Abstract
Starch is a natural stored carbohydrate resource for the human diet, synthesized and stored in amyloplasts in higher plants. Starch synthase III is directly linked to the amylopectin chain’s elongation in wheat grains, but global warming is a critical barrier to its activity. The present investigation includes the characterization of the TaSS-IIIa1D gene in two wheat genotypes, viz. PBW-343 (heat susceptible) and IC252874 (heat tolerant). The genomic DNA of the above genotypes was used for PCR amplification of TaSS-IIIa1D gene using overlapping gene-specific primers and cloning the gene using the pJET1.2/blunt cloning vector, followed by sequencing and detection of SNPs. In-silico analysis revealed that the TaSS-III gene contains two homologs, TaSS-IIIa and TaSS-IIIb, which were found on the plus strand of chromosome 1 (1A, 1B and 1D) and minus strand of chromosome 2 (2A, 2B and 2D), respectively. The genomic size of the TaSS-IIIa1D gene was approximately 10,529 bp. All homeologous copies of the gene contained 16 exons. Twenty-nine specific SNPs were identified in PBW343 and twenty in IC252874 genotypes for the gene vis-a-vis with the reference copy of Chinese spring cultivar. There were 14 intronic and 15 exonic SNPs contributing to 18 transitions and 9 transversions in the genotype PWB-343, reflecting the transition bias, while in genotype IC252874, nine transitions, nine transversions, and two deletions contribute to 6 intronic and 14 exonic SNPs showing no such bias. Eighteen SNPs detected between the two genotypes for the gene consisting of 11 transitions and 7 transversions also showed transition bias. Third exon found most variable with the maximum number of SNPs. Seven SNPs were found associated with SBD-1, SBD-2, and SS-CD domains of the TaSS-IIIa1D protein. SNPs identified between the heat-sensitive and heat-tolerant genotypes for the heat-sensitive enzyme coding gene could be utilized for wheat improvement after proper validation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Starch is the second most abundant carbohydrate after cellulose, synthesized inside amyloplasts of higher plants. The carbohydrate comprises two glucose biopolymers of amylose and amylopectin, consisting of α-1, 4-linked gluten chains connected with α-1, 6-branch points, but differ in structure and biosynthesis (Pérez et al. 2010; Jeon et al. 2010). Amylopectin constitutes 75–90% of wild-type starch has a degree of polymerization (DP) of ~ 105 and an α-1, 6-branch level of 4–5%, and made up the structural framework and a semi-crystalline nature of starch. In comparison, amylose is smaller, less soluble in water, and lightly branched. It is considered to fill in the gap in a semi-crystalline branched matrix of amylopectin, possibly triggering denser packing of the starch granule (Manners 1989). Several enzymes coordinately catalyze the starch biosynthesis process. The ADP-glucose pyrophosphorylase (AGPase) catalyzes the first step and produces the activated glycosyl donor ADP-glucose (James et al. 2003). The starch synthase (SS) uses the activated donor ADP-glucose for the chain elongation via α-1,4-glycosidic linkages. Subsequently, starch branching enzyme (SBE) catalyzes the α-1,6-glycosidic linkages, and starch debranching enzyme (DBE) breaks the branch to adjust the starch structure (James et al. 2003; Keeling and Myers 2010; Tetlow 2011; Jeon et al. 2010; Brust et al. 2013). Out of the enzymes involved, starch synthase (SS) plays an important role during starch biosynthesis by extending the α-1,4 glucan chains by catalyzing the glucosyl moiety transfer from ADP-glucose to the non-reducing end of a pre-existing glucan chain (Thompson 2000). Five distinct SS isoforms, SSI, II, III, IV, and granule bound SS, have been identified. Each class has unique roles in the starch formation that arise from their physicochemical properties and substrate specificities (Ball and Morell 2003; Jeon et al. 2010). A new SS isoform in maize, designated SSV, was identified and suggested that SSIV and SSV resulted from gene duplication events (Liu et al. 2015). SSVI has been reported in potato starch but has not yet been functionally characterized (Helle et al. 2018). SSI forms the short chains of amylopectin (Commuri and Keeling 2001; Li et al. 2003). SSII plays a distinct role in catalyzing the formation of intermediate chains of amylopectin. Starch synthase III plays a pivotal role in extending the amylopectin fraction’s average chain length (Wang et al. 1993). Its loss of function is associated with reducing the very long chains’ proportion and slightly reduced gelatinization temperature in maize and barley (Yang et al. 2018; Li et al. 2011). SSIV has a role in priming the starch granule formation; moreover, its functions are partially supported by SSIII, depending on the plant species (Szydlowski et al. 2009). Palopoli et al. (2006) reported that SSIII from Arabidopsis thaliana has a putative N-terminal transit peptide followed by SSIII-specific domain (SSIII-SD) with starch binding function and a C-terminal catalytic domain. The endosperm of hexaploid wheat (Triticum aestivum L.) contains a high molecular weight starch synthase (SS) analogous to the maize du1 gene’s product. The starch synthase III (SSIII; DU1) cDNA sequences encoding wheat SSIII were isolated and characterized. The cDNA is 5346 bp long and contains an open reading frame that encodes a 1628-amino acid polypeptide with a putative N-terminal transit peptide, a central 470-amino acid SSIII-specific domain containing three regions of repeat, and a 436-amino acid C-terminal catalytic domain (Li et al. 2000). SS-III gene from the Arabidopsis genome shows a strongly conserved exon structure to that of the wheat SS-III gene, except for the N-terminal region (Li et al. 2000). Mishra et al. (2017) reported true orthologs of the well-characterized maize SSIII (ZmSSIII) among monocots and dicot species with nucleotide similarity ranging from 56 to 81%. They predicted the protein size of the SSIII orthologs with sequence identity ranging from 60 to 89%. The SSIII, SSIV, and SSV isoforms contained one or two coiled-coil domains in the N terminus involved in regulating protein–protein interactions (Hennen-Bierwagen et al. 2009). Moreover, three conserved carbohydrate-binding modules (CBM53 domain) of the N-terminal regions of SSIII isoforms play important roles in substrate binding (Valdez et al. 2008). The SS isoforms have also undergone gene duplication to different degrees (Qu et al. 2018). The SSIII protein has a C-terminal domain of glycosyltransferase family 1. It catalyzes the transfer of sugar moieties from activated donor molecules to specific acceptor molecules, forming glycosidic bonds. All living organisms share this highly conserved catalytic domain (Sinnott 1990; Campbell et al. 1997; Coutinho et al. 2003). Glycogen synthase (GSs) of archeal and eubacteria are the nearest relevance to starch synthase (SSs) of the plant in the GT5 family (Ball et al. 2011; Cenci et al. 2014), hinting that this family is ancient.
In wheat, starch synthesis activity was optimal between 20 and 25 °C, and 97% activity was lost at 40° C (Keeling et al. 1994). Heat stress led to a reduction of starch content, accumulation of starch, and grain dry weight at maturity (Yang et al. 2018). Heat and drought stress significantly decline the activity of the enzymes involved in the starch synthesis in wheat (Lu et al. 2019). High temperature applied from anthesis to maturity reduces the expression profiling of starch biosynthesis genes; particularly of soluble starch synthase (SSS), which is extremely sensitive to high temperature, as a result, starch accumulation period decreases and also reduces activities of starch biosynthesis enzymes (Rijven 1986; Hurkman et al. 2003; Keeling et al. 1994; Hawker and Jenner 1993; Zhao et al. 2008). Temperature stress also changes starch’s functional properties by altering the amylose-amylopectin composition ratio (Panozzo and Eagles 1998; Ball et al. 2011). Out of all SS genes, SSIII is most susceptible to heat stress; maize SSIII activity declined by approximately 50% after brief incubation at 45 °C, suggesting a role in reduced starch accumulation under the stress (Huang et al. 2016). Though there are few reports on the characterization of SSIII in higher plants, the full length of SSIII has not been cloned yet in wheat. The possible critical regulatory role and sensitivity to heat stress make the SSIII ideal for the detailed study. Considering the facts above, the present investigation is the first attempt in cloning the full length of the SSIII gene in wheat. It was carried out using the genomic DNA of heat stress-sensitive (PBW-343) and tolerant genotypes (IC252874) for in vitro cloning using a gene-based genome-specific primer of TaSS-IIIa1D gene and vector-mediated cloning using pJET1.2/blunt cloning vector followed by their sequencing to detect the gene-specific SNPs with a putative thermotolerance activity.
Materials and methods
Plant material
Two wheat genotypes PBW-343 (heat stress-sensitive) and IC252874 (heat stress-tolerant) were used for the present investigation based on their sensitivity toward heat stress. The experimental site was situated at Agricultural Farm of Dr. Rajendra Prasad Central Agricultural University, Pusa, Bihar, India. In-silico analysis and molecular characterization were carried out in the Genetic Transformation Laboratory of the university. Further primer validation, in vitro, in vivo cloning, and sequencing-related work was carried out in Plant Mediator Laboratory, National Institute of Plant Genome Research, New Delhi.
Genomic sequence retrieval using URGI-IWGSC BLAST
Reported accession ID of the SS-III gene of the wheat genome was used as a query sequence for searching reference DNA sequences of the SS-III gene by using NCBI-BLAST tool, and FASTA sequence of the cDNA was used for searching genomic sequences with the help of URGI-IWGSC BLAST of the wheat genome database.
Manual designing of genome-specific primers
Overlapping Genome-specific primers of the gene were designed manually to cover the gene of interest completely. For this, the nucleotide sequence of the target template was blasted using URGI-IWGSC BLAST (a wheat genome database) to access the sequence of homeologous (corresponding A B and D genome) gene. The homeologous sequences were aligned using Clustal Omega (multiple sequence alignment tool) (Fig. S1). Homeologous SNPs (single nucleotide polymorphism) were identified among aligned sequences. The SNPs that were specific to the genome of interest were considered in designing genome-specific primers. Using a primer design program (e.g., Primer3plus), forward (left), and reverse (right) primers designed separately, keeping the genome-specific SNP at the 3′ end of the primers. Primer designing parameters were set to optimize primer length (18–25), Tm (55–62 °C), and GC content (40–60%). Enlisted the selected primers with the genome-specific SNP at the 3′ end or close to (2–3 bases) the 3′ end (Table 1).
Molecular characterization of starch synthase III (TaSS-IIIa1D) gene
Molecular characterization of the TaSS-IIIa1D gene was done by including genomic DNA isolation (Porebski et al. 1997) method adapted from Doyle and Doyle (1990), PCR amplification, gel elution of the amplified product, pJET1.2/blunt-end cloning, and DNA ligation with vector. Preparation of LB broth & LB agar media, preparation of calcium competent cell, heat-shock transformation of E. coli (DH5-a Strain), selection of transformed cells, E. coli colony PCR, plasmid isolation using alkaline lysis method, sequencing of plasmid, and identification of SNPs are with respect to reference SS-III gene sequence.
Sequencing using automated DNA sequencer
Purified amplified products (TaSS-IIIa1D gene) obtained from both PBW-343 and IC252874 were sequenced using the ABI3730xI DNA analyzer at the sequencing facility NIPGR, New Delhi (Principle:Sanger method). For sequencing, 2 µl eluted amplified DNA (Conc. 20 ng) and 2 µl respective genome-specific sequencing primers (Conc. 3 µm/µl) were added into the 96-well plates, and then the rest of the 6 µl sequencing mixtures were added.
pJET1.2/blunt-end cloning and ligation
Out of 15 primers pairs, randomly two primer pairs (1 and 4) were selected for vector cloning, colony PCR, and plasmid sequencing, respectively. Then a comparative analysis of sequences of both vector clones and PCR amplicons of the TaSS-IIIa1D gene was aligned, and confirmation of exact sequencing was validated. In total, 20 µl DNA ligation reaction with pJET1.2/blunt vector was set up, following 10 µl (2X T4 DNA ligase buffer), 3 µl PCR insert (50 ng/µl), 1 µl pJET1.2/blunt vector, 1 µl Ligase enzyme, and 5 µl nuclease-free water. For cloning, two primers from the exonic region were selected and used for PCR amplification. The purified blunt-end PCR products (insert) were used in a 3:1 molar ratio with pJET1.2/blunt-end vector. The ligation mixture was incubated at room temperature (22 °C) for 2 h. 65 °C temperature was used for heat inactivation of ligase enzyme activity for 10 min. The ligation mixture was directly used for transformation.
Preparation of competent cells
For the preparation of the chemical-competent cells, a stationary overnight culture was diluted 1:100 in fresh LB broth media. After the cells were grown up to the logarithmic phase (OD600 0.4–0.6) at 37 °C, they were harvested by centrifugation for 10 min at 3500 rpm at 4 °C. Then, 10 ml CaCl2 (75 mM CaCl2 + 15% glycerol) were added in the pellet and resuspended. The cells were put on ice for 10 min and then centrifuged at 4500 rpm for 5 min, and the supernatant was discarded. Again 10 ml of CaCl2 was added in pellets and re-suspended.
Heat-shock transformation of E. coli (DH5-a strain)
Competent cells were mixed with the ligated product (vector ligated with PCR product) immediately before thawing. The content of the tubes was mixed by swirling gently and stored on ice for 30 min. All tubes were placed in a circulating water bath that had been preheated to 42 °C. The tubes were left in the water bath for precisely 90 s without shaking. Rapidly the tubes were transferred to an ice bath and were kept there for 1–2 min. One milliliter of LB broth media was added in all the tubes, and tubes were incubated at 37 °C for 1 h in a rotary shaker with 200 rpm. Then tubes were centrifuged at 4500 rpm for 5 min. The supernatant was decanted in such a way that only a small amount of supernatant was left with the pellets. The pellet was mixed well with the remaining supernatant. The pellet was placed in Petri plates having LB agar with ampicillin, using a sterile glass spreader. Then Petri plates were inverted and were incubated at 37° for 12–16 h.
Selection of transformed cells
Re-circularized pJET1.2/blunt vector molecules lacking an insert express a lethal restriction enzyme, which kills the host E. coli cell after transformation. As a result, only recombinant clones (colonies) containing the insert appeared on culture plates.
E. coli colony PCR
Colony PCR was used for screening of positive clones. Individual colonies were picked and re-suspended in 10 µl of distilled H2O. One microliter of this suspension and the respective primers (genome-specific primers) were added to the PCR mixture. After completing the PCR program, the amplicon size was verified by gel electrophoresis with respect to the 1 Kb DNA ladder.
Sequencing of plasmids with desired inserts
Plasmid isolation was done using the alkaline lysis method (Birnboim and Doly 1979). The isolated plasmids containing the insert of interest were sequenced by automated sequencer at the sequencing facility (Sanger platform) of the National Institute of Plant Genome Research, New Delhi. For sequencing, 2 µl of the isolated plasmid DNA (Conc. 5–10 ng), 2 µl of respective genome-specific primer (Conc. 3 µm/µl), and 6 µl sequencing mixtures were added to the 96-well plates.
Identification of SNPs
The sequencing data of the TaSS-III1D gene obtained from the samples taken from the wheat genotypes PBW343 and IC252874 were aligned against each other and also with reference sequence using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) with default parameters for the identification of SNPs.
Results
Retrieval of genomic sequence using URGI-IWGSC BLAST
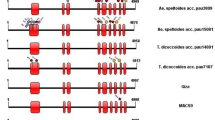
Reported accession ID no. AF258608 of 5346 bp cDNA sequence of wheat was retrieved from the NCBI database and used as a query sequence for searching and retrieving the TaSS-III gene’s genomic sequence IWGSC (Wheat Genomic database). BLAST report exhibited that the query sequence of SS-III cDNA has aligned maximum with the chromosome no.1 of each A, B, and D genome at a particular site with 98%, 96%, and 96% of aligned scores and 0.0 E-value. It also aligned with chromosome no. 2 of each of the A, B, and D genome at a particular site having 76%, 76%, and 76% of aligned scores, with a very high, 3e−152, 1e−157, and 8e−153, E-values, respectively. Full length of the three homeologous copies of the TaSS-IIIa gene with length 12,168 bp, 12,839 bp, and 10,529 bp was found at specific locations, i.e., from 84,002,521 to 84,014,688 (+ strand), 141,283,445 to 141,296,283 (+ strand), and 87,756,557 to 87,767,085 (+ strand) on chromosome 1A, 1B, and 1D in forwarding direction, respectively. Similarly three homeologous copies of the TaSS-IIIb gene of size 8244 bp, 8440 bp, and 9194 bp were identified on chromosome 2A, 2B, and 2D but in reverse orientation, at location from 712,573,140 to 712,581,384 (− strand), 690,026,216 to 690,034,656 (− strand) and 574,104,017 to 574,113,211 (− strand), respectively. Out of all homeologous, we selected the TaSS-IIIa1D gene for cloning and sequencing purposes. Bioinformatics finding further showed the details information of the TaSS-IIIa1D gene, for example, the TaSS-IIIa1D gene (ID no. TraesCS1D02G100100) present on chromosome 1D, has 3 transcripts (splice variants), 87 orthologs, and 37 paralogs. The three splice variants, viz. TraesCS1D02G100100.1, TraesCS1D02G100100.2, and TraesCS1D02G100100.3, contain 5348 bp, 5322 bp, and 5493 bp length of the cDNA sequence, encoding 1613, 1554, and 1611 amino acid sequence, respectively. Fully expressed, the first transcript has 16 exons. It is annotated with 24 domains and features, associated with 468 variant alleles and maps to 17 oligo probes. The second transcript has 15 exons (exon ten is spliced out). It is annotated with 26 domains and features, associated with 468 variant alleles and maps to 17oligo probes. The third transcript has 16 exons and is annotated with 25 domains and features. It is associated with 468 variant alleles and maps to 20 oligo probes.
Amplification of DNA using TaSS-IIIa1D gene-specific primers
Out of all copies of the TaSS-III genes, molecular characterization was carried out for the TaSS-IIIa1D gene. Fifteen genome-specific primer sets were designed to cover the complete length of the TaSS-IIIa1D gene. The primer pairs were designed in such a way that their amplifiable length has overlapping ends so that the entire length of the gene is covered without any gap. Except for the primer pair 15, rest all, 1–14 primer pairs were designed to have an average length of 700–750 bp. All the primer pairs except the 10th and 11th were amplified with desired lengths (Table 1). A modified touchdown or two-step PCR program was used to amplify the entire length of the TaSS-IIIa1D gene using genome-specific overlapping primer pairs. The primer pairs from 1st to 9th and 12th were amplified at 64 °C and 60° annealing temperature, and the rest 13th, 14th, and 15th were amplified at 66 °C and 62 °C annealing temperatures. The amplicon size of the primer pairs 1–9 and 12–14th was found comparable to the 750 bp band of the DNA ladder, which was the desirable length. For the primer pair 15, a very bright amplicon of size 416 bp, comparable to the 500 bp band of the DNA ladder, was obtained. No desirable size of amplicon could be obtained for the 10th and 11th primers from an extensive range of annealing temperatures (55–66 °C). The desired amplicons with approximately sharp band resolved on a 2% gel were cut and purified using QIAquick Qiagen Kit (Fig S2, S3, and S4).
Blunt-end cloning and selection of transformed cells
PCR products of selected primer pairs 1 and 4 were extracted and purified from agarose gel and ligated into the pJET1.2/blunt cloning vector. A 3:1 molar ratio of the PCR product (insert) and pJET1.2/blunt-end vector was used, which increased the probability of ligation. The ligated vector was mixed with E. coli (DH5α strain) culture and incubated at 37 °C for 2–3 h and then poured onto ampicillin containing the LB plate. An ampicillin-containing plate was used as a control (Fig S5). Two single positive colonies of transformed cells were picked for each pair of primer and cultured in LB broth for the multiplication of more clones. Transformed bacterial colonies were obtained in sufficient numbers (Fig S6). Re-circularized pJET1.2/blunt vector molecules lacking an insert express a lethal restriction enzyme, which kills the host E. coli cell after transformation. As a result, only recombinant clones (colonies) containing the insert appeared on culture plates. No contamination was observed in the control plate, which proved no contamination in transformed colonies with other bacterial strains.
E. coli colony PCR
Colony PCR was used to determine the presence or absence of the inserted DNA in the plasmid vector directly using bacterial colonies. The primer pairs 1 (748 bp, amplicon length) and 4 (745 bp, amplicon length) were used to amplify the insert containing plasmid vector, and positive results were found (Fig S7).
Comparative analysis of the data obtained from the sequencing of direct PCR and vector cloning products of primer pairs 1 and 4
Plasmid with the insert of interest was isolated and sequenced. The sequencing was carried out only with the forward primer of pairs 1 and 4 for validating the sequences of both the products through a comparative analysis of the sequencing data of both direct PCR products and indirect vector cloned products. The plasmid vectors with insert were sequenced using already designed forward primer and compared with the sequenced PCR products for validation. No significant variations were observed in the nucleotide sequences obtained from the sequencing of direct PCR products and the cloning vector (Fig S8 and S9). Variations in nucleotide sequences or SNPs were found only against reference sequences.
Identification of SNPs with respect to reference SS-III gene sequence
The TaSS-IIIa1D gene’s amplicons obtained from PBW-343 and IC252874 were sequenced with forward and reverse primer separately in two reads. Sequenced reads were aligned against the gene sequence of TSS-IIIa1D obtained from the reference genome of the bread wheat variety Chinese Spring, using Clustal Omega, and SNPs of individual genotypes were marked against the reference sequence (Table 2). A total of 29 SNPs were detected in genotype PBW343 vis-a-vis the reference copy, out of which 15 were found in the exonic region, and 14 were present in the intronic region. In total, 80% of the exonic SNPs were detected in Exon 3, and 50% of the intronic SNPs were found in Intron 8. Among exonic SNPs, 66.67% were due to transition, whereas 57.14% of intronic SNPs were transition types. Genotype IC252874 exhibited 20 SNPs compared to the reference copy. All 14 exonic SNPs were detected in Exon 3, whereas 6 SNPs were found in the intronic region with a maximum of 3 SNPs that were present in Intron 5. In total, 57.14% of the exonic and 16.67% of the intronic SNPs were of transition type. A comparison of the gene sequences of the two wheat genotypes under consideration also revealed 18 SNPs (Table 3). Out of the total SNPs detected, 12 were found in the exonic region, whereas 6 were revealed in the intronic region. Exon 3 inhabited 91.67% of total exonic SNPs; on the other hand, 50% of intronic SNPs were located in Intron 5. Out of total exonic SNPs, 66.67% were due to transition; on the other hand, 50% of the total intronic SNPs were transition type.
SNPs detection in domains of TaSS-IIIa1D based on sequencing data
A total of 7 SNPs were detected in the Starch Binding Domain and SS-Catalytic Domain of the TaSS-IIIa1D gene of wheat genotypes PBW-343 and IC252874. In SBD-1, SNPs (4004th) of the gene was a cause of a non-synonymous mutation, leucine in place of phenylalanine (phe8) already evolutionarily conserved in SBD-1 domains of both TaSS-IIIa1A and TaSS-IIIa1B. In contrast, SNPs (4100th) of both genotypes were found to be a cause of a synonymous mutation, histidine to histidine in SBD-1. Furthermore, SNPs (4596th) in the genotype IC252874 were found to be a cause of one non-synonymous mutation, serine to threonine in SBD-2. SNP (7180th) was found to be a cause of a synonymous mutation, leucine to leucine, and SNP (7202th), a cause of non-synonymous mutation serine to cysteine in starch synthase catalytic domain (SS-CD) of PBW-343.
Discussion
Starch is the major reserved food energy source globally, mainly depending on the processes of starch synthesis and accumulation. It is a polymer of amylose and amylopectin, with 20–30% and 70–80% ratios, respectively. Both polymers are made of α-1, 4-linked glucan chains connected with α-1, 6-branch points. However, amylopectin is highly branched than amylose. It has a high degree of polymerization (DP) of ~ 105 parts of the molecules (Thompson 2000; Gao et al. 2003; Bresolin et al. 2006; Pflister and Zeeman 2016). Starch biosynthesis is carried out in a cascade manner through ADP-glucose pyrophosphorylase (AGPase), starch synthase (SS), starch branching enzyme (SBE), and starch debranching enzyme (DBE). To date, seven distinct classes of starch synthase have been reported, i.e., granule bound SS (GBSSI) and SS classes I, II, III, and IV, V, VI (SSI, SSII, SSIII, SSIV, SSV, SSVI) (Ball and Morell 2003; Jeon et al. 2010). Among them, SS-III plays an important role in elongating amylopectin’s length and helping in forming the starch granule. However, it is susceptible to heat stress, ultimately leading to yield losses. Considering the significant role of starch synthase III enzyme in starch biosynthesis and incomplete information about the TaSS-III gene and its homologs in wheat, the current study was designed to cover this gap through cloning and detailed study of TaSS-III gene sequences at the SNPs level. The study takes into account recently published information of reference wheat genome sequence and uses the genomic DNA of PBW-343 (heat susceptible) and IC252874 (heat tolerance) genotypes. Bioinformatics analysis shows that the wheat genome has two variants of TaSS-III, TaSS-IIIa, and TaSS-IIIb located on chromosome 1 and chromosome 2, respectively. The gene TaSS-IIIa is highly expressed in the endosperm of the grains. Fujita et al. (2007) also reported two similar forms OSS-IIIa and OSS-IIIb in rice. The OSS-IIIa expresses late during grain filling phage, five days after fertilization (DAF), and plays a major role in elongating the relatively long amylopectin chains of starch (Hirose and Terao 2004; Ohdan et al. 2005), whereas TaSS-IIIb expresses early at different development stages in the five tissues, viz. leaf, stem, root, spike, and grain, but highly expresses in leaves and during the grain filling stage (1–5 DAF) (Dian et al. 2005). Among all, we selected TaSS-IIIa1D located on the D genome of wheat for our cloning and sequencing purposes. It is based on the fact that among the ancestral species of hexaploid wheat, the diploid D genome progenitor has the widest geographic distribution from Turkey on the West to Afghanistan and Central Asia in the East adapted to diversified environmental conditions. This wide geographic distribution is coupled with an overall greater genetic variation (diversity) that helps in wheat improvement programs (Mizuno et al. 2010; Wang et al. 2013; Majka et al. 2017) as Ae. tauschii, the D genome progenitor of wheat, is an excellent source of novel genes to various biotic and abiotic stresses (Valkoun et al. 1985; Gill et al. 1986; Cox et al. 1992; Assefa and Fehrmann 2004). The pJET1.2/blunt cloning vector was used for in vivo cloning into E. coli (DH5α strain) because it is re-circularized into pJET1.2/blunt vector molecules lacking an insert express a lethal restriction enzyme, which kills the host E. coli cell after transformation. As a result, only recombinant clones (colonies) containing the insert appeared on culture plates. Also, the 5′ end of the plasmid is already phosphorylated, so no extra step of phosphorylation is needed for ligation (Fig S6). Colony PCR was performed to screen and validate the inserted DNA using a pre-designed primer, specific to the TSS-IIIa1D gene (Fig S7). It was followed by plasmid isolation and sequencing, respectively. Comparative analysis of the sequencing information of the products of direct PCR and indirect vector cloning using primer pairs 1 and 4 showed (Fig S8 and S9) no variation. Hence, Phusion polymerase may be used to amplify long strands of DNA for sequencing and SNPs detection purposes. Findings of the Dolgova and Stukolova (2017) and McInerney et al. (2014) also showed that Phusion polymerase contains high fidelity and very less mismatches of nucleotides during PCR amplification, with an error rate > 50 times lower than that of Taq polymerase and sixfold lower than that of pfu polymerase. Because it possesses both 5´ → 3´ DNA polymerase and 3´ → 5´ exonuclease activity and generates blunt-end PCR products. SNPs are third-generation molecular marker techniques that came after RFLP and SSR. It is used successfully to examine genetic variations in the genome sequence of individuals of one population, different species and breeds, respectively (The Bovine HapMap Consortium 2009; Brooks et al. 2010). SNPs are present in three different forms: transitions (C/T or G/A), transversions (C/G, A/T, C/A, or T/G), and small insertions–deletions (indels). They are used as direct markers, as the sequence information provides the exact nature of the allelic variants. The base substitutions are the most common mutations and create individual variations, community diversity, and species evolution. Single base substitution can be divided into two classes: transitions (C/T or G/A), transversions (C/G, A/T, C/A, or T/G). Since there are 4 types of transitions and 8 types of transversions, the expected ratio or the probability of the occurrence of a transition to transversions (Ts/Tv) is 0.5 (Denver et al. 2009, 2012).
In the current study, we identified a total of 49 SNPs in 10,529 bp of the TSS-IIIa1D gene. Twenty-nine SNPs were revealed in heat-sensitive genotype PBW-343, and 20 specific SNPs were identified in the heat-tolerant genotype IC252874 (Table 2) vis-a-vis with the retrieved reference gene copy of the Chinese Spring bread wheat variety. In comparison with the reference genome, there were 14 intronic and 15 exonic SNPs contributing to 18 transitions and 9 transversions in the PWB-343 genotype, suggesting that the expected ratio or the probability of the occurrence of a transition to transversions (Ts/Tv) is 2.0, reflecting the transition bias in PBW-343. It is often observed that transitions occur more than transversion, called transition bias (Stoltzfus and Norris 2016), while in IC252874, 9 transitions, 9 transversions, and two deletions contribute to 6 intronic and 14 exonic SNPs showing the Ts/Tv ratio to be 1, suggesting that no bias was found. However, as per trend, the exonic region exhibited a transition bias with 57.14% transition, but contrary to trend intronic region exhibited a transversion bias may be due to the small sample size. Comparing the gene sequence from the two genotypes understudy revealed 18SNPs consisting of 11 transitions and 7 transversions reflecting the transition bias in conformity with the previous study. This transition bias may occur due to the relatively high rate of mutation of methylated cytosine to thymine. The cytosine-guanine (CpG) dinucleotides exhibit the high transition frequencies expected of methylated sites (Keller et al. 2007). The bias observed in wheat is higher than that observed in barley using a similar method (Duran et al. 2009), revealing the higher level of DNA methylation in the wheat genome. Transition bias may also be due to the biasedness of DNA polymerization to transitions over transversion. This transitional bias is supported for both coding and non-coding regions (Zhang and Gerstein 2003; Jiang and Zhao 2006; Pauly et al. 2017) or natural selection favoring transition than transversions because it has been found that transversion mutations are more lethal than transition mutations. This hypothesis is based on the observation that, depending on codon usage, non-synonymous transitions are more likely to conserve important biochemical properties of the original amino acid (Vogel et al. 1997; Zhang 2000). For example, a mutation that changes an amino acid’s charge is a “radical” change, whereas a “conservative” change does not favor this. Stoltzfus and Norris found a statistically significant difference after combining the data, which was considered valuable biological significance (Stoltzfus and Norris 2016). Exon 3 was found as the most variable region in the gene with the maximum number of SNPs. Mishra et al. (2017) also reported Exon 3 to be evolutionarily highly variable among all monocots and dicots taxa. A comparison of the sequence information of the TaSS-IIIa1D gene from the two genotypes under study led to the detection of 7 SNPs associated with SBD-1, SBD-2, and SS-CD domains of the TaSS-IIIa1D protein (Appendix II and III). The 4004th SNPs of TaSS-IIIa1D were found to be a non-synonymous mutation, which replaces phenylalanine (phe8) with leucine, in already evolutionarily conserved SBD-1 domains of both TaSS-IIIa1A and TaSS-IIIa1B. On the other hand, the 4596th SNP in IC252874 was identified as a non-synonymous mutation, serine to threonine in SBD-2. Threonine in place of serine may more efficiently be involved in the catalytic activity because threonine, as a nucleophile, may donate an electron pair to form a chemical bond in the chemical reaction (Kisselev et al. 2000). Deleting the amino-terminal threonine by alanine in archaeal, proteasomes abolish their proteolytic activity (Seemüller et al. 1995). However, it is still unclear how an additional methyl group in threonine affects ester hydrolysis in precisely the opposite manner to its effects on natural substrates, where the N-terminal serine is insufficient to support rapid rates of protein breakdown. Besides, 7202nd SNP of TaSS-IIIa1D was a non-synonymous substitution, serine to cysteine. Both serine and cysteine differ only in the swap of a sulfur atom with oxygen and retain their hydrophilic properties. The serine to cysteine substitution may have import bearing on the catalytic role of the enzyme. Skryhan et al. (2015) also experimented to know that specific thiol-disulfide exchange in starch synthase 1 (AtSS1) of Arabidopsis thaliana influences its catalytic function and role of conserved Cys residue on AtSS1 catalysis. The results indicated that both cysteines play important roles in enzyme catalysis; Cys545 is involved in ADP–glucose binding and cys164 in acceptor binding, respectively. Besides, Cys265 and Cys164 could be involved in proper protein folding, whereas Cys442 could play an important role in enzyme stability upon oxidation. Hence it indicates that the substitution may lead to enhanced stress tolerance. Therefore the SNPs identified between the heat-tolerant and heat-susceptible genotypes for TaSS-IIIa1D gene coding for the heat-sensitive enzyme could be validated further for linking them for heat tolerance to be utilized in the development of heat-tolerant wheat variety.
Availability of data and materials
All data generated or analyzed during this study are included in this article.
References
Assefa S, Fehrmann H (2004) Evaluation of Aegilopstauschii Coss. for resistance to wheat stem rust and inheritance of resistance genes in hexaploid wheat. Genet Resour Crop Evol 51:663–669
Ball SG, Morell MK (2003) From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu Rev Plant Biol 54:207–233
Ball S, Colleoni C, Cenci U, Raj JN, Tirtiaux C (2011) The evolution of glycogen and starch metabolism in eukaryotes gives molecular clues to understand the establishment of plastid endosymbiosis. J Exp Bot 62:1775–1801
Birnboim HC, Doly J (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res 7(6):1513–1523
Bresolin NS, Li Z, Kosar-Hashemi B, Tetlow IJ, Chatterjee M, Rahman S, Morell MK, Howitt CA (2006) Characterisation of disproportionating enzyme from wheat endosperm. Planta 224:20–31
Brooks SA, Gabreski N, Miller D, Brisbin A, Brown HE, Streeter C, Mezey J, Cook D, Antczak DF (2010) Whole-genome SNP association in the horse: identification of a deletion in myosin va responsible for lavender foal syndrome. PLoS Genet 6:e1000909
Brust H, Orzechowski S, Fettke J, Steup M (2013) Starch synthesizing reactions and paths: in vitro and in vivo studies. J Appl Glycosci 60:3–20
Campbell JA, Davies GJ, Bulone V, Henrissate B (1997) A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J 326:929–939
Cenci U, Nitschke F, Steup M, Minassian BA, Colleoni C, Ball SG (2014) Transition from glycogen to starch metabolism in Archaeplastida. Trends Plant Sci 19:18–28
Commuri PD, Keeling PL (2001) Chain-length specificities of maize starch synthase I enzyme: studies of glucan affinity and catalytic properties. Plant J 25:475–486
Coutinho PM, Deleury E, Davies GJ, Henrissat B (2003) An evolving hierarchical family classification for glycosyltransferases. J Mol Biol 328:307–317
Cox TS, Raupp WJ, Wilson DL, Gill BS, Leath S, Bockus WW (1992) Resistance to foliar diseases in a collection of T. tauschii germplasm. Plant Dis 76:1061–1064
Denver DR, Dolan PC, Wilhelm LJ, Sung W, Lucas-Lledó JI, Howe DK, Lewis SC, Okamoto K, Thomas WK, Lynch M, Baer CF (2009) A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. PNAS USA 106:16310–16314
Denver DR, Wilhelm LJ, Howe DK, Gafner K, Dolan PC, Baer CF (2012) Variation in base-substitution mutation in experimental and natural lineages of Caenorhabditis nematodes. Genome Biol Evol 4(4):513–522
Dian W, Jiang H, Wu P (2005) Evolution and expression analysis of starch synthase III and IV in rice. J Exp Bot 56:623–632
Dolgova AS, Stukolova OA (2017) High-fidelity PCR enzyme with DNA-binding domain facilitates de novo gene synthesis. 3Biotech 7:128–136
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:11–15
Fujita N, Yoshida M, Kondo T, Saito K, Utsumi Y, Tokunaga T, Nishi A, Satoh H, Park JH, Jane JL (2007) Characterization of SSIIIa-deficient mutants of rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol 144:2009–2023
Gao SJ, Wang WJ, Gu TC, Han JF (2003) C –N metabolic characteristics in flag leaf and starch accumulating developments in seed during grain filling stage in two winter wheat cultivars with different Spike Type. Acta Agron Sin 29:427–431
Gill BS, Raupp WJ, Sharma HC, Browder LE, Hatchett JH, Harvey TL (1986) Resistance in Aegilops squarrosa to wheat leaf rust, wheat powdery mildew, greenbug and Hessian fly. Plant Dis 70:553–556
Hawker JS, Jenner CF (1993) High temperature affects the activity of enzymes in the committed pathway of starch synthesis in developing wheat endosperm. Funct Plant Biol 20:197–209. https://doi.org/10.1071/PP9930197
Helle S, Bray F, Verbeke J, Devassine S, Courseaux A, Facon M, Tokarski C, Rolando C, Szydlowski N (2018) Proteome analysis of potato starch reveals the presence of new starch metabolic proteins as well as multiple protease inhibitors. Front Plant Sci 9:746
Hennen-Bierwagen TA, Lin Q, Grimaud F, Planchot P, Keeling PL, James MG, Myers AM (2009) Proteins from multiple metabolic pathways associate with starch biosynthetic enzymes in high molecular weight complexes: a model for regulation of carbon allocation in maize amyloplasts. Plant Physiol 149(3):1541–1559
Hirose T, Terao T (2004) A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.). Planta 220:9–16
Huang B, Keeling PL, Hennen-Bierwagen TA, Myers AM (2016) Comparative in vitro analyses of recombinant maize starch synthases SSI, SSIIa, and SSIII reveal direct regulatory interactions and thermosensitivity. Arch Biochem Biophys 596:63–72
Hurkman WJ, Mccue KF, Altenbach SB, Korn A, Tanaka CK, Kothari KM (2003) Effect of temperature on expression of genes encoding enzymes for starch biosynthesis in developing wheat endosperm. Plant Sci 164(5):873–881
James MG, Denyer K, Myers AM (2003) Starch synthesis in the cereal endosperm. Curr Opin Plant Biol 6:215–222
Jeon JS, Ryoo N, Hahn TR, Walia H, Nakamura Y (2010) Starch biosynthesis in cereal endosperm. Plant Physiol Biochem 48(6):383–392
Jiang C, Zhao Z (2006) Mutational spectrum in the recent human genome inferred by single nucleotide polymorphisms. Genomics 885:527–534
Keller I, Bensasson D, Nichols RA (2007) Transition-transversion bias is not universal: a counter example from grasshopper pseudogenes. PLoS Genet 3(2):e22
Keeling PL, Myers AM (2010) Biochemistry and genetics of starch synthesis. Annu Rev Food Sci Technol 1:271–303
Keeling PL, Bacon PJ, Holt DC (1994) Elevated temperature reduces starch deposition in wheat endosperm by reducing the activity of soluble starch synthase. Planta 191(3):342–348
Kisselev AF, Songyang Z, Goldberg AL (2000) Why does threonine, and not serine, function as the active site nucleophile in proteasomes? J Biol Chem 275:14831–14837
Li Z, Mouille G, Kosar-Hashemi B, Rahman S, Clarke B, Gale KR, Appels R, Morell MK (2000) The structure and expression of the wheat starch synthase III gene. Motifs in the expressed gene define the lineage of the starch synthase III gene family. Plant Physiol 123:613–624
Li Z, Sun F, Xu S, Chu X, Mukai Y, Yamamoto M, Ali S, Rampling L, Kosar-Hashemi B, Rahman S, Morell MK (2003) The structured organization of the gene encoding class II starch synthase of wheat and barley and the evolution of the genes encoding starch synthase in plants. Funct Integr Genom 3:76–85
Li Z, Li D, Du X, Wang H, Larroque O, Jenkins CLD, Jobling SA, Morell MK (2011) The barley amo1 locus is tightly linked to the Starch Synthase IIIa gene and negatively regulates expression of granule-bound starch synthetic genes. J Exp Bot 62(14):5217–5231
Liu H, Yu G, Wei B, Wang Y, Zhang J, Hu Y, Liu Y, Yu G, Zhang H, Huang Y (2015) Identification and phylogenetic analysis of a novel starch synthase in maize. Front Plant Sci 6:1013
Lu H, Hu Y, Wang C, Liu W, Ma G, Han Q, Ma D (2019) Effects of high temperature and drought stress on the expression of gene encoding enzymes and the activity of key enzymes involved in starch biosynthesis in wheat grains. Front Plant Sci 10:1414
Majka M, Kwiatek MT, Majka J, Wiśniewska H (2017) Aegilopstauschii accessions with geographically diverse origin show differences in chromosome organization and polymorphism of molecular markers linked to leaf rust and powdery mildew resistance genes. Front Plant Sci 8:1149
Manners DJ (1989) Recent developments in our understanding of amylopectin structure. Carbohyd Polym 11(2):87–112
McInerney P, Adams P, Hadi MZ (2014) Error rate comparison during polymerase chain reaction by DNA polymerase. Mol Biol Int 287–430
Mishra BP, Kumar R, Mohan A, Gill KS (2017) Conservation and divergence of Starch Synthase III genes of monocots and dicots. PLoS ONE 12(12):e0189303. https://doi.org/10.1371/journal.pone.0189303
Mizuno N, Yamasaki M, Matsuoka Y, Kawahara T, Takumi S (2010) Population structure of wild wheat D-genome progenitor Aegilops tauschii Coss.: implications for intraspecific lineage diversification and evolution of common wheat. Mol Ecol 19(5):999–1013
Ohdan T, Francisco PB, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y (2005) Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J Exp Bot 56:3229–3244
Palopoli N, Busi MV, Fornasari MS, Gomez-Casati D, Ugalde R, Parisi G (2006) Starch-synthase III family encodes a tandem of three starch-binding domains. Proteins 65:27–31
Panozzo JF, Eagles HA (1998) Cultivar and environmental effects on quality characters in wheat. I. Starch. Aust J Agri Res 49:757–766
Pauly MD, Procario MC, and Lauring AS (2017) A novel twelve class fluctuation test reveals higher than expected mutation rates for influenza A viruses. eLife 6:e26437
Pérez S, Bertoft E (2010) The molecular structures of starch components and their contribution to the architecture of starch granules: a comprehensive review. Starch‐Stärke 62(8):389–420
Pflister B, Zeeman SC (2016) Formation of starch in plant cells. Cell Mol Life Sci 73:2781–2807
Porebski S, Grant B, Boum BR (1997) Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep 15:8–15
Qu J, Xu S, Zhang Z, Chen G, Zhong Y, Liu L, Zhang R, Xue J, Guo D (2018) Evolutionary, structural and expression analysis of core genes involved in starch synthesis. Nature 8:12736. https://doi.org/10.1038/s41598-018-30411-y
Rijven AHGC (1986) Heat inactivation of starch synthase in wheat endosperm tissue. Plant Physiol 81:448–453. https://doi.org/10.1104/pp.81.2.448
Seemüller E, Lupas A, Stock D, Löwe J, Huber R, Baumeister W (1995) Proteasome from Thermoplasma acidophilum: a threonine protease. Science 268(5210):579–582
Sinnott ML (1990) Catalytic mechanisms of enzymatic glycosyl transfer. Chem Rev 90:1171–1202
Skryhan K, Cuesta-Seijo JA, Nielsen MM, Marri L, Mellor SB, Glaring MA (2015) The role of cysteine residues in redox regulation and protein stability of arabidopsis thaliana starch synthase 1. PLoS ONE 10(9):e0136997
Stoltzfus A, Norris RW (2016) On the causes of evolutionary transition: transversion bias. Mol Biol Evol 333:595–602
Szydlowski N, Ragel P, Raynaud S, Lucas MM, Roldán I, Montero Muñoz FJ, Ovecka M, Bahaji A, Planchot V, Pozueta-Romero J, D'Hulst C, Mérida A (2009) Starch granule initiation in Arabidopsis requires the presence of either class IV or class III starch synthases. Plant cell 21:2443–2457
Tetlow IJ (2011) Starch biosynthesis in developing seeds. Seed Sci Res 21:5–32
The Bovine HapMap Consortium (2009) Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science 324:528–532
Thompson DB (2000) On the non-random nature of amylopectin branching. Carbohydr Polym 43:223–239
Valdez HA, Busi MV, Wayllace MZ, Parisi Z, Ugalde RA, Gomez-Casati DF (2008) Role of the N-terminal starch-binding domains in the kinetic properties of starch synthase III from Arabidopsis thaliana. Biochemistry 47(9):3026–3032
Valkoun J, Hammer K, Kucerova D, Bartos P (1985) Disease resistance in the genus Aegilops L. stem rust, leaf rust, stripe rust, and powdery mildew. Kulturpflanze 33:133–153
Vogel J, Hubschmann T, Börner T, Hess WR (1997) Splicing and intron-internal RNA editing of trnKmatK transcripts in barley plastids: support for MatK as an essential splicing factor. J Mol Biol 270:179–187
Wang J, Luo MC, Chen Z, You FM, Wei Y, Zheng Y, Dvorak J (2013) Aegilopstauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol 198(3):925–937
Yang H, Gu X, Ding M, Lu W, Lu D (2018) Heat stress during grain filling affects activities of enzymes involved in grain protein and starch synthesis in waxy maize. Nature 8:15665. https://doi.org/10.1038/s41598-018-33644-z
Zhang J (2000) Rates of conservative and radical nonsynonymous nucleotide substitutions in mammalian nuclear genes. J Mol Evol 501:56–68
Zhang Z, Gerstein M (2003) Patterns of nucleotide substitution, insertion and deletion in the human genome inferred from pseudogenes. Nucleic Acids Res 3118:5338–5348
Zhao H, Dai T, Jiang D, Cao W (2008) Effects of high temperature on key enzymes involved in starch and protein formation in grains of two wheat cultivars. J Agron Crop Sci 194:47–54. https://doi.org/10.1111/j.1439-037X.2007.00283.x
Acknowledgements
The author sincerely thanks the Department of Biotechnology (DBT) for the studentship, Biotechnology Industry Research Assistance Council, India, for project funding (BIRAC/TG/USAID/08/2014), United States Agency for International Development; Feed the Future Innovation Lab-Climate Resilient Wheat (Grant No. AID-OAA-A-13-00008) for HRD, and Dr. Rajendra Prasad Central Agricultural University, Pusa, Samastipur, Bihar, for the logistics. I would also like to thank Dr. K. S. Gill, WSU, Pullman, USA, and Dr. Jitendra K. Thakur, NIPGR, New Delhi, India, for their valuable support for the investigation.
Author information
Authors and Affiliations
Contributions
RK conceived the idea and RK and VK designed experiments. VK performed the experiments and RK, VK analyzed the results. VK, SP, and YKP wrote the manuscript. RK supervised and complemented the manuscript writing. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Communicated by F. Békés.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kumar, V., Kumar, R., Panigrahi, S. et al. Molecular cloning and characterization of the wheat (Triticum aestivum L.) starch synthase III gene sheds light on its structure. CEREAL RESEARCH COMMUNICATIONS 50, 199–209 (2022). https://doi.org/10.1007/s42976-021-00182-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42976-021-00182-w