2009).
Utilization of genetic variation available in the gene pool and targeting the important traits for attaining stress tolerance is one of the key principles for crop improvement. The diversity of cultivated wheat has been eroded due to continuous pressure of domestication and selection, leaving bleak chances for any improvement (Donini et al. 2005; Peleg et al. 2011; Longin et al. 2014). On the other hand, wild germplasm of wheat including Triticum and Aegilops species, old wheat varieties, landraces, and exotic wheats have numerous unknown and useful alleles for plants to resist, tolerate, or avoid extreme temperatures, drought, or flooding, as well as resist against pests and diseases. Identification and transfer of valuable alleles from variable germplasm are like hitting goldmine to introgress novel alleles for wheat improvement (Dwivedi et al. 2008; Pajkovic et al. 2014; Ceoloni et al. 2017; Olivera et al. 2018; Dhillon et al. 2020). The present study was undertaken to look for diversity of SSI-7B isoform of SS gene wild progenitor and cultivated tetraploid and hexaploid wheats and its comparison with their counterpart from A genome (SSI-7A) and D genome (SSI-7D) wild species.
Materials and methods
Plant material
A set of 13 different variable wheat genotypes, including wild and cultivated wheat germplasm, were selected for this study. Wild genotypes included two accessions of diploid Aegilops speltoides (SS) and two accessions of tetraploid Triticum dicoccoides (AABB). Besides, one accession each of T. monococcum (AmAm) and T. boeoticum (AbAb) and two accessions of Ae. tauschii (DD) were selected to amplify and compare the SSI-7A and SSI-7D homeologue of the gene. Cultivated wheat genotypes include one tetraploid (T. durum, AABB) MACS9 and ten hexaploid wheat (T. aestivum AABBDD) cultivars, consisting of two winter wheat and eight spring wheat genotypes. Among the spring wheat, two were old tall Indian wheats and six dwarf wheats. The list of these selected genotypes is given in Table S1.
Primer designing for homoeologous SSI gene and in vitro DNA amplification
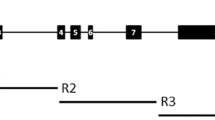
Partial nucleotide sequence (5156 bp) of SSI-7B (Traes_7BS_6135B1D85.1) gene was used to design gene-specific primers using the PerlPrimer tool (Marshall 2004). Two sets of different overlapping primers were designed (Table S2). One primer pair SSL1 amplifies 2289 bp sequence from nucleotide positions 130–2419, while the second primer pair SSL2 amplifies 2677 bp sequence between nucleotide positions 2229 and 4906. These primers could not amplify the expected fragments in wild wheat. Thus, the second set of seven overlapping primer pairs was designed for the amplification of gene in seven small fragments as SSOL1 (130–879), SSOL2 (700–1555), SSOL3 (1320–2164), SSOL4 (1970–2864), SSOL5 (2726–3598), SSOL6 (3410–4290) and SSOL7 (4152–4906) with the amplicon length ranging from 830 to 900 bp. The same set of seven overlapping primers was also used for sequencing PCR amplicons. Two additional sequencing primers, SSOL4A, and SSOL4B were also designed for sequencing two fragments of size 2289 bp and 2419 bp.
The genomic DNA was isolated from young leaves of field-grown plants sown in the month of November with standard practices for wheat cultivation and quantified using 0.8% agarose gel electrophoresis. A PCR reaction of 25 μL volume containing 100 ng template DNA (4 μL), 5X PCR buffer plus Mg2+ (2.5 μL), 10 mM dNTPs (2.0 μL), 5 pM forward and reverse primers (1.25 μL), 5 units/μL Takara Ex Taq DNA Polymerase (0.25 μL), and sterile water (13.75 μL) was used for amplification. Amplification for 2289 bp and 2677 bp amplicons was done as initial denaturation at 94 °C for 5 min, followed by denaturation and primer annealing. The first step included 14 cycles of 94 °C for 1 min, 60 °C for 1 min and 68 °C for 3 min followed by the second step of 16 cycles of 94 °C for 1 min, 60 °C for 1 min, and 68 °C for 3 min with 20 s increment per cycle. The last step included a final extension of 10 min at 68 °C. The PCR reaction was loaded and separated on a 0.8 percent (w/v) agarose gel.
Molecular cloning of SSI gene
The PCR products were purified using QIAquick Gel Extraction Kit (Qiagen®) and cloned into a pGEM®-Teasy vector (Promega) and transformed into E. coli, DH-5α host strain. The white recombinant clones were selected on LB-Amp-Xgal agar plates using blue/white screening. Positive clones were identified with colony PCR using the gene-specific primers, and the product was resolved on 1.2 percent agarose gel. Plasmid DNA was isolated from positive clones using R.E.A.L® prep 96 (QIAGEN). Cloning of candidate gene fragments was done in tetraploid and hexaploid genotypes only, while in diploids, namely T. monococcum, T. boeoticum, Ae. speltoides and Ae. tauschii, the PCR product was sequenced directly after purification.
Sequencing of SSI gene
The sequencing of PCR products was done for both the complementary strands using overlapping primers. The sequences were extracted from chromatogram files using CHROMAS Lite 2.1.1 (http://technelysium.com.au/), and a full-length sequence of SSI gene was completed by manual aligning of the sequence of one strand with that of the reverse complementary sequence and high-quality contigs were generated using DNA Baser v4.23.0 (http://www.dnabaser.com/). Multiple sequence alignment of cloned sequences and reference sequence was done using ClustalX 2.1 (Larkin et al. 2007) and based on the alignment, candidate SNPs and InDels were predicted. Exonic and intronic boundaries of the SSI gene were predicted using the Artemis tool (Carver et al. 2008). The sequence of SSI genes obtained from T. monococcum and T. boeoticum were aligned with SSI-7A as reference (Traes_7AS_70ED86B15.1), sequences of two accessions Ae. tauschii aligned with SSI-7D as reference (Traes_7DS_5159E3934.1), while sequences from all other genotypes of Ae. speltoides, T. dicoccoides, T. durum, and T. aestivum, were aligned using SSI-7B as reference (Traes_7BS_6135B1D85.1).
Phylogenetic analysis
Primary structure and amino acid sequences were predicted using the Artemis tool (Carver et al. 2008). The amino acid substitutions in SSI between different wheat genotypes were identified visually with respect to the reference, using the multiple alignment viewer JalView 2.0 (www.jalview.org). Phylogenetic and molecular evolutionary analysis of the multiple sequence alignment of protein sequences from selected genotypes and the monocot and dicot SSI sequences were carried out using the MAFFT tool (Katoh and Standley 2013). The phylogenetic tree of these sequences was constructed using Phylip software using the neighbor-joining method (Felsenstein 2005).
Results
In-silico candidate gene identification
Nucleotide sequences of SSI genes selected from NCBI (https://www.ncbi.nlm.nih.gov/) and EMBL (https://www.ebi.ac.uk/ena/browser/home) databases were clustered to remove redundant sequences using the CD-HIT Suite tool (Fu et al. 2012) at 95% identity, shortlisting ten representative sequences. Three full-length coding sequences (CDS) of SSI gene from the TriFLDB database (Mochida et al 2009) were clustered into these ten representative sequences at 99% identity narrowing down to seven sequences which then used as a query for standalone (offline) blast (Tao 2010) against cDNA and gDNA IWGSC (Ensembl Plants; https://plants.ensembl.org/Triticum_aestivum/) T. aestivum databases. A total of 40 and 56 hits were obtained in cDNA and gDNA databases, respectively. Based on the common hits, maximum query coverage, and bit score, three sequences on homoeologous chromosomes 7A, 7B, and 7D, were selected. The expression of these three homoeologous sequences was more in grain and spike compared to leaf, root, and stem (http://wheat.pw.usda.gov/WheatExp/) (Pearce et al. 2015). Expression of homoeologue 7B (gene ID Traes_7BS_6135B1D85.1) specific transcript was much higher in grains along with a higher expression of this transcript under heat and drought stress. (Fig. S1, S2).
Characteristic of SSI gene
A partial sequence of SSI-7B gene was amplified from all the 13 genotypes, including two di1ploid (SS), three tetraploid (AABB), and eight hexaploid (AABBDD) wheat genotypes. Similarly, partial sequence of SSI-7A and SSI-7D was also amplified from two AA genome and two DD genome-specific diploid accessions, respectively. None of the sequenced loci contained more than one SSI gene (Fig. S3, S4, S5). Thus, the SSI sequence amplified from all the 17 different genotypes has a similar structure with nine exons and eight introns as reference gene. The sequences were deposited in the GenBank with accession numbers MN75891 to MN75907.
Exonic/intronic variation
Length of partial SSI-7B gene in 13 diploid, tetraploid and hexaploid genotypes varied from 3626 to 3709 bp. Total exon length varied from 1059 to 1146 bp, with the first exon being the largest (281-360 bp), followed by the fifth (173 bp) and third (124 bp) exon. Exon-2, 4, 6, 7, and 8 were smaller with 62–91 bp length (Table S3). The total length of eight introns varied from 2563 to 2575 bp, with the third intron being the largest (1079–1083 bp) and the eighth being smallest (70–82 bp). Intron-1, 3, and 8 showed some length variation, while intron-2, 5, 6, and 7 were conserved across diploid, tetraploid, and hexaploid. Within hexaploid genotypes, SSI-7B had similar sizes (3708–3709 bp) with length variations in intron-1 (C306), intron-8 (Arbon and WH542) and exon-9 (WH542). Among three tetraploid (AABB) wheat genotypes, T. durum and T. dicoccoides, length variations were found in intron-1, exon-2, and intron-3. In the case of diploids (SS), intron-1, 3, and 8 showed length variations which were also prominent across the three species used in this study. The differences in the first and last exons were due to differences in the position of the start and stop codons in the amplified sequence as these codons were identified manually.
The partially amplified sequence of SSI-7A gene from T. monococcum and T. boeoticum was longer (3854 bp) than SSI-7B with 1122 bp exonic length and 2732 bp intronic length. Nine exons and eight introns were of similar length in both A-genome species. The exonic length was also identical to SSI-7B amplified sequences, although the seventh intron (293 bp) showed significant length variation. Exonic and intronic lengths in SSI-7D sequence of two Ae. tauschii accessions had a total gene length of 3704–3705 bp with an exon length of 1118 bp and intron length of 2586–2587 bp, respectively. These two accessions were similar except for one base pair difference in the third intron. Exonic regions of the SSI gene amplified from three homoeologues of chromosome 7 showed more proximity than intronic regions as exon 2–7 were conserved across three homoeologues. At the same time, intron-6 was the only intron that had conserved length among three homeologues.
SNPs/indel in genomic sequences
In total, 44 exonic SNPs and 282 intronic SNPs were identified in the partly amplified SSI-7B gene in 13 wheat genotypes (Fig. 1, Table S4). Maximum exonic SNPs were detected from two diploid S-genome accessions of Ae. speltoides, while there was no exonic SNP in T. dicoccoides acc. pau14801 and four hexaploid wheats (Giza, Arbon, C306, PBW 343). Exon-1, being the longest, had a maximum of 18 SNPs, with 2–5 SNPs in the rest of the exons (Fig. 1). C ↔ T (transition) was the most commonly occurring exonic SNP detected across all the genotypes. Several intronic SNPs outnumbered exonic SNPs irrespective of genotype. The first and third introns were highly variable with 99 and 156 SNPs, respectively, as expected from their comparatively longer size. Intron 2, 6, and 8 were conserved without any SNP. No insertions were found in the sequenced SSI-7B, although deletions were present in the intronic regions only.
There were nine exonic and 24 intronic SNPs in the eight hexaploid wheats with an average of 1SNP/824 bp (Fig. 2; Table 1; Table S4). No indel was detected in any of the hexaploid lines. Of the 21 SNPs representing 20 haplotypes in three tetraploid wheats, there were five exonic and 16 intronic SNPs with the frequency of 1SNP/523 bp in 10974 bp total gene length (Table 1). T. dicoccoides acc pau14801 also had one single base pair deletion in intron-3.
In two Ae. speltoides accessions, maximum exonic (21), and intronic (126) SNPs were identified with the frequency of 1SNP/50 bp, representing 124 haplotypes (Table 1). The first exon had the maximum number of SNPs (7), while other exons had 2–4 SNPs except for exon-2, which did not have any SNP. Intron-1 (54 SNPs) and intron-3 (62 SNPs) have a large number of SNPs as compared to introns-4, 5, and 7 with 3 SNPs only (Table S4).
There were six exonic and 89 intronic SNPs in two A-genome species with the frequency of one exonic SNP per 374 bp (Fig. 2; Table 1) and one intronic SNP/61 bp, respectively, representing 58 haplotypes. Exonic SNPs were concentrated only in exon-1 and 4, while intronic SNPs were more concentrated in intron-1 and 3, with no SNPs in intron-2,6,7 and 8. Four deletions were also detected in intron-1, 3, and 8. A total of 22 haplotypes representing 25 SNPs, including three exonic and 22 intronic SNPs, were found in D-genome species. These SNPs were found only in exon-1 and intron-1, and 3. The frequency of exonic SNPs was lower with 1SNP/745 bp than intronic 1SNP/235 bp. It represented the second-lowest frequency of exonic SNPs after the 1/1009 SNP of SSI-7B in hexaploid (AABBDD) genotypes followed by 1/651 SNP of tetraploid (AABB), 1/374 SNP of AA, and 1/104 SNP of SS genome species (Fig. 2; Table 1). No Indels were detected in D-genome species.
Thus, the frequency of SNPs was lower in hexaploids than in tetraploids and diploids. Overall, introns were more diverse than exons. Among diploids, the highest diversity was observed in Ae. speltoides (SS) compared to A-genome or D-genome species in both the intronic and exonic region.
Transitions and transversions
SNPs involve both transition (Tr), change between the purine-purine/pyrimidine-pyrimidine, or transversions (Tv), changes between the purines and pyrimidines. In SSI-7B amplified sequences, more transitions than transversions were detected with Tr/Tv ratio of 0.6:1, 0.3:1, and 0.42:1 in diploid, tetraploid, and hexaploid (Table 2). Maximum Tr (80) and Tv (44) were found in Ae. speltoides followed by A-genome species with 37 Tr and 21 Tv and hexaploid with 31 Tr and 4 Tv. G ↔ T Tv was completely absent in tetraploids, while G ↔ C Tv was missing in hexaploid. Exonic regions also lack C ↔ A type Tv in tetraploid and T ↔ A type Tv in tetraploid and hexaploid. Hexaploid genotypes (MACS 9, Giza, Arbon, PBW343, and Halna) had Tr only, C306 has Tv only, while C591, Impala, and WH542 had both Tr and Tv.
In SSI-7A and SSI-7D homeologue, specific amplified sequences, 37 and 13 Tr and 21 and 9 Tv were found, respectively, with a Tv/Tr ratio of 0.57:1 and 0.69:1 in A- and D-genome diploid species (Table 2).
Phylogenetic and molecular evolutionary analysis
The sequence of full-length SSI gene obtained from 17 genotypes was clustered into three groups based on phylogenetic analysis conducted using Phylip software (Fig. 3). T. monococcum and T. boeoticum accessions were clustered with A-genome reference (Traes_7AS_70ED86B15.1). While, Ae. tauschii acc. pau3747 and Ae. tauschii acc. pau14102 were clustered with D-genome reference (Traes_7DS_5159E3934.1). Rest of the genotypes, Ae. speltoides, T. dicoccoides, T. durum, and T. aestivum were grouped with B-genome reference (Traes_7BS_6135B1D85.1).
To understand the relationship between SSI proteins examined in the current study, a brief phylogenetic analysis of SSI protein was done in the major monocot crops (wheat, rice, maize, barley) along with Glycine max and Arachis hypogaea. The analysis clustered dicots and monocots into two different clusters (Fig. 4, Table S5). As expected, closely related species showed fewer sequence variations than the distantly related species and were clustered together. Among the monocots, the SSI gene also diverged into three major clusters first one with maize (Zm) and sorghum (Sb), the second with rice (Os), and the third with wheat (Ta), barley (Hv), and Brachypodium (Bd) along with few genotypes from T. urartu (Mishra et al. 2017). All the genotypes of the current study were grouped into a third cluster (Figs. 4, 5).
Neighbor-joining phylogenetic tree of SSI proteins corresponding to amplified gene sequences from 17 different diploid, tetraploid and hexaploid wheats in the present study along with representative SSI protein sequences from monocots and dicots. Predicted amino acid sequences of the SSI genes were obtained from the NCBI (List of Accession Numbers is given in Supplementary Table 4) Bootstrap values are shown as percentages for 100 replicates
Detection of variations in protein sequences
Out of 34 exonic SNPs in SSI-7B amplified sequence, 24 showed synonymous substitutions (SSt) while 10 were non-synonymous substitutions (NSSt). SSt were absent in T. dicoccoides acc. pau7107, MACS9, and C591, as all the exonic SNPs contribute to non-synonymous substitution. On the other hand, NSSt was absent in hexaploid wheats WH542 and Halna. T. dicoccoides acc. pau7107 genotype contributed maximum NSSt (3) followed by Ae. speltoides acc. pau15081 (2), Impala (2) C591 (2), and MACS9 (1) (Table
3).
In SS1-7D, one NSSt was detected in each of two D genome accessions of Ae. tauschii while no NSSt was detected in SS1-7A of A-genome species. The rate of SSt and intronic substitutions were more in the partially amplified SSI gene. The maximum number of NSSt (3) was identified for amino acid D-Aspartic acid, followed by two NSSt for amino acid A-Alanine and E-Glutamic acid. While one for L-Leucine, Q-Glutamine, T-Threonine, N-Asparagine, and P-Proline (Table 3).
Structure prediction of SSI protein
Starch synthase catalytic domain belonging to the glycosyltransferase 5 (GT-5) family was also found in the SSI gene sequences from cultivated and wild genotypes under study. Analysis results using the Pfam program suggested that in the selected genotypes, the alignment of the GT-5 domain starts from amino acid on 35th position and ends at amino acid 293. The predicted GT-5 domain was found to be of similar length in all the selected wheat genotypes. This domain lies from exons 1- 6, and six SNPs corresponding to non-synonymous substitutions fall in this region (Fig. 5; Table S6).
Discussion
Four different isoforms of the SS gene and its chromosomal locations have been reported as SSI and SSII (homeologous group 7) (Li et al. 1999a, b; Gao and Chibbar 2000; Peng et al. 2001; Shimbata et al. 2005; Huang and Brlé-Babel 2010; Li et al. 2013; McMaugh et al. 2014), SSIII and SSIV (homeologous group 1) (Li et al. 2000; Pan et al. 2011). SSI is a major gene in wheat starch synthesis and contributes 60–70% of the SS activity (Cao et al. 1999; Fujita et al. 2006; Zeeman et al. 2010). It is constitutively expressed at all stages of seed development with a higher transcript level than SSII and SSIII during the early seed development stage (6 DAP) and steady expression thereafter throughout endosperm development (15 DAP) (Park and Nishikawa 2012; Kumar et al. 2021).
Polyploid wheat underwent a strong differentiation compared to wild ancestral species, significantly decreasing the genetic diversity on major loci. The progenitors of common wheat are a rich reservoir of various economically important traits, and diverse alleles of these species can be utilized in wheat improvement (Lopes et al. 2015). We compared the diversity and genetic differentiation of the most important isoform SSI-7B among hexaploid (spring wheat, winter wheat, tall wheat), tetraploid (T. dicoccoides and T. durum), and diploid wheats and their comparison with SSI-7A and SSI-7D isoforms from A- and D- genome species. There is an apparent reduction in the diversity of SSI-7B in hexaploid wheat, which is visible in the number of SNPs with the progression from diploids (Ae. speltoides) to tetraploids and hexaploids. The frequency of SNPs (exon + intron) varied from 1SNP/50 bp in Ae. speltoides to 1SNP/523 bp in tetraploid and 1SNP/824 bp in hexaploids. These frequency estimations are clear signs of loss of diversity in both intronic and exonic regions at different ploidy levels.
Similarly, the number of haplotypes was reduced from 124 in Ae. speltoides to 20 in tetraploids and 35 in hexaploids. SNP frequency in A-genome was 1SNP/81 bp, which is more than that of the D-genome species (1SNP/296 bp). Introns were found to be more diversified in terms of size and nucleotide sequences (Breathnach and Chambon 1981; Haga et al. 2002; Ramakrishna et al. 2002) than exons as the exons are under intense selection pressure and eliminate the deleterious changes, whereas introns have a high buffering rate for any change. In SSI high frequency of variations were found in intronic regions than in exonic regions. These variations were more pronounced between the different species.
Similarly, all the deletions observed were in diploid and tetraploid and in the intronic region, while no Indel was found in hexaploid wheat. In another isoform of starch synthase genes, SSIIa represented more allelic variation in the intronic regions (Huang and Brûlé-Babel 2012). Similarly, the sequenced AGP-L-B allele of AGP-L genes had 13 indels and 58 SNPs in the introns compared to only nine SNPs in the exon (Rose et al. 2016).
The rate of transition was more than transversion in all three homologous sequences. Transitions are usually favored over transversions (Sankoff et al. 1976) as transition mutations do not alter the 3D structure of the protein. These mutations result in synonymous substitution because selection pressure acts to conserve the chemical properties of amino acids (Vogel and Kopun 1977). Transitions are about twice as frequent as transversions in rice (Hayashi et al. 2004) and maize (Batley et al. 2003). A higher number of transversions in diploid and tetraploid species than hexaploid indicated clear preservation of diversity, absent in cultivated hexaploids making wild non-cultivated germplasm an important source of new alleles. Luo et al. (2016) provided evidence that universal bias occurs in favor of transitions over transversions. This bias results as the process of transversion mutations, including size conformation, is more complicated than transition. Leterrier et al. (2008) studied homology modeling for protein structure prediction of SS gene using Tool for Incremental Threading Optimization (TITO) and have suggested that valine residue in the K-X-G-G-L conserved motif in SSIII and SSIV isoforms could be an important factor for protein specificity as compared to SSI and SSII isoforms.
SS gene has a conserved function
SS gene is known to encode a protein with a glucosyltransferase domain. In most known SS isoforms, the GT-1 domain occupies the C-terminal half, and the GT-5 domain occupies the N-terminal half of catalytic active motifs. The GT-5 domain is responsible for binding with glucosyl donor, i.e., ADPglucose (Keeling and Myers 2010). Leterrier et al. (2008) studied starch synthase glycosyl transferase (GT-5) domain homology, which was compared to prokaryotic SS and revealed that GT-5 domain (Pfam PF08323) was conserved among all SS (SSI-SSIV) isoforms.
Exon 1–6 corresponds to the GT-5 domain of the N-terminal half, and six crucial non-synonymous SNPs have been identified in this region. Of these six SNP-based alleles, four were from wild progenitor species of Ae. tauschii acc. pau14102, Ae. tauschii acc. pau3747, Ae. speltoides acc. pau15081 and T. dicoccoides acc. pau7107, each present in exon-1 and two from cultivated wheats Impala and C591 present in exon 2 and 4, respectively. SSI involved in starch synthesis losses half of its activity when a temperature ~ 35 °C is experienced by the crop (Keeling et al. 1994) but a higher average starch deposition at high temperature is required to combat terminal heat stress. Waines (1994), Pradhan et al. (2012), and Awlachew et al. (2016) reported that Ae. speltoides could serve as a source of genetic variability for improved thermotolerance in wheat.
PAU, Ludhiana has a collection of > 1500 wild progenitor and non-progenitor species and different germplasm of hexaploid wheat that have been evaluated for the past many years for different biotic and abiotic stress-related parameters and agronomic traits. Wild and cultivated wheats of different ploidy were selected for the current study, as these species carry tremendous variation for biotic and abiotic stresses (Kaur et al. 2018). Ae. tauschii accessions were selected for having stay green and bold grain traits while T. dicoccoides accessions were selected for their bold grain, expecting a better process of starch deposition in these accessions. On the other hand, Ae. speltoides accessions have been identified to be better performing under high temperatures (Awlachew et al. 2016). Impala is a winter wheat collection, and our studies showed it has a stay-green trait under North Indian environmental conditions. C591, on the other hand, derived from local landrace material, belongs to a group of tall traditional cultivars grown under rainfed conditions in Punjab before the advent of semi-dwarf wheat in the 1960s. These tall wheats were found to be resistant to abiotic stress tolerance (Almeselmani et al. 2012). The six haplotypes corresponding to synonymous substitutions, identified in the present study could serve as new haplotypes contributing to better grain filling and starch deposition though only phenotypic confirmation has yet been done.
Conclusion
Starch is the major component of mature wheat grain and significantly determines the ultimate yield of the wheat crop. However, the increments in the global temperature especially during the seed development have raised some serious concerns across the world. The elevated temperatures during these stages affect the major biosynthetic pathways/enzyme (s) involved in starch deposition and grain development. One of these enzymes, Starch Synthase I, was targeted in the present study with an aim to understand the allelic diversity available in the cultivated and wild germplasm of wheat. In total, six SNPs (four from wild species; two from cultivated species) present in the Exon-1, -2, and -4 were identified in the SSI gene and were associated with the GT domain of the starch synthase protein. Exploitation and transfer of allelic variation have proven to be a useful resource for wheat improvement. A number of allelic variants have been identified for various candidate genes in wheat such as Pm3 for powdery mildew resistance (Kaur et al. 2008), Wx-A1 for waxy gene and amylose biosynthesis (Saito and Nakamura 2005), Ap1 and PhyC for vernalization response (Beales et al. 2005), SSII for endosperm starch biosynthesis (Shimbata et al., 2005), Wx-B1 for waxy protein (Monari et al. 2005), PSY-1 and PSY-2 for grain yellow pigment content (Zhang and Dubcovsky 2008), Viviparous-1 for pre-harvest sprouting tolerance (Xia et al. 2008) and TaALMT1 for aluminum resistant (Raman et al. 2008). Identification of diversity and novel variants in the present study indicated that wild and cultivated germplasm is a good resource for useful alleles and the positive effects of these haplotypes could be further validated through molecular and phenotypic evaluation.
Availability of data and material
The sequences deposited in gene bank with accession numbers MN75891 to MN75907.
Code availability
Not applicable.
Abbreviations
- SS :
-
Soluble starch synthase
- SNP:
-
Single Nucleotide Polymorphism
- GT5:
-
Glycosyltransferase 5
- dp:
-
Degree of polymerization
- bp:
-
Base pairs
References
Almeselmani M, Deshmukh PS, Chinnusamy V (2012) Effects of prolonged high temperature stress on respiration, photosynthesis and gene expression in wheat (Triticum aestivum L.) varieties differing in their thermotolerance. Plant Stress 6:25–32
Awlachew ZT, Singh R, Kaur S, Bains NS, Chhuneja P (2016) Transfer and mapping of the heat tolerance component traits of Aegilops speltoides in tetraploid wheat Triticum durum. Mol Breed 36:78–94
Batley J, Mogg R, Edwards D, O’Sullivan H, Edwards KJ (2003) A high-throughput SNuPE assay for genotyping SNPs in the flanking regions of Zea mays sequence tagged simple sequence repeats. Mol Breed 11:111–120
Beales J, Laurie DA, Devos KM (2005) Allelic variation at the linked AP1 and PhyC loci in hexaploid wheat is associated but not perfectly correlated with vernalization response. Theor Appl Genet 110:1099–1107
Breathnach R, Chambon P (1981) Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem 50:349–383
Cao H, Inparl-Radosevich J, Guan H, Keeling PL, James MG, Myers AM (1999) Identification of the soluble starch synthase activities of maize endosperm. Plant Physiol 120:205–215
Carver T, Berriman M, Tivey A, Patel C, Böhme U, Barrell BG, Parkhill J, Rajandream MA (2008) Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24(23):2672–2676. https://doi.org/10.1093/bioinformatics/btn529
Ceoloni C, Kuzmanovi L, Ruggeri R, Rossini F, Forte P, Cuccurullo A, Bitti A (2017) Harnessing genetic diversity of wild gene pools to enhance wheat crop production and sustainability: challenges and opportunities. Diversity 9:55–76
Crofts N, Nakamura Y, Fujita N (2017a) Critical and speculative review of the roles of multi-protein complexes in starch biosynthesis in cereals. Plant Sci 262:1–8
Crofts N, Sugimoto K, Oitome NF, Nakamura Y, Fujita N (2017b) Differences in specificity and compensatory functions among three major starch synthases determine the structure of amylopectin in rice endosperm. Plant Mol Biol 94:399–417
Delvallé D, Dumez S, Wattebled F, Roldán I, Planchot V, Berbezy P, Colonna P, Vyas D, Chatterjee M, Ball S, Mérida A, D’Hulst C (2005) Soluble starch synthase I: a major determinant for the synthesis of amylopectin in Arabidopsis thaliana leaves. Plant J 43:398–412
Dhillon GS, Kaur S, Das N, Singh R, Poland J, Kaur J, Chhuneja P (2020) QTL mapping for stripe rust and powdery mildew resistance in Triticum durum–Aegilops speltoides backcross introgression lines. Plant Genet Resour 18:211–221
Donini P, Law JR, Koebner RMD, Reeves JC, Cooke RJ (2005) The impact of breeding on genetic diversity and erosion in bread wheat. Plant Genet Resour 3:391–399
Dubey R, Pathak H, Chakrabarti B, Singh S, Gupta DK, Harit RC (2020) Impact of terminal heat stress on wheat yield in India and options for adaptation. Agric Syst 181:102826–102832
Dwivedi SL, Upadhyaya HD, Stalker HT, Blair MW, Bertioli DJ, Nielen S, Ortiz R (2008) Enhancing crop gene pools with beneficial traits using wild relatives. In: Jules J (ed) Plant breeding reviews. Wiley, Hoboken, NJ, pp 179–230
FAO (2020) FAOSTAT statistical database. FAO, Rome
Felsenstein J (2005) PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genome Sciences, University of Washington, Seattle. https://evolution.genetics.washington.edu/phylip.html
Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28(23):3150–3152. https://doi.org/10.1093/bioinformatics/bts565
Fujita N, Yoshida M, Asakura N, Ohdan T, Miyao A, Hirochika H, Nakamura Y (2006) Function and characterization of starch synthase I using mutants in rice. Plant Physiol 140:1070–1084
Fujita N, Yoshida M, Kondo T, Saito K, Utsumi Y, Tokunaga T, Nishi A, Satoh H, Park J-H, Jane J-L, Miyao A, Hirochika H, Nakamura Y (2007) Characterization of SSIIIa-deficient mutants of rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol 144:2009–2023
Gao M, Chibbar RN (2000) Isolation, characterization, and expression analysis of starch synthase IIa cDNA from wheat (Triticum aestivum L.). Genome 43:768–775
Haga H, Yamada R, Ohnishi Y, Nakamura Y, Tanaka T (2002) Gene-based SNP discovery as part of the Japanese Millennium Genome Project: identification of 190 562 genetic variations in the human genome. J Hum Genet 47:605–610
Hayashi K, Hashimoto N, Daigen M, Ashikawa I (2004) Development of PCR-based SNP markers for rice blast resistance genes at the Piz locus. Theor Appl Genet 108:1212–1220
Huang XQ, Brlé-Babel A (2010) Development of genome-specific primers for homoeologous genes in allopolyploid species: the waxy and starch synthase II genes in allohexaploid wheat (Triticum aestivum L.) as examples. BMC Res Notes 3:140–151
Huang X-Q, Brûlé-Babel A (2012) Sequence diversity, haplotype analysis, association mapping and functional marker development in the waxy and starch synthase IIa genes for grain-yield-related traits in hexaploid wheat (Triticum aestivum L.). Mol Breed 30:627–645
Hurkman WJ, McCue KF, Altenbach SB, Korn A, Tanaka CK, Kothari KM, Johnson EL, Bechtel DB, Wilson JD, Anderson OD, DuPont FM (2003) Effect of temperature on expression of genes encoding enzymes for starch biosynthesis in developing wheat endosperm. Plant Sci 164:873–881
James MG, Denyer K, Myers AM (2003) Starch synthesis in the cereal endosperm. Curr Opin Plant Biol 6:215–222
Jeon J-S, Ryoo N, Hahn T-R, Walia H, Nakamura Y (2010) Starch biosynthesis in cereal endosperm. Plant Physiol Biochem 48:383–392
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780. https://doi.org/10.1093/molbev/mst010
Kaur N, Street K, Mackay M, Yahiaoui N (2008) Allele mining and sequence diversity at the wheat powdery mildew resistance locus Pm3. Mol Breed 78:1–3
Kaur S, Jindal S, Kaur M, Chhuneja P (2018) Utilization of wild species for wheat improvement using genomic approaches. In: Gosal SS, Wani SH (eds) Biotechnologies of crop improvement. Springer, Cham, pp 105–150
Kaur A, Chhuneja P, Srivastava P, Singh K, Kaur S (2021) Evaluation of Triticum durum–Aegilops tauschii derived primary synthetics as potential sources of heat stress tolerance for wheat improvement. Plant Genet Resour: Charact Util 19(1):74–89. https://doi.org/10.1017/S1479262121000113
Keeling PL, Myers AM (2010) Biochemistry and genetics of starch synthesis. Annu Rev Food Sci Technol 1:271–303
Keeling PL, Bacon PJ, Holt DC (1993) Elevated temperature reduces starch deposition in wheat endosperm by reducing the activity of soluble starch synthase. Planta 191:342–348
Keeling P, Banisadr R, Barone L, Wasserman B, Singletary G (1994) Effect of temperature on enzymes in the pathway of starch biosynthesis in developing wheat and maize grain. Funct Plant Biol 21:807–827
Kumar V, Kumar R, Panigrahi S, Pankaj YK (2021) Molecular cloning and characterization of the wheat (Triticum aestivum L.) starch synthase III gene sheds light on its structure. Cereal Res Commun. https://doi.org/10.1007/s42976-021-00182-w
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Leterrier M, Holappa LD, Broglie KE, Beckles DM (2008) Cloning, characterisation and comparative analysis of a starch synthase IV gene in wheat: functional and evolutionary implications. BMC Plant Biol 8:98–119
Li Z, Chu X, Mouille G, Yan L, Hashemi BK, Hey S, Napier J, Shewry P, Clarke B, Appels R, Morell MK, Rahman S (1999a) The localization and expression of the class II starch synthases of wheat. Plant Physiol 120:1147–1155
Li Z, Rahman S, Kosar-Hashemi B, Mouille G, Appels R, Morell MK (1999b) Cloning and characterization of a gene encoding wheat starch synthase I. Theor Appl Genet 98:1208–1216
Li Z, Mouille G, Hashemi BK, Rahman S, Clarke B, Gale KR, Appels R, Morel MK (2000) The structure and expression of the wheat starch synthase III gene. Motifs in the expressed gene define the lineage of the starch synthase III gene family. Plant Physiol 123:613–624
Li W, Liu A, Sheng YZ, Pu Z, Liu Y, Cheng GY (2013) Molecular characterization of SSI gene in Triticum L. and Aegilops tauschii. J Biol Sci 13:528–533
Longin C, Friedrich H, Reif JC (2014) Redesigning the exploitation of wheat genetic resources. Trends Plant Sci 19:631–636
Lopes MS, El-Basyoni I, Baenziger PS, Singh S, Royo C, Ozbek K, Aktas H, Ozer E, Ozdemir F, Manickavelu A, Ban T, Vikram P (2015) Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J Exp Bot 66:3477–3486
Luo G-H, Li XH, Han ZJ, Zhang ZC, Yang Q, Guo HF, Fang JC (2016) Transition and transversion mutations are biased towards GC in transposons of Chilo suppressalis (Lepidoptera: Pyralidae). Genes 7:72–82
MacLeod L, Duffus C (1988) Reduced starch content and sucrose synthase activity in developing endosperm of barley plants grown at elevated temperatures. Funct Plant Biol 15:375–390
Marshall OJ (2004) PerlPrimer: Cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics 20:2471–2472
McMaugh SJ, Thistleton JL, Anschaw E, Luo J, Konik-Rose C, Wang H, Huang M, Larroque O, Regina A, Jobling SA, Morell MK, Li Z (2014) Suppression of starch synthase I expression affects the granule morphology and granule size and fine structure of starch in wheat endosperm. J Exp Bot 65:2189–2201
Miralles DJ, Slafer GA (2007) Sink limitations to yield in wheat: How could it be reduced? J Agric Sci 145:139–149
Mishra BP, Kumar R, Mohan A, Gill KS (2017) Conservation and divergence of Starch Synthase III genes of monocots and dicots. PLoS ONE 12(12):e0189303
Mochida K, Yoshida T, Sakurai T, Ogihara Y, Shinozaki K (2009) TriFLDB: a database of clustered full-length coding sequences from Triticeae with applications to comparative grass genomics. Plant Physiol 150(3):1135–1146. https://doi.org/10.1104/pp.109.138214
Monari AM, Simeone MC, Urbano M, Margiotta B, Lafiandra D (2005) Molecular characterization of new waxy mutants identified in bread and durum wheat. Theor Appl Genet 110:1481–1489
Olivera PD, Rouse MN, Jin Y (2018) Identification of new sources of resistance to wheat stem rust in Aegilops spp. in the tertiary genepool of wheat. Front Plant Sci 9:1719
Pajkovic M, Lappe S, Barman R, Parisod C, Neuenschwander S, Goudet J, Alvarez N, Fran C (2014) Wheat alleles introgress into selfing wild relatives: empirical estimates from approximate Bayesian computation in Aegilops triuncialis. Mol Ecol 23:5089–5101
Pan X, Yan H, Li M, Wu G, Jiang H (2011) Evolution of the genes encoding starch synthase in sorghum and common wheat. Mol Plant Breed 2:60–67
Park Y, Nishikawa T (2012) Characterization and expression analysis of the starch synthase gene family in grain amaranth (Amaranthus cruentus L.). Genes Genet Syst 87:281–289
Pearce S, Gross HV, Herin SY, Hane D, Wang Y, Gu YQ, Dubcovsky J (2015) WheatExp: an RNA-seq expression database for polyploid wheat. BMC Plant Biol 15:299–307
Peleg Z, Fahima T, Korol AB, Abbo S, Saranga Y (2011) Genetic analysis of wheat domestication and evolution under domestication. J Exp Bot 62:5051–5061
Peng M, Hucl P, Chibbar RN (2001) Isolation, characterization and expression analysis of starch synthase I from wheat (Triticum aestivum L.). Plant Sci 161:1055–1062
Phan TTT, Ishibashi M, Miyazaki HT, Tran K, Okamura S, Tanaka J, Yuasa NT, Iwaya-Inoue M (2013) High temperature-induced repression of the rice sucrose transporter (OsSUT1) and starch synthesis-related genes in sink and source organs at milky ripening stage causes chalky grains. J Agron Crop Sci 199:178–188
Pradhan GP, Prasad PVV, Fritz AK, Kirkham MB, Gill BS (2012) High temperature tolerance in Aegilops species and its potential transfer to wheat. Crop Sci 52:292–304
Prakash P, Sharma-Natu P, Ghildiyal MC (2004) Effect of different temperature on starch synthase activity in excised grains of wheat cultivars. Indian J Exp Biol 42:227–230
Rahman S, Li Z, Batey I, Cochrane MP, Appels R, Morell M (2000) Genetic alteration of starch functionality in wheat. J Cereal Sci 31:91–110
Ramakrishna W, Dubcovsky J, Park YJ, Busso C, Embereton J, SanMiguel P, Bennetzen JL (2002) Different types and rates of genome evolution detected by comparative sequence analysis of orthologous segments from four cereal genomes. Genet 162:1389–1400
Raman H, Ryan PR, Raman R, Stodart BJ, Zhang K, Martin P, Wood R, Sasaki T, Yamamoto Y, Mackay M (2008) Analysis of TaALMT1 traces the transmission of aluminum resistance in cultivated common wheat (Triticum aestivum L.). Theor Appl Genet 116:343–354
Rose MK, Huang X-Q, Brûlé-Babel A (2016) Molecular characterization and sequence diversity of genes encoding the large subunit of the ADP-glucose pyrophosphorylase in wheat (Triticum aestivum L.). J Appl Genet 57:15–25
Saito M, Nakamura T (2005) Two point mutations identified in emmer wheat generate null Wx-A1 alleles. Theor Appl Genet 110:276–282
Sankoff D, Cedergren RJ, Lapalme G (1976) Frequency of insertion-deletion, transversion, and transition in the evolution of 5S ribosomal RNA. J Mol Evol 7:133–149
Sehgal A, Kumari S, Siddique KHM, Kumar R, Bhogireddy S, Varshney RK, Rao BH, Nair RM, Prasad PVV, Nayyar N (2018) Drought or/and heat-stress effects on seed filling in food crops: impacts on functional biochemistry, seed yields, and nutritional quality. Front Plant Sci 871:1705–1715
Shimbata T, Nakamura T, Vrinten P, Saito M, Yonemaru J, Seto Y (2005) Mutations in wheat starch synthase II genes and PCR-based selection of a SGP-1 null line. Theor Appl Genet 111:1072–1079
Tao T (2010) Standalone BLAST setup for Windows PC. In: BLAST® Help. National Center for Biotechnology Information (US), Bethesda, MD. 2008. https://www.ncbi.nlm.nih.gov/books/NBK52637/
Tuncel A, Okita TW (2013) Improving starch yield in cereals by over-expression of ADP-glucose Pyrophosphorylase: expectations and unanticipated outcomes. Plant Sci 211:52–60
Vogel F, Kopun M (1977) Higher frequencies of transitions among point mutations. J Mol Evol 9:159–180
Waines J (1994) High temperature stress in wild wheats and spring wheats. Funct Plant Biol 21:705–725
Xia LQ, Ganal MW, Shewry PR, He ZH, Yang Y, Roder MS (2008) Exploiting the diversity of Viviparous-1 gene associated with pre-harvest sprouting tolerance in European wheat varieties. Euphytica 159:411–417
Yamakawa H, Hakata M (2010) Atlas of rice grain filling-related metabolism under high temperature: Joint analysis of metabolome and transcriptome demonstrated inhibition of starch accumulation and induction of amino acid accumulation. Plant Cell Physiol 51:795–809
Yang H, Gu X, Ding M, Lu W, Lu D (2019) Activities of starch synthetic enzymes and contents of endogenous hormones in waxy maize grains subjected to post-silking water deficit. Sci Rep 9:7059–7067
Yin X, Guo W, Spiertz JH (2009) A quantitative approach to characterize sink-source relationships during grain filling in contrasting wheat genotypes. Field Crops Res 114:119–126
Zeeman SC, Kossmann J, Smith AM (2010) Starch: Its metabolism, evolution, and biotechnological modification in plants. Annu Rev Plant Biol 61:209–234
Zhang W, Dubcovsky J (2008) Association between allelic variation at the Phytoene synthase 1 gene and yellow pigment content in the wheat grain. Theor Appl Genet 116:635–645
Zhenlin W, Mingrong H, Jinmin F (1999) Effects of source sink manipulation on production and distribution of photosynthate after flowering in irrigated and rainfed wheat. Acta Agron Sin 25:162–168
Funding
Funding received from the Department of Science and Technology (DST), Govt. of India in the form of Project No. DST/INT/Aus/P-66/2015 is highly acknowledged.
Author information
Authors and Affiliations
Contributions
KS and SK conceived and designed the research. MS conducted the experiments and collected the data. ISY and AK reviewed the in-silico analysis. SK, PC, PS, and AK produced the final draft of the manuscript. SK and PC reviewed the experiments and manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that there is no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All the authors have agreed to the publication of this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Legends to Supplementary Figures: Fig. S1:
Dynamic expression profiles of SSI homoeologous (7A, 7B and 7D) gene in different wheat tissues. Fig. S2: Dynamic expression profiles of SSI homoeologous (7A, 7B and 7D) gene under heat and drought stress conditions. Fig. S3: PCR amplification profile of tetraploid and hexaploid wheat genotypes with SSL1 primer amplifying the SSI gene from nucleotides position 130 to 2419 L refers to 1 kb ladder (Promega, Cat. No. G5711); NC—negative control; 1-MACS9, 2-Arbon, 3-Impala, 4-C306, 5-C591, 6-WH542, 7-PBW343, 8-Halna. Fig. S4: PCR amplification of tetraploid and hexaploid wheat genotype using SSL2 primer amplifying the SSI gene from nucleotides position 2229 to 4906 (L refers to 1 kb ladder (Promega, Cat. No. G5711); NC- negative control; 1-MACS9, 2-Arbon, 3-Impala, 4-C306, 5-C591, 6-WH542, 7-PBW343, 8-Halna. Fig. S5: PCR amplification profile of wild wheat genotypes using SSOL3 primer pair amplifying the SSI gene from nucleotides position 1320 to 2164 (L refers to 1 kb ladder (Promega, Cat. No. G5711); NC—negative control; 1-T. monococcum acc. pau14087, 2-T. boeoticum acc. pau5088, 3-Ae. speltoides acc. pau3809, 4-Ae. speltoides acc. pau15081, 5-Ae. tauschii acc. pau3747, 6-Ae. tauschii acc. pau14102, 7-T. dicoccoides acc. pau14801, 8-T. dicoccoides acc. pau7107). Fig. S6: Neighbor-joining phylogenetic tree of SSI proteins corresponding to amplified gene sequences from 17 different diploid, tetraploid and hexaploid wheats in the present study with representative SSI protein sequences from monocots and dicots. Predicted amino acid sequences of the SSI genes were obtained from the NCBI (List of Accession Numbers is given in Supplementary Table 4). Bootstrap values are shown as percentages for 100 replicates. Fig. S7: Non-synonymous substitutions (with position) lying in the GT-5 domain region (1–6 exons); SSI amplified sequences from six different wheat genotypes. The coloured box represents exon; bar line (-) represents intron. Supplementary file1 (DOCX 703 kb)
Legends to Supplementary Tables: Table S1:
Plant material of wild and cultivated wheats used in this study. Table S2: Sequence of primers designed to amplify SSI gene from selected genotypes. Table S3: Lengths of different exon and Introns in SSI partial sequences as amplified from different wheat genotypes (length of each feature given in bp). Table S4: Detailed information of SNPs identified in Exon and Intron of SSI gene in 17 different wheat genotypes. Table S5: List of Accession Numbers of predicted amino acid sequences of the SSI genes obtained from the NCBI. Table S6: Non-synonymous substitutions in SSI gene (with position) lying in the GT-5 domain and corresponding exon. Supplementary file2 (DOCX 44 kb)
Rights and permissions
About this article
Cite this article
Singh, M., Kaur, S., Kaur, A. et al. Nucleotide diversity and molecular characterization of soluble starch synthase I gene in wheat and its ancestral species. J. Plant Biochem. Biotechnol. 32, 92–105 (2023). https://doi.org/10.1007/s13562-022-00785-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-022-00785-2