Abstract
A better understanding of the genetic factors controlling grain yellow pigment content (GYPC) is important for both pasta (high GYPC) and bread wheat (low GYPC) quality improvement. Quantitative trait loci (QTL) for GYPC have been mapped repeatedly on the distal regions of chromosome arms 7AL and 7BL in wheat, and the Phytoene synthase 1 (PSY-1) gene located in this region has been proposed as a candidate gene. We show here that PSY-E1, the tall wheatgrass orthologue, is completely linked to differences in GYPC, and that selection for white endosperm mutants in recombinant lines carrying this gene resulted in the identification of a mutation in a conserved amino acid of PSY-E1. These results, together with the association between GYPC and allelic differences in PSY-1 in hexaploid wheat, suggest that this gene plays an important role in the determination of GYPC. However, a second white endosperm mutant previously mapped to chromosome arm 7EL showed no mutations in PSY-E1 suggesting the existence of additional gene(s) affecting GYPC in this chromosome region. This hypothesis was further supported by the mapping of QTL for GYPC on 7AL proximal to PSY-1 in a cross between pasta wheat varieties UC1113 and Kofa. Interestingly, the Kofa PSY-B1 allele showed unusually high levels of polymorphisms as a result of a conversion event involving the PSY-A1 allele. In summary, our results support the hypothesis that allelic differences in PSY-1 and at least one additional gene in the distal region of the long arm of homoeologous group 7L are associated with differences in GYPC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The main carotenoid pigment in the grains of durum wheat (Triticum turgidum L. ssp. durum) is lutein (Hentschel et al. 2002), which confers a natural yellow color to pasta products. This is a desirable quality trait for pasta and, therefore, an important target in durum wheat breeding programs. In contrast, white flour varieties are usually selected in common wheat (T. aestivum L.) breeding programs because yellow pigments are considered a detrimental quality factor for breadmaking. Therefore, both pasta and common wheat breeders can benefit from a better understanding of the genetic factors controlling grain yellow pigment content (GYPC). Such knowledge also will find use in future attempts to engineer the nutritionally important carotenoid pathway in cereals.
A major locus affecting GYPC is known to be present in the distal regions of the long arms of chromosomes from homoeologous group 7 in several Triticeae species (Atienza et al. 2007; Elouafi et al. 2001; Pozniak et al. 2007; Zhang et al. 2005). In wheat, Quantitative Trait Loci (QTL) for GYPC have been mapped to the distal region of chromosome arm 7AL in three hexaploid (Mares and Campbell 2001; Parker et al. 1998) and one tetraploid mapping populations (Elouafi et al. 2001). QTL for the same trait have also been mapped on the colinear region of chromosome 7BL in hexaploid (Mares and Campbell 2001) and tetraploid wheat (Elouafi et al. 2001; Pozniak et al. 2007).
The enzyme phytoene synthase (PSY) appears to catalyze a rate-controlling step in the synthesis of phytoene, one of the initial products in the carotenoid biosynthetic pathway (Fig. 1). Pozniak et al. (2007) found the Phytoene synthase 1 (PSY-B1) gene to be linked to a 7BL QTL for GYPC and suggested that it might be a good candidate gene for this QTL. The PSY gene is duplicated in the grasses, but only the PSY-1 paralogue shows a correlation between transcript accumulation and GYPC (Gallagher et al. 2004). High levels of PSY-1 transcripts have been observed in the endosperm of yellow maize but not in white maize or in the white endosperm of rice (Gallagher et al. 2004).
Simplified representation of the lutein and zeaxanthin biosynthetic pathway (based on DellaPenna and Pogson 2006). The phytoene synthase (PSY) is responsible for the synthesis of phytoene (black arrow). The conversion of phytoene to lycopene is catalyzed by the phytoene desaturase (PDS), the z-carotene desaturase (ZDS) and the carotenoid isomerase (CRTISO). The pathway then bifurcates in two branches one leading to α-carotene and lutein (the main carotenoid in the wheat grain) and the other to β-carotene and zeaxanthin. The main enzymes of these last steps are the β-carotene cyclase (β-LCY) and ε-cyclases (ε-LCY) and the β-carotene hydroxylases (β-OHase) and ε-carotene hydroxylases (ε-OHase)
The distal region of chromosome arm 7EL from tall wheatgrass [Lophopyrum ponticum (Podp.) Löve] is also known to affect GYPC (Knott 1968; Zhang et al. 2005). This segment was initially transferred from L. ponticum to common wheat by irradiation in an attempt to introgress the leaf rust resistance gene Lr19. The resulting 7E/7D translocation line, designated “Agatha” (Sharma and Knott 1966), found limited use in bread wheat breeding programs due to the association between the presence of Lr19 and a significant increase in GYPC. The locus for increased yellow pigment was designated “Y” and mapped distal to Lr19 on the distal region of chromosome arm 7EL (Prins et al. 1996; Prins and Marais 1998; Zhang et al. 2005). Agatha was later mutagenized with ethylmethane sulfonate (EMS) to eliminate the negative effect of the Y locus (Knott 1980). Significantly lower GYPC was detected in white endosperm mutants Agatha-28-4 (3.7 ppm) and Agatha-235-6 (4.9 ppm) relative to the non-mutagenized line Agatha (8.7 ppm). The values in the white endosperm mutants were similar to those in the white endosperm control wheat Neepawa (3.4–3.7 ppm) (Knott 1980). The different levels of GYPC observed in these mutants could be interpreted as the result of mutations in different regions of a gene affecting GYPC, or of mutations in two different genes. Interestingly, a preliminary report based on the detection of tetraploid 7EL/7AL recombinant lines with intermediate GYPC suggested the existence of more than one gene affecting GYPC on 7EL (Ceoloni et al. 2000).
In this study, we used the two white endosperm mutant Agatha lines to test the role of PSY-E1 on the regulation of GYPC. We also investigated the association between GYPC and natural allelic differences in both PSY-A1 and PSY-B1 in tetraploid and hexaploid wheat to expand the conclusions based on the Agatha white endosperm mutants. If the hypothesis that GYPC is regulated by multiple genes (including PSY-1) located on the distal region of homoeologous group 7 is correct, it should be possible to find frequent exceptions to the association between sequence diversity in PSY-1 and GYPC natural and induced variation. We will refer hereafter to this hypothesis as the “More-than-one-gene-on-7L hypothesis.” Alternatively, if PSY-1 is the only gene in the distal region of chromosome arm 7L affecting GYPC, we would expect a good association between GYPC differences and PSY-1 natural or induced sequence polymorphisms. We will refer to this hypothesis as the “PSY-1 hypothesis” hereafter. Finally, if a different gene is responsible for the differences in GYPC (“alternative gene hypothesis”), we would expect to find many exceptions to the association between GYPC differences and PSY-1 natural or induced polymorphisms.
Materials and methods
Plant materials
The durum segregating population used to map the PSY-B1 locus was produced from the cross between breeding line UC1113 (low GYPC) and WestBred variety “Kofa” (high GYPC). This population includes 93 recombinant inbred lines (RILs) that have been characterized for GYPC in a previous study (Carrera et al. 2006).
The 7DS·7DL-7EL chromosome translocation no 1 from L. ponticum developed by Sears (1972a, b) was the original source of the PSY-E1 gene. The translocated 7EL segment, which represents approximately 70% of the long arm, was recombined with the wheat chromosomes 7A or 7D using the ph1b mutation. A total of 97 recombinant chromosomes were identified by Genomic In Situ Hybridization (GISH) (Zhang et al. 2005). Since the yellow pigment Y was mapped on the distal end of 7EL, only the 37 recombinant chromosomes with the most distal recombination events were characterized using several molecular markers (Zhang et al. 2005). These same recombinant chromosomes were used in this study to map PSY-E1.
Tetraploid wheat recombinant line 1–23 carrying a distal segment of the Agatha 7E translocation (Zhang et al. 2005) was used to sequence the wild type PSY-E1 gene. We also sequenced the PSY-E1 gene from white endosperm EMS mutant lines Agatha-28-4 and Agatha-235-6, which were generously provided by Dr D. R. Knott (Knott 1980), as well as from the common wheat variety “Wheatear” derived from the Agatha-28-4 mutant. The lower GYPC of the two mutants relative to the original Agatha was confirmed by spectrophotometric analysis.
Sequencing and molecular markers for different PSY-1 genes
Based on published wheat PSY-1 and PSY-2 partial sequences (DQ642439-46) and related EST sequences, we designed conserved primers PSY1_F3/PSY1_R3 (Table 1). The conserved primers were used to sequence partially the three wheat homoeologous copies of PSY-1 using DNA from diploid wheat ancestors T. urartu (A genome), Aegilops tauschii (D genome), and A. speltoides (S genome, closely related to the B genome). We also sequenced several PSY-1 sub-clones from the tetraploid wheat variety Langdon (A and B genomes) and PSY-E1 from L. ponticum from tetraploid wheat 7E/7A recombinant line 1–23 (Zhang et al. 2005). These sequences were compared and inter-genomic polymorphisms were used to develop genome-specific primers and locus-specific primers (Table 1). The specificity of these primers was confirmed using nulli-tetrasomic lines for homoeologous group 7.
PSY-A1 and PSY-B1 genome-specific primers (Table 1) were used to screen the tetraploid wheat BAC library from variety “Langdon” (Cenci et al. 2003). BAC clones 313-J21 (PSY-A1) and 474-K06 (PSY-B1) were used to obtain the complete genomic sequences of the PSY-1 genes. Primers designed from the BAC sequences and covering the complete gene were then used to sequence the PSY-A1 and PSY-B1 alleles from tetraploid UC1113 and Kofa, as well as from white endosperm hexaploid wheat using Chinese Spring (CS) nulli-tetrasomic lines for homoeologous group 7 (Sears 1954).
Mapping and statistical analyses
Genetic maps for the distal regions of chromosome arms 7AL and 7BL were constructed using Mapmaker (Lander et al. 1987) with distances calculated using the Haldane mapping function. GYPC was measured for the two parents and the 93 RILs from the UC1113 × Kofa population using a modified version of the AACC 14–50 procedure, as described before (Zhang et al. 2005). Briefly, pigments were extracted from 0.5 g of integral flour using water-saturated n-butanol. The spectrophotometric determinations of carotenoid pigments (c) were carried out at 448 nm and converted to parts per million using the formula c = [E (extinction at 448 nm) × Volume (ml) × 1,000]/[251 × g (sample weight) × s (optical path length)]. Color determinations were made from samples collected from each RIL in five different field experiments (UC Davis, CA 2003, 2004, 2006; and Imperial Valley, CA 2005, and 2006), using 3–4 m2 plots as experimental units. Growing conditions were as described by Carrera et al. (2006). For the statistical analysis we used SAS version 9.1 (SAS Institute 2006) and the 5-year location combinations were used as blocks.
Results
Mapping of PSY-E1
To develop a marker for PSY-E1, we first sequenced the orthologous PSY-1 genes from the different genomes. A PCR fragment from PSY-1 was obtained from a tetraploid wheat 7E/7A translocation (Zhang et al. 2005) using the PSY-1 conserved primers PSY1_F3 and PSY1_R3 developed from available EST sequences (Table 1). The products, which included a mixture of B and E genome copies, were cloned and sequenced. The PSY-B1 sequence was identified using nulli-tetrasomic lines and the other sequence was assigned to PSY-E1. This partial sequence was used to generate E genome-specific primers, which were then used to amplify and sequence the rest of the PSY-E1 gene using additional conserved primers and inverse-PCR. The PSY-E1 sequence, including 3,021-bp from start to stop codon, 1,316-bp upstream from the start codon, and 304-bp downstream from the stop codon was deposited as GenBank accession EU096095.
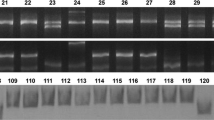
The E genome-specific primers PSY1_EF2 and PSY1_ER4 (Table 1) amplified a PCR product of 191-bp when the 7E allele was present and no product when it was absent (Fig. 2a). This dominant marker was complemented with markers specific for PSY-D1 (Table 1, Fig. 2b) and PSY-A1 (Table 1, Fig. 2d) to generate a codominant marker system for PSY-E1. Using this marker system, we mapped PSY-E1 completely linked to the Y locus in the same 37 recombinant chromosomes used in our previous study to map GYPC (Zhang et al. 2005). These 37 recombinant chromosomes were pre-selected from a larger set of 97 recombinant chromosomes for recombination events in the distal region of the chromosome.
Combined codominant marker system for PSY-E1 based on dominant markers for a PSY-E1, b PSY-D1, and c PSY-A1. 1 Nulli-tetrasomic line N7AT7B, 2 N7BT7D, 3 N7DT7A, 4 Sear’s 7E/7D translocation line, 5 tetraploid 7E/7A recombinant, 6 hexaploid 7E/7A recombinant, and 7 hexaploid 7E/7D recombinant (7EL recombinant lines include the PSY-E1 distal region). d PSY-A1 codominant marker for intron 4 insertion (1–4) T. turgidum ssp. durum (5–6) T. turgidum ssp. dicoccon, and (7–9) T. turgidum ssp. dicoccoides. 1 UC1113, 2 Kofa, 3 UC1112, 4 Vitron, 5 PI 352352, 6 PI 319868, 7 HU10, 8 HU40, 9 PI 428047 (see Supplementary Table 1 for geographic origins). e PSY-B1 codominant marker for the Kofa allele in durum wheat varieties 1 UC1113, 2 Kofa, 3 Langdon, 4 Appio, 5 Appulo, 6 Capitti-8, 7 Cappelli, 8 Cappelli ph1c, 9 UC1112, and 10 Kronos. The 200-bp band is characteristic of the PSY-B1 Kofa allele
Among the recombinant chromosomes used in our previous study 1–32 carried the shortest 7EL segment (less than 3% of the chromosome length) based on GISH results (Zhang et al. 2005). This 7EL segment was distal to the most distal RFLP marker in the genetic map (Xpsr687) but was still associated with high GYPC, indicating that the Y locus was distal to Xpsr687 and close to the telomeric region of 7EL (Zhang et al. 2005). The presence of the L. ponticum PSY-E1 marker in line 1–32 confirmed the existence of a recombination event between Xpsr687 (wheat allele) and PSY-E1 (L. ponticum allele), and the distal location of Y proposed in our previous study (Zhang et al. 2005).
PSY-E1 mutant alleles
The analysis of the Agatha white endosperm mutants was expected to provide more conclusive information than the mapping results because, except for the different sets of mutations, the two mutant lines are isogenic with Agatha.
Comparison of the PSY-E1 sequences from the wild 7EL segment with the white endosperm mutant Agatha-28-4 (and the derived Wheatear) revealed a single mutation event in the last exon of PSY-E1. The observed cytosine to thymine mutation predicts an amino acid change from the small cyclic amino acid proline (P) to the aliphatic amino acid leucine (L). This amino acid substitution has a very low score (−3) in the Block Substitution Matrix BLOSUM 62 (Henikoff and Henikoff 1992), indicating that the interchanged amino acids have very different biochemical properties. The proline amino acid substituted in the Agatha-28-4 mutation is conserved in wheat, Lophopyrum, rice, and maize (Fig. 3).
Comparison of predicted PSY-1 protein sequences from mutant line Agatha 28-4 (LpPSY-E1_mut) and wild type alleles (LpPSY-E1, EU096095), tetraploid wheat (TtPSY-A1, TtPSY-B1_Kofa, TtPSY-B1_UC with UC = UC1113), maize (ZmPSY-1, AAR08445.1) and rice (OsPSY-1, AAS18307.1). Only the last 61 amino acids are shown
In contrast, the 4,641-bp sequence of PSY-E1 from the other white endosperm EMS mutant Agatha-235-6 was found to be no different than the wild type PSY-E1 sequence. This result demonstrates that the reduction in GYPC in the Agatha-235-6 mutant is due neither to mutations in the coding region or introns of PSY-E1 nor in the 1,316-bp upstream from the start codon or the 304-bp downstream from the stop codon. Although we cannot rule out the possibility of mutations in PSY-E1 regulatory elements outside the regions sequenced in this study, the most likely explanation for the Agatha-235-6 result is a mutation in a different gene affecting GYPC within the translocated 7EL segment.
Taken together, the results from the two mutants support the existence of more than one gene in the distal region of chromosome arm 7EL affecting GYPC. To test if this was also the case in tetraploid and hexaploid wheat, we analyzed the allelic variation at the PSY-A1 and PSY-B1 loci in several wheat accessions in which differences in GYPC were previously mapped to the distal region of the long arm of homoeologous group 7.
PSY-A1 in tetraploid wheat
We sequenced the PSY-A1 genomic region from UC1113 and Kofa including 2,967 bp between the start and stop codons, 996 bp upstream from the start codon, and 625 bp downstream from the stop codon. Since the two sequences were 100% identical, only the Kofa sequence was deposited in GenBank (EU096090). Although the lack of polymorphisms precluded the mapping of PSY-A1 in the Kofa × UC1113 segregating population, we were able to map PSY-A1 to the distal 7AL bin 7AL18-0.90-1.00 using CS deletion lines (Fig. 3a). Using the same deletion lines, we assigned the linked SSR markers Xcfa2293-7A and Xwmc116-7A to the adjacent proximal bin 7AL16-0.84-0.90 and Xgwm276-7A to the 7AL21-0.74-0.86 bin (Fig. 4a).
Genetic maps of the distal regions of chromosome arms 7AL (a) and 7BL (b). Distances to the left are in centiMorgan, calculated using the Haldane formula. Vertical lines indicate deletion bins (7AL-18-0.90-1.00, 7AL16-0.86-0.90, 7AL-21-0.74-0.86, and 7BL10-0.78-1.00). ANOVA F values were calculated from average values of spectrophotometric determinations at 448 nm from 5 year-location combinations treated as blocks. Peak F values are indicated in bold. c Comparison of critical recombinant line RIL-36 with parental lines and with the average of selected RILs with identical genotype for loci at the 6A and 7AL QTLs for GYPC (five with the Kofa allele and five with the UC1113 allele at both PSY-B1 and Xbarc340-7B). K represents the Kofa allele and U represents the UC1113 allele at the specified markers. Different letters to the right of the averages indicate significant mean differences using Least Significant Difference test (P < 0.05)
A highly significant QTL for GYPC (P < 0.0001) was associated with the linked SSR markers Xcfa2293-7A and Xwmc116-7A. The spectrophotometer values for GYPC were 16% higher in the lines carrying the UC1113 markers (6.85 ± 0.07 ppm) than those from the lines carrying the Kofa markers (5.91 ± 0.07 ppm) at the peak of the QTL. These differences, as well as the F values associated to the adjacent markers, decrease on both sides of the Xcfa2293-7A-Xwmc116-7A locus (Fig. 4a).
PSY-B1 in tetraploid wheat
In contrast to the highly conserved PSY-A1 sequences, the PSY-B1 genomic sequences revealed an unusually high number of polymorphisms between Kofa (EU096092) and UC1113 (EU096093). The differences were concentrated in the distal 2,000 bp (93.2% identity), but showed normal levels in the first 1,000 bp (99.9% identity). This unusually high level of polymorphism seems to be the result of the introgression of PSY-A1 sequences within the PSY-B1 Kofa allele (Fig. 5, and Supplementary Material). For the 122 polymorphisms present between PSY-A1 and PSY-B1 in the distal 2,000 bp of the genomic sequence, the PSY-B1 sequences from the Kofa allele are identical to the A genome variants in 85 cases (70%) and to the B genome variants in only 37 cases (30%, Fig. 5). This differs significantly from the first 1,000 bp, where 99% of the A-B genome polymorphisms are represented by B genome variants in the Kofa allele.
Concatenated indels and SNPs between PSY-A1 and PSY-B1. CS = hexaploid Chinese Spring (PSY-A1: EU096091, PSY-B1: EU096094), UC = tetraploid UC1113 (PSY-B1: EU096093), Kf = Kofa, (PSY-B1: EU096092), UK = UC1113 and Kofa (PSY-A1: EU096090). The 160 SNPs and 51 indels (represented with their actual size) found between PSY-A1 and PSY-B1 are shown without the intermediate non-polymorphic sequences. The bottom row indicates if the sequence in the Kofa PSY-B1 allele corresponds to an A or B genome polymorphism. Note the difference in the proportion of A genome polymorphisms in the Kofa PSY-B1 allele in the first third of the gene (1%) and the distal part of the gene (70%)
The PSY-B1 allele from Kofa was not detected in tetraploid wild wheat (T. turgidum ssp. dicoccoides) or domesticated emmer (T. turgidum ssp. dicoccon), but was present in approximately 30% of the durum wheat varieties (Table 2, and Supplementary Table 1). Using pedigree analysis we were able to trace back the source of this unusual Kofa allele to the Cappelli ph1c mutant (Supplementary Figure 3). The Kofa PSY-B1 allele is not present in the original Cappelli variety but is present in the Cappelli ph1c mutant (100% identical to Kofa).
A codominant marker was developed for PSY-B1 using primers PSY1_BF3 and PSY1_BR2 (Table 1) flanking a polymorphic 17 bp deletion in intron 2 (Fig. 2e). This is the same polymorphisms used by Pozniak et al. (2007) to develop an EcoRI CAP marker for PSY-B1. The PSY-B1 17-bp polymorphism was mapped in the RIL population completely linked to SSR marker Xgwm146-7B and between flanking SSR markers Xbarc340-7B and Xcfa2257-7B (Fig. 4b). The statistical analyses showed the presence of a highly significant QTL for GYPC associated with these markers (Fig. 4b), with the Kofa alleles associated with higher GYPC. Surprisingly, the peak of the QTL did not coincide with the PSY-B1 locus but with the closely linked proximal marker Xbarc340-7B (Fig. 4b). This marker was separated from PSY-B1 by a single recombination event on RIL-36, which carries Kofa alleles at the proximal Xbarc340-7B and barc1073 loci and UC1113 alleles at the distal PSY-B1 and Xcfa2257-7B 7BL loci, but still shows relatively high levels of GYPC (Fig. 3c).
Given the importance of RIL-36 in determining the location of the gene affecting GYPC, we re-analyzed seed samples from the field experiments used in the QTL analyses and confirmed the presence of correct RIL-36. This excludes planting errors in the field experiments as an explanation for the unexpectedly high GYPC values found in this line. We also performed an additional statistical test comparing this line with the two parental lines, and also with the average of RILs carrying the same alleles as RIL-36 for other QTL affecting GYPC (Fig. 4c). The rationale for this second analysis was to consider the confounding effect of other QTL for GYPC segregating in this population. The selected RILs, as well as RIL-36, all have Kofa alleles at the 7AL QTL (Xwmc116-7A, see above) and at an additional highly significant QTL on 6AL (Xbarc113-6A-Xgwm570-6A, P < 0.0001, W. Zhang and J. Dubcovsky, unpublished). This analysis showed that RIL-36 has intermediate GYPC values, which were significantly different from both parental lines (P < 0.05). In addition, RIL-36 GYPC values were also significantly higher than the average of the selected RILs carrying the UC1113 allele at the distal markers on chromosome arm 7BL (P < 0.05, Fig. 4c), and slightly lower (but not significantly) than the average of the selected RILs carrying the Kofa alleles for PSY-B1 and Xbarc340-7B.
We used RFLP (restriction fragment length polymorphisms) to test if the differences in GYPC associated with the distal regions of chromosome arms 7AL and 7BL in the UC1113 × Kofa segregating population could be the result of differences in PSY-A1 or PSY-B1 copy number. Comparison of the RFLP profiles of the two parental lines with five different restriction enzymes showed no differences in the number of bands or in their relative intensity, suggesting no differences in PSY-I copy number between Kofa and UC1113 (Supplementary Figure 1).
PSY-A1 in hexaploid wheat
Since common wheat varieties generally have very low GYPC (white endosperm) and durum wheat varieties have high GYPC, we speculated that the comparison of the PSY-A1 and PSY-B1 allelic differences between these two species could provide some useful information.
Alignment of the PSY-A1 protein from CS (EU096091) with the one from UC1113 and Kofa revealed a single amino acid difference at position 283 (S to T) (Supplementary Figure 2). The mutated amino acid seems to be the one present in tetraploid wheat because the S amino acid in this position is conserved in Lophopyrum, maize, and rice. This result suggests that the PSY-A1 protein from hexaploid wheat is as functional as the one present in tetraploid wheat.
One interesting difference between the DNA sequences of the hexaploid and tetraploid PSY-A1 alleles was a 676 bp region flanked by perfect direct repeats TCCCTTGTAAA in the fourth intron of the PSY-A1 allele from CS. This insertion is absent in the tetraploid PSY-A1 sequences and in PSY-B1 suggesting an insertion in hexaploid wheat rather than a deletion in the other alleles. This is further supported by the discovery of a very similar element (92% identical) upstream of the CBF14 gene in T. monococcum (AY951948), suggesting that this is likely a repetitive element.
We used primers PSY1_AF1 and PSY1_R3 (Table 1) flanking the insertion to survey a collection of tetraploid and hexaploid wheat accessions. The 12 bp host duplication + the 676 bp insertion resulted in a length difference of 688 bp between the amplification products (Fig. 2d). This analysis showed that the insertion in intron four is present in approximately 40% of the T. dicoccoides and 91% of the T. dicoccon accessions tested, indicating that the insertion allele was very frequent after the first step of tetraploid wheat domestication (Table 2). Even though both forms are present in modern tetraploid and hexaploid varieties, the PSY-A1 allele with the intron insertion shows a high frequency among the bread wheat accessions (94%) and a low frequency among the pasta wheat accessions (12%, Table 2). Among the 17 US hexaploid wheat lines for which we obtained amplification products, only “Penawawa” showed the PSY-A1 allele without the intron insertion. However, this variety does not seem to carry an allele for high GYPC, since yellow (b*) values from 18 independent Penawawa quality tests for white salted noodles are similar to those from other soft wheat varieties (Doug Engle personal communication, Wheat Quality Laboratory, WSU, WA, USA).
In addition to the set of 17 US common wheat varieties, we analyzed the yellow endosperm Australian wheat varieties Dundee (PI 89424, PI106125), Raven (PI 303633, PI 330959), and Aroona (PI 464647). These three related varieties have an STS marker characteristic of a QTL for high GYPC on the distal region of chromosome arm 7AL, where PSY-A1 is located (Parker and Langridge 2000). These three common wheat varieties all have the PSY-A1 allele lacking the intron insertion.
The relatively good association between GYPC and the PSY-A1 variants for the intron repetitive element insertion suggests that this marker may be linked to differences affecting GYPC either in the regulatory regions of PSY-A1 or in a closely linked gene.
PSY-B1 in hexaploid wheat
Alignment of the PSY-B1 protein from CS (EU096094) with that from UC1113 and Kofa show three amino acid differences at positions 95 (V to L), 128 (E to K), and 324 (F to V) (Supplementary Figure 2). The comparative sequence analysis suggests that the first two mutations may not have a large impact on PSY-1 protein function. The first (V to L, BLOSUM 62 score = 1) and second mutations (E to K, BLOSUM 62 score = 1) are between amino acids with similar biochemical properties. Furthermore, the L in the first mutation is also present in this position in both maize and rice, and the second mutation is also in a position that is variable in rice. The last mutation (F to V, BLOSUM62 score = −1), however, yields an amino acid with different biochemical properties in a position that is otherwise conserved in all the species tested in this study. Based on a comparative sequence analysis, this last mutation seems to have a higher probability of affecting PSY-B1 function in CS than the first two mutations; though this hypothesis requires experimental validation.
Discussion
Natural and induced variation in PSY-E1
The complete genetic linkage observed between the differences in GYPC and PSY-E1 in the 7AL/7EL and 7DL/7EL recombinant lines suggests that allelic difference in this gene may be responsible for the observed differences in GYPC. However, this result is not sufficient to rule out the possibility of a closely linked gene affecting this trait. The same limitation applies to a previous QTL study showing linkage between PSY-B1 and GYPC in tetraploid wheat (Pozniak et al. 2007). A more formal validation of PSY-1 as the cause of the differences in GYPC requires either independent mutations or complementation experiments using transgenic plants.
Fortunately, two Agatha white endosperm mutants (Agatha-28-4 and Agatha 235-6) were available from previous EMS studies (Knott 1980). Crosses between these two mutants and the common wheat varieties “Neepawa” and “Manitou” demonstrated that both white endosperm mutations were completely linked with the leaf rust resistance genes Lr19 (Knott 1984). Since these crosses were made in the presence of the functional Ph1b allele, which precludes recombination between the 7EL translocated segment and the homoeologous wheat chromosomes, the previous results cannot be used to estimate the genetic distances between the mutations and Lr19. However, these experiments are still useful in demonstrating that both mutations occurred within the 7EL segment. Based on these results, Knott (1984) concluded that the white endosperm EMS mutations affected the Y pigment locus or a locus so close to the gene that no crossovers were obtained.
We show here that Agatha-28-4 has a mutation in the PSY-E1 gene that results in a P to L amino acid change in the predicted protein. The simplest explanation for this result is that this mutation was found as a result of the selection for white endosperm. This hypothesis is supported by the independent observation of complete linkage between PSY-E1 and GYPC and is consistent with the knowledge that this gene affects the accumulation of carotenoid pigments in the grains of other grass species. In maize, transcription profiles of PSY-1 (but not of PSY-2) correlate with carotenoid pigment accumulation in the grains (Gallagher et al. 2004), and over-expression of PSY in Arabidopsis seeds results in increased levels of carotenoids (Lindgren et al. 2003).
The alternative hypothesis is that the PSY-E1 mutation is a random event and that the mutation affecting GYPC in Agatha-28-4 occurred in a different and yet unknown gene located within the 7EL segment. This alternative hypothesis, requires several simultaneous assumptions: (1) that the PSY-E1 mutation has no effect on the function of the protein, (2) that the second gene affecting GYPC is so close to PSY-E1 that no recombination events occurred in our mapping population, and (3) that the PSY-E1 mutation was found by chance rather than as a result of selection for white endosperm.
The assumption that the PSY-E1 mutation in Agatha-28-4 has no effect on GYPC seems more unlikely than the alternative one, considering that this mutation resulted in a change between amino acids with very contrasting biochemical properties. The large and negative BLOSUM62 score (−3) indicates that this substitution is very infrequent in conserved protein blocks. In addition, this change occurred at a position that is conserved in widely different grass species, spanning the tribes Triticeae (wheat), Oryzeae (rice), and Paniceae (maize) (Fig. 3), which is usually considered as indirect evidence for an important role in the function of a protein. However, a final test of this assumption will require additional biochemical tests or transgenic experiments.
The second assumption of the alternative hypothesis is also unlikely since we could not find a single recombination event between PSY-E1 and GYPC among 97 recombinants chromosomes selected for segments smaller than the 7EL original translocation (Zhang et al. 2005). This result limits the hypothetical second gene to a small region close to PSY-E1. The mapping of PSY-A1 and the gene affecting GYPC on 7AL in two different physical bins, and the identification of recombination events between two genes affecting GYPC in 7E/7A tetraploid recombinant lines (Ceoloni et al. 2000), suggest that the two genes affecting GYPC on this chromosome region might not be that closely linked.
The third assumption of the alternative hypothesis is probably the most unlikely one, as indicated by the following calculations. It has been reported before that an EMS concentration of 1% generates a mutation density of one mutation every 24,000 bp in hexaploid wheat (Slade et al. 2005). The two Agatha white endosperm mutants were selected from a population produced using less than one-fifth of the EMS dosage indicated above. Half of the population was mutagenized using 0.15% v/v and the other half 0.20% v/v of EMS (Knott 1980). Assuming an inverse proportionality between EMS concentration and mutation density, the average mutation density in the mutagenized Agatha lines can be estimated to be approximately 1 in 140,000 bp. Using this value, we calculated the probability of finding at least one mutation within the coding region of PSY-E1 (1,293 bp) to be approximately 0.009 {[1 − (1/140,000)]1293}. In addition, the probability that one of these nucleotide mutations would result in an amino acid change is less than half of the number calculated above (P ≈ 0.003), because 60% of the codon changes are synonymous. Therefore, the discovery of a mutation in PSY-E1 just by chance is expected to happen less than three times in one thousand plants.
Although we can not completely rule out this alternative hypothesis, the probability that all its assumptions occur simultaneously is sufficiently low to consider it an unlikely hypothesis (P < 0.003). On the contrary, the hypothesis that the P to L mutation in PSY-E1 found in Agatha-28-4 is responsible for the white endosperm only requires assuming that the mutant amino acid affects the ability of this enzyme to perform its function. This is a likely assumption given the complete linkage of PSY-E1 with the observed GYPC differences and it is known to affect GYPC in other grass species.
We sequenced PSY-E1 from the second white endosperm mutant Agatha-235-6 with the expectation of finding an additional mutation in this gene. To our surprise, Agatha-235-6 did not show any mutation within the coding region or intron sequences of PSY-E1 or in the 1,316 bp upstream from the start codon. Although at this point we cannot rule out the possibility of a mutation in a regulatory element outside the sequenced region, the fact that most of the regulatory elements in plants are located within the first 1,000 bp suggests that this line carries a mutation in a different gene affecting GYPC also located in the 7EL segment. This additional gene(s) on 7EL could be either an enzyme involved in the synthesis of lutein or a transcription factor affecting the regulation of any of the steps of this pathway. Since the Agatha-235-6 mutation was mapped within the 7EL translocation (Knott 1984), we can exclude as candidate genes the enzymes mapped on different chromosomes such as the phytoene synthase 2 (PSY-2, homoeologous group 5), the phytoene desaturase (PDS, homoeologous group 4), and the ζ-carotene desaturase (ZDS, homoeologous group 2) (Cenci et al. 2004).
Taken together, the results from the two mutants are consistent with the hypothesis that PSY-1 and at least one additional gene located on the distal end of the long arm of homoeologous group 7 affect GYPC. This hypothesis was previously suggested by Ceoloni et al. (2000) based on the observation of intermediate levels of GYPC in some tetraploid 7EL/7AL recombinant lines. Interestingly, the two white endosperm Agatha mutants also differ in their levels of GYPC, with that of Agatha-235-6 mutant (4.9 ppm) being 30% higher than in the Agatha-28-4 mutant (3.7 ppm) (Knott 1980). These differences are consistent with the observation that these two mutations likely occurred in different genes. Additional evidence supporting the existence of a second gene in the distal region of chromosome arm 7L affecting GYPC was obtained from the characterization of PSY-1 allelic diversity in tetraploid wheat.
Allelic diversity in PSY-A1 and PSY-B1 in tetraploid wheat
The absence of any sequence difference in the coding and promoter regions of PSY-A1 between the Kofa and UC1113 alleles, together with the physical mapping of PSY-A1 in a bin located distal to the peak of the QTL, suggests that the differences in GYPC associated with the distal region of chromosome arm 7AL in the Kofa × UC1113 population are the result of allelic differences in a gene different from PSY-A1. The absence of polymorphisms in PSY-A1 (the 4,588 bp), together with the lack of polymorphisms in two additional SSR loci present in the distal deletion bin suggest the possibility that Kofa and UC1113 have identical-by-descent chromosome segments at the distal region of chromosome arm 7AL. Therefore, the most likely explanation for the 7AL QTL for GYPC is the presence of an additional gene affecting GYPC proximal to PSY-A1. However, we can not completely rule out the possibility of a polymorphism in a regulatory element outside the sequenced PSY-A1 region.
The mapping of the peak of the 7BL QTL for GYPC at the Xbarc340-7B locus rather than at the more distal PSY-B1 locus also provides indirect evidence supporting the existence of a second gene affecting GYPC in the distal region of chromosome arm 7BL. The proximal location of the QTL peak relative to PSY-B1 was further confirmed by comparing the critical RIL-36 with average RILs with identical alleles at two other QTL for GYPC known to segregate in this population. Both results, however, are based on a single recombination event and require validation by additional recombination events. Currently, we cannot dismiss the possibility that the relatively high GYPC observed in RIL-36 is caused by allelic differences in other unknown loci segregating in this population.
The Kofa PSY-B1 allele is very interesting because of the alternation of A and B genome sequences (Fig. 5). Pedigree analysis of this hybrid allele indicates that it likely originated in the Cappelli ph1c mutant, since the allele was not detected in the original non-mutagenized Cappelli seeds (Supplementary Figure 3). The observed hybrid allele could have originated by a chromosome break at the PSY-B1 locus produced by the mutagenesis treatment, followed by DNA repair using the homoeologous PSY-A1 gene as template, or by an intergenomic conversion event favored by the presence of the ph1c mutation. It is interesting to point out that the borders of this inter-genomic conversion event are not perfect, showing alternation of A and B polymorphisms. If this observation is confirmed in other inter-genome conversion events, it may indicate some particular characteristics of molecular mechanisms involve in inter-genome conversions or in the mode of action of the ph1c mutation.
In spite of the numerous differences at the DNA level, the predicted PSY-B1 protein from Kofa differs in only two amino acids from the predicted protein from UC1113 (Supplementary Figure 2). The first difference at amino acid 240 (K to T) is in a position that was also polymorphic in maize PSY-1, whereas the second mutation at position 390 (K to R) resulted in a replacement of a conserved amino acid, but the replacement involved two similar amino acids (BLOSUM62 positive score 2). The fact that the Kofa allele was associated with high values of GYPC suggests that these mutations do not negatively affect the PSY-B1 protein activity.
Allelic diversity in PSY-A1 and PSY-B1 in hexaploid wheat
The comparison of the PSY-A1 and PSY-B1 alleles from tetraploid pasta wheat varieties (selected for high GYPC) with those from hexaploid bread wheat varieties (selected for white endosperm) provides additional indirect evidence for the important role played by the PSY-1 gene in the determination of GYPC.
The PSY-B1 protein from white endosperm CS differs from the orthologous protein in pasta varieties UC1113 and Kofa in three amino acids. One of them (F 324 V) is in a position that is conserved across the different grass species sequenced in this study and involves amino acids with different biochemical properties. The effect of this mutation on PSY-1 activity remains to be tested experimentally.
In contrast with the differences in PSY-B1 described above, only one amino acid difference was found at the PSY-A1 locus between tetraploid and hexaploid wheat (S 283 T). This difference is unlikely to be important for PSY-A1 function in common wheat, since the common wheat allele is the one conserved with rice and maize (Supplementary Figure 2). In addition the mutation found in UC1113 and Kofa is polymorphic in other tetraploid accessions (e.g., ABG29736).
At the DNA level, the tetraploid and hexaploid PSY-A1 gene regions differ in the presence of an insertion of a repetitive element in the fourth intron. Although this insertion has frequencies close to 50% in the wild T. turgidum ssp. dicoccoides accessions, its frequency increased drastically in the white endosperm common wheat germplasm and decreased in the cultivated durum germplasm. The hexaploid lines carrying a 7AL locus associated with a QTL for high GYPC on the distal region of chromosome arm 7AL (Parker and Langridge 2000) also exhibited no insertion in the fourth intron. The association between the PSY-A1 allelic variants with differences in GYPC suggests that the insertion in the fourth intron may be linked to a mutation in regulatory regions of PSY-A1 or in a closely linked gene affecting GYPC. However, this association is not perfect since the common wheat variety Penawawa has white flour and no insertion.
An additional 37-bp insertion in the second exon of PSY-A1 has been recently reported in several Chinese common wheat varieties (He et al. 2008). This insertion, which is absent in CS or the durum wheat varieties analyzed here, is associated with reduced levels of GYPC. The two sequences reported by He et al. (EF600063 and EF600064) both have the 676-bp insertion in the fourth intron.
Taken together, the results from tetraploid and hexaploid wheat provide additional support to the hypothesis that PSY-1 and at least one additional gene located in the distal region of the long arm of homoeologous group 7 are responsible for the natural differences in GYPC. This conclusion agrees with the one obtained from the white endosperm Agatha mutants. We are currently generating high density tetraploid mapping populations in isogenic genetic backgrounds to further test this hypothesis.
References
Atienza SG, Avila CM, Martin A (2007) The development of a PCR-based marker for PSY1 from Hordeum chilense, a candidate gene for carotenoid content accumulation in tritordeum seeds. Aust J Agric Res 58:767–773
Carrera A, Echenique V, Zhang W, Helguera M, Manthey F, Schrager A, Picca A, Cervigni G, Dubcovsky J (2006) A deletion at the Lpx-B1 locus is associated with low lipoxygenase activity and improved pasta color in durum wheat (Triticum turgidum ssp. durum). J Cereal Sci 45:67–77
Cenci A, Chantret N, Kong X, Gu Y, Anderson OD, Fahima T, Distelfeld A, Dubcovsky J (2003) Construction and characterization of a half million clone BAC library of durum wheat (Triticum turgidum ssp. durum). Theor Appl Genet 107:931–939
Cenci A, Somma S, Chantret N, Dubcovsky J, Blanco A (2004) PCR identification of durum wheat BAC clones containing genes coding for carotenoid biosynthesis enzymes and their chromosome localization. Genome 47:911–917
Ceoloni C, Forte P, Ciaffi M, Nenno M, Bitti A, De Vita P, D’Egidio MG (2000) Chromosomally engineered durum wheat: the potential of alien gene introgressions affecting disease resistance and quality. In: Royo C, Nachit MM, Di Fonzo N, Araus JL (eds) Durum wheat improvement in the Mediterranean region: new challenges, vol 40. International centre for advanced Mediterranean agronomic studies, Zaragoza, Spain, pp 363–371
DellaPenna D, Pogson BJ (2006) Vitamin synthesis in plants: tocopherols and carotenoids. Annu Rev Plant Biol 57:711–738
Elouafi I, Nachit MM, Martin LM (2001) Identification of a microsatellite on chromosome 7B showing a strong linkage with yellow pigment in durum wheat (Triticum turgidum L. var. durum). Hereditas 135:255–261
Gallagher CE, Matthews PD, Li FQ, Wurtzel ET (2004) Gene duplication in the carotenoid biosynthetic pathway preceded evolution of the grasses. Plant Physiol 135:1776–1783
He XY, Zhang YL, He ZH, Wu YP, Xiao YG, Ma CX, Xia XC (2008) Characterization of Phytoene synthase 1 gene (Psy1) located on common wheat chromosome 7A and development of a functional marker. Theor Appl Genet 116:213–221
Henikoff S, Henikoff JG (1992) Amino-acid substitution matrices from protein blocks. Proc Natl Acad Sci USA 89:10915–10919
Hentschel V, Kranl K, Hollmann J, Lindhauer MG, Bohm V, Bitsch R (2002) Spectrophotometric determination of yellow pigment content and evaluation of carotenoids by high-performance liquid chromatography in durum wheat grain. J Agric Food Chem 50:6663–6668
Knott DR (1968) Translocations involving Triticum chromosomes and Agropyron chromosomes carrying rust resistance. Can J Genet Cytol 10:695–696
Knott DR (1980) Mutation of a gene for yellow pigment linked to Lr19 in wheat. Can J Genet Cytol 22:651–654
Knott DR (1984) The genetic nature of mutations of a gene for yellow pigment linked to Lr19 in ‘Agatha’ wheat. Can J Genet Cytol 26:392–393
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Lindgren LO, Stalberg KG, Hoglund AS (2003) Seed-specific overexpression of an endogenous Arabidopsis Phytoene synthase gene results in delayed germination and increased levels of carotenoids, chlorophyll, and abscisic acid. Plant Physiol 132:779–785
Mares DJ, Campbell AW (2001) Mapping components of flour and noodle colour in Australian wheat. Aust J Agric Res 52:1297–1309
Parker GD, Langridge P (2000) Development of a STS marker linked to a major locus controlling flour colour in wheat (Triticum aestivum L.). Mol Breed 6:169–174
Parker GD, Chalmers KJ, Rathjen AJ, Langridge P (1998) Mapping loci associated with flour colour in wheat (Triticum aestivum L.). Theor Appl Genet 97:238–245
Pozniak CJ, Knox RE, Clarke FR, Clarke JM (2007) Identification of QTL and association of a Phytoene synthase gene with endosperm colour in durum wheat. Theor Appl Genet 114:525–537
Prins R, Marais GF (1998) An extended deletion map of the Lr19 translocation and modified forms. Euphytica 103:95–102
Prins R, Marais GF, Janse BJH, Pretorius ZA, Marais AS (1996) A physical map of the Thinopyrum-derived Lr19 translocation. Genome 39:1013–1019
SAS Institute I (2006) SAS user’s guide, version 9.1. SAS Institute Inc., Cary
Sears ER (1954) The aneuploids of common wheat, vol 572. Research bulletin, University of Missouri, Agricultural Experiment Station, pp 1–58
Sears ER (1972a) Agropyron-wheat transfers through induced by homoeologous pairing. Can J Genet Cytol 14:736
Sears ER (1972b) Chromosome engineering in wheat, vol 4. Stadler Symposia, Columbia, pp 23–38
Sharma D, Knott DR (1966) The transfer of leaf rust resistance from Agropyron to Triticum by irradiation. Can J Genet Cytol 8:137–143
Slade AJ, Fuerstenberg SI, Loeffler D, Steine MN, Facciotti D (2005) A reverse genetic, nontransgenic approach to wheat crop improvement by TILLING. Nat Biotechnol 23:75–81
Zhang W, Lukaszewski A, Kolmer J, Soria M, Goyal S, Dubcovsky J (2005) Molecular characterization of durum and common wheat recombinant lines carrying leaf rust resistance (Lr19) and yellow pigment (Y) genes from Lophopyrum ponticum. Theor Appl Genet 111:573–582
Acknowledgments
The authors thank Dr D.R. Knott for the seeds of the white endosperm EMS mutants, Dr J. Dvorak for the “Cappelli mutant” seeds, Dr M. Maccaferri and Dr R. Tuberosa for the durum germplasm, Dr B. Gill for the Chinese Spring deletion lines, Dr A. Lukaszewski for the 7E recombinant lines, and Dr H. Bockelman at the National Small Grain Collection for all the additional germplasm used in this study. The authors also thank Dr C. Uauy and PhD student I. Lowe for their valuable suggestions and careful review of the manuscript. The project was supported by the National Research Initiative of USDA’s Cooperative State Research, Education and Extension Service, CAP grant number 2006-55606-16629.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by B. Keller.
An erratum to this article can be found at http://dx.doi.org/10.1007/s00122-011-1596-6
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, W., Dubcovsky, J. Association between allelic variation at the Phytoene synthase 1 gene and yellow pigment content in the wheat grain. Theor Appl Genet 116, 635–645 (2008). https://doi.org/10.1007/s00122-007-0697-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-007-0697-8