Abstract
The aim of present study was to evaluate the relationship between vascular endothelial growth factor (VEGF) −2578C/A, −2549I/D, −460T/C and −7C/T and VEGFR1 −710C/T polymorphisms with risk to breast cancer in North Indians. A total of 204 sporadic breast cancer patients and 204 controls were recruited for this case-control study. Significantly increased frequency of II genotype of −2549I/D polymorphism was observed in patients as compared to control individuals (odds ratio (OR) = 2.76, 95 % confidence interval (CI), 1.55–4.92; p = 0.0005). VEGF −2578AA genotype (OR = 2.87; 95 % CI, 1.61–5.10; p = 0.0003) and A allele (OR = 1.65, 95 % CI, 1.25–2.18; p = 0.0004) were found to be associated with increased risk for breast cancer. Individuals carrying CC genotype (OR = 2.23, 95 % CI, 1.25–3.97) and C allele (OR = 1.42, 95 % CI, 1.07–1.87) of VEGF −460T/C polymorphism were at higher risk of breast cancer. There was no significant difference in genotype and allele distribution of VEGF −7C/T and VEGFR1 −710C/T polymorphisms between cases and control individuals (p > 0.05). Linkage disequilibrium analysis showed a strong linkage between VEGF −2549I/D and −2578C/A polymorphisms (Lewontin’s \( \overset{^{\prime }}{D} \) = 0.99; r 2 = 0.97), −2549I/D and −460T/C (\( \overset{^{\prime }}{D} \) = 0.94; r 2 = 0.84), and −2578C/A and −460T/C polymorphisms (\( \overset{^{\prime }}{D} \) = 0.93; r 2 = 0.83). In the present study, we concluded that VEGF −2549I/D, −2578C/A and −460T/C polymorphisms are associated with risk to breast cancer in Punjab, North India.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is a heterogeneous disease encompassing multiple subgroups with different molecular signatures, prognosis and responses to therapies and involves lymphangiogenesis [1, 2]. In solid tumours, angiogenic switch is a critical step and is mediated by production of excess of pro-angiogenic molecules over anti-angiogenic factors [3–5]. Vascular endothelial growth factor (VEGF), a potent angiogenic cytokine, plays a pivotal role from tumour proliferation to inflammatory and ischemic processes [6, 7]. It is also a survival factor for endothelial cells during physiological and tumour angiogenesis with vasodilatation, vascular permeability and anti-apoptosis functions [7–9].
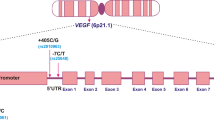
The human VEGF (OMIM 192240) mapped to 6p21.1 is highly polymorphic with multiple polymorphisms in the promoter 5′ untranslated region (5′-UTR) and 3′ UTR [10–12]. The single-nucleotide polymorphisms (SNPs) in the promoter and 5′ UTR have been reported to regulate VEGF expression via alternative initiation of transcription and internal initiation of translation [13, 14]. Some of the variants of VEGF including −460T/C, −1154G/A, −2549I/D, −2578C/A and +405C/G in the promoter or 5′-UTR and +936C/T in the 3′-UTR have been reported to be associated with variation in VEGF protein production [11, 12, 15–18]. Overexpression of VEGF has been reported in breast cancer [19, 20], and its higher level in tumour tissue has been associated with both increased microvessel density and breast cancer recurrence [21–24]
VEGF −2578C/A polymorphism (rs699947) has been shown to functionally affect VEGF messenger RNA (mRNA) levels [25]. Prognostic importance of VEGF −2578C/A polymorphism has been documented in several cancer types including breast [26–28], hepatocellular [29], nasopharyngeal [30], colorectal [31] and colon [32]. Several studies have reported the association of −2549I/D polymorphism (rs35569394) with susceptibility to diseases like bladder cancer [33], renal cell carcinoma [34], diabetic nephropathy [35], diabetic retinopathy [36], Bechet’s disease [37, 38], giant cell arteritis [39], Kawasaki disease [40], prostate cancer [41] and end-stage renal disease [42]. The correlation of 18-bp deletion at position 2549 has been reported with 1.95-fold increased transcriptional activity compared with those containing the insert [35]. A number of molecular epidemiological studies have been conducted to examine the association between VEGF −460T/C polymorphism (rs833061) and cancer susceptibility with disparate results [43–52]. The association of TT genotype of VEGF −460T/C has been reported with poor tumour differentiation in gastric cancer patients from Oman [50].
VEGF participates in the stimulation of both migration and survival of malignant cells by distinct signalling pathways [53]. VEGF and its receptors VEGFR1, VEGFR2 and VEGFR3 are essential in vascular development and maintenance of the adult vasculature [54]. VEGFR1 is one of the important receptors of VEGF angiogenesis signalling and has a relevant role in process of normal vessel formation [55]. It has been documented that breast cancer patients with high sVEGFR1/VEGF-A ratio have a markedly favourable prognosis as compared to patients with low ratio [56]. There is only one published study on VEGFR1 −710C/T polymorphism in Spanish breast cancer patients showing significant association of VEGFR1 −710CT + TT genotype with reduced breast cancer risk [57].
VEGF is an important target in anti-cancer therapy, and different VEGF polymorphisms have been separately reported to regulate VEGF expression. Therefore, the present study was an attempt to evaluate the relationship between four polymorphisms in VEGF (−2578C/A, −2549I/D, −460T/C, −7C/T) and one in VEGFR1 (−710C/T) with risk to breast cancer in North Indian breast cancer patients. In the Punjab state of North India, the cancer incidence is reportedly increasing [58]. In spite of an increasing incidence of breast cancer in Amritsar city of Punjab state in North West part of India, there is no reported study on VEGF polymorphisms in breast cancer from this population.
Methods
Study design and selection of subjects
In the present case-control study, 408 subjects consisting of 204 sporadic breast cancer patients (6 males and 198 females) and 204 unrelated healthy, gender- and age-matched control individuals (6 males and 198 females) were investigated. The breast cancer patients were recruited from Sri Guru Ram Das Institute of Medical Sciences and Research, Vallah, Amritsar, Punjab. The classification of tumour stages was performed by pathologists as per American Joint Committee on Cancer’s TNM staging system [59]. The patients included were those who had not received chemotherapy, radiotherapy or blood transfusion before surgery. The control subjects were recruited from the same geographical area as that of patients. The controls had no history of any cancer or other chronic disease for the last three generations and were not on regular medications for at least 2 years from the date of sampling. The demographic characteristics, detailed family history, reproductive history and disease history of all the subjects were recorded on the pretested structured questionnaire. After informed consent, 5 ml venous blood was collected from each subject in 0.5 M EDTA. The study was approved by ethical committee of Guru Nanak Dev University, Amritsar, Punjab, India.
Analysis of VEGF (−2578C/A, −2549I/D, −460T/C, −7C/T) and VEGFR1 −710C/T polymorphisms
Genomic DNA was extracted from peripheral blood leucocytes by the standard phenol chloroform method [60]. VEGF −2549I/D, −7C/T, −2578C/A and −460T/C and VEGFR1 −710C/T polymorphisms were analyzed using direct PCR, ARMS PCR and PCR-RFLP method. The details of the screening condition have been described in Table 1. To ensure quality control, genotyping was performed without knowledge of case/control status.
Statistical analyses
The power calculations were done using Quanto software, version 1.0 (available from: http://hydra.usc.edu/gxe) using minor allele frequency data from HapMap (http://www.hapmap.org/). The present study was designed to have a statistical power of over 80 % for detection of an association of SNPs and breast cancer at significance level of 0.05. The characteristics of patients and controls were compared using t test for continuous variables and chi-square test (χ 2) for categorical variables. The allele frequencies were tested for the Hardy-Weinberg equilibrium (HWE) for both patients and controls using the chi-square test. This test was also used to evaluate the differences in the VEGF genotypes and allele frequencies between the patient and control groups. Odds ratio (OR) and its 95 % confidence interval (CI) were used to assess the association between genotypes and alleles with the breast cancer risk. Chi-square analysis was also performed for the correlation of genotype and allele frequencies with various parameters including age, gender, menstrual status and pathological stage of the cancer. Probability value <0.05 was considered statistically significant. The analysis was done using SPSS (version 16, SPSS Inc, Chicago, IL, USA). Lewontin’s standardized disequilibrium coefficient (\( \overset{^{\prime }}{D} \)) among the two SNPs was estimated using Haploview version 4.2 [61].
Results
Characteristics of subjects
In the present case-control study, 204 sporadic breast cancer patients (198 females and 6 males) and 204 healthy unrelated normal individuals (198 females and 6 males) were analyzed. The characteristics of patients and controls are listed in Table 2. The mean age of breast cancer patients and controls was 49.70 ± 12.17 and 49.68 ± 12.19 years, respectively. There was no significant difference in age, gender, menstrual status, mean age at menarche, mean age at first child birth and number of full-term pregnancies between patients and controls (p > 0.05). Among the female patients, 32 had stage I, 98 had stage II, 50 had stage III, and 18 had stage IV tumour. In male patients, two patients had stage I and four patients had stage II cancer.
Genotype frequencies of VEGF, VEGFR1 polymorphisms and breast cancer risk
The genotype and allele frequencies of VEGF polymorphisms in the breast cancer patients and control individuals are shown in Table 3. The genotype distributions of VEGF −2578C/A, −2549I/D and −460T/C polymorphisms were in Hardy-Weinberg equilibrium (HWE) in the patients and control groups (p > 0.05).
The frequencies of CC, CA and AA genotypes for VEGF −2578C/A polymorphism were 26.96 vs 37.25 %, 45.59 vs 49.51 % and 27.45 vs 13.24 % in patients and controls, respectively. The frequency of AA genotype was significantly higher in patients as compared to control individuals (27.45 vs 13.24 %; p = 0.0003) and showed 2.87-fold increased risk to breast cancer (OR = 2.87; 95 % CI, 1.61–5.10). Significantly higher frequency of −2578A allele was observed in breast cancer patients (p = 0.0004), and greater than 1.5-fold risk to breast cancer was associated with −2578A allele (OR = 1.65, 95 % CI, 1.25–2.18) (Table 3). Genetic model analysis revealed significantly higher risk for breast cancer (OR = 1.61, 95 % CI, 1.06–2.45; p = 0.026 in dominant model; OR = 2.48, 95 % CI, 1.49–4.12; p = 0.0004 in recessive model) (Table 4).
For VEGF −2549I/D polymorphism, the frequencies of DD, ID and II genotypes were 26.96 vs 35.29 %, 45.10 vs 51.47 % and 27.94 vs 13.24 %, in patients and controls, respectively. There was a significant increase in the frequency of II genotype in patients as compared to control individuals (p = 0.0005), and 2.76-fold risk to breast cancer was associated with II genotype (OR = 2.76, 95 % CI, 1.55–4.92). The frequency of −2549I allele in patients and controls was 50.49 vs 38.97 %, respectively, and presence of I allele revealed greater than 1.5-fold risk to breast cancer (OR = 1.60, 95 % CI, 1.21–2.11) (Table 3). In the recessive genetic model, II genotype was associated with higher breast cancer risk as compared to combined DD + ID genotype (OR = 2.54, 95 % CI, 1.53–4.22; p = 0.0002) (Table 4).
For VEGF −460T/C polymorphism, the frequency of TT, TC and CC genotypes was 29.90 vs 35.29 %, 45.10 vs 51.47 % and 25 vs 13.24 % in cases and controls, respectively. A significant increased risk for breast cancer was observed with CC genotype (OR = 2.23, 95 % CI, 1.25–3.97; p = 0.006) and C allele (OR = 1.42, 95 % CI, 1.07–1.87; p = 0.012) of −460T/C polymorphism (Table 3). VEGF −460CC genotype was associated with increased breast cancer risk as compared to TT + TC genotype in recessive genetic model (OR = 2.19, 95 % CI, 1.31–3.65; p = 0.002) (Table 4). There was no significant difference in genotype and allele frequencies of VEGF −7C/T and VEGFR1 −710C/T polymorphisms between cases and controls (p > 0.05) (Table 3).
Additionally, we stratified our study subjects to investigate the relationship of the studied polymorphisms with various parameters including gender, age, menstrual status and tumour stage. We observed significant difference in genotype distribution of VEGF −2578C/A (p = 0.006), −2549I/D (p = 0.006), −460T/C (p = 0.032) and −7C/T (p = 0.026) polymorphisms in breast cancer patients aged ≤40 years and cases aged >40 years (Table 5). Lewontin’s standardized disequilibrium coefficient (D′) was calculated as a measure for linkage disequilibrium between the studied VEGF polymorphisms (Fig. 1). A strong linkage was observed between VEGF −2549I/D and −2578C/A polymorphisms (Lewontin’s \( \overset{^{\prime }}{D} \) = 0.99; correlation coefficient, r 2 = 0.97), −2549I/D and −460T/C (\( \overset{^{\prime }}{D} \) = 0.94; r 2 = 0.84) and −2578C/A and −460T/C polymorphisms (\( \overset{^{\prime }}{D} \) = 0.93; r 2 = 0.83). We also analyzed the distributions of combined genotypes and observed that genotype combinations VEGF −2549II/−2578AA (p = 0.0006), −2549II/−460CC (p = 0.003), −2549II/−7CC (p = 0.002), −2549II/−7CT (p = 0.009), −2549II/VEGFR1 −710CC (p = 0.001), −2578AA/−460CC (p = 0.002), −2578CA/−460TT (p = 0.013), −2578AA/VEGFR1 −710CC (p = 0.0007), −2578AA/−7CC (p = 0.0009), −2578AA/−7CT (p = 0.009), −460CC/−7CC (p = 0.010), −460CC/−7CT (p = 0.037) and −460CC/VEGFR1 −710CC (p = 0.015) were more common among breast cancer patients as compared to control individuals. However, after Bonferroni correction of multiple variables, significant association to increased breast cancer risk remained with VEGF −2549II/−2578AA (p c = 0.0024), −2549II/−460CC (p c = 0.024), 2549II/−7CC (p c = 0.012), 2549II/VEGFR1 −710CC (p c = 0.006), −2578AA/−460CC (p c = 0.018), −2578AA/VEGFR1 −710CC (p c = 0.0042) and −2578AA/−7CC (p c = 0.0054) genotype combinations (Table 6).
Discussion
Angiogenesis not only is essential for tumour growth but also plays a critical role in the invasion and metastasis. It is regulated by many growth factors, among which VEGF is one of the most important activators of tumour-associated angiogenesis [62]. The human VEGF is highly polymorphic, and there is considerable variation between individuals in VEGF expression [18]. VEGF plasma levels are highly predictive for tumour growth and survival rate of breast cancer patients [4, 22]. Elevated serum VEGF levels have been reported in several cancers including breast cancer [63–65]. In our previous study, we reported significantly higher serum VEGF levels in breast cancer patients as compared to controls [66]. In vitro and in vivo data suggested that genetic variability affects the activity and expression of VEGF [16, 18, 67]. A number of functional polymorphisms in the VEGF have been reported and have been associated with increased risk for several tumours [68].
In the present study, we observed significant association of VEGF −2549I/D, −2578C/A and −460T/C polymorphisms with the risk to breast cancer. For −2549I/D polymorphism, we observed significant association of II genotype (p = 0.0005) and I allele (p = 0.0009) with the increased risk to breast cancer. VEGF −2549II genotype has been correlated with higher VEGF protein production in lipopolysaccharide-stimulated peripheral blood mononuclear cells [37, 39]. Significant association of −2549D allele has been reported with the reduced risk for bladder cancer in North Indians [33]. Contrary to our findings, significant association of −2549D allele and ID genotype has been reported with renal cell carcinoma [34] and prostate cancer, respectively [41]. No correlation of −2549I/D polymorphism has been observed in Chinese hepatocellular carcinoma [69] and Swedish colorectal cancer cases [70].
For −2578C/A polymorphism, homozygous AA genotype revealed 2.87-fold risk of breast cancer (OR = 2.87; 95 % CI, 1.61–5.10). Our results are concordant with few other studies that also showed significant risk association. Association of AA genotype has been reported with higher risk to breast cancer [71], colon cancer [72] and lung cancer [73]. In Italian population, significant reduced risk for the colorectal cancer has been reported with the −2578CC and CA genotypes [51]. We also observed significantly higher frequency of A allele in cases in comparison to controls (50.25 vs 37.99 %, p = 0.0004). Association of −2578A allele with increased VEGF expression has been reported in lung cancer cells [74]. Correlation of A allele has been reported with an increased risk to nasopharyngeal carcinoma [75] and thyroid cancer [76]. In North Indian population, significant association of CA genotype has been reported with the increased risk for bladder cancer [33]. Contrary to our findings, association of C allele has been reported with increased risk for invasive breast [77] and nasopharyngeal carcinoma [30]. A case-control study including patients with familial breast cancer from Germany and Poland and patients with sporadic breast cancer from Sweden failed to find a relation between −2578C>A polymorphism and risk of breast cancer [26]. However, −2578C/A polymorphism has not been associated with different cancers including skin [78], gastric [79, 80], prostate [47], colorectal [31, 70, 81] and epithelial ovarian cancers [82] and renal carcinoma [83].
For VEGF −460T/C polymorphism, we observed significant association of CC genotype (p = 0.006) and C allele (p = 0.012) with increased risk of breast cancer. Similarly, an association of CC genotype has been documented with colorectal cancer in Italian population [51]. Another study from India has also documented significant association of −1498C allele with type 1 diabetic retinopathy [73]. In smoker oesophageal adenocarcinoma cases, increased cancer risk has been reported with −460CT and combined CT + CC genotype [49]. No association of VEGF −460T/C polymorphism has been reported in lung [43], breast [44, 46], ovarian, cervical and endometrial [84], colon [45], prostate [47, 48], pancreatic [52] and renal cell carcinoma [83].
There was no significant difference in genotype and allele distribution of VEGF −7C/T polymorphism in breast cancer patients and controls (p > 0.05). Similarly, no correlation of VEGF −7C/T polymorphism has been documented in Austrian breast cancer patients [46], Caucasian prostate cancer patients [47] and North Indian bladder cancer patients [33]. Significant association of C allele of −7C/T polymorphism has been reported with neuropathy in British-Caucasian type 1 diabetic subjects [85].
VEGFR1 is one of the important receptors of VEGF angiogenesis signalling and has a relevant role in process of normal vessel formation [55]. In the present study, we did not observe any association between VEGFR1 −710C/T polymorphism and breast cancer risk. Significant association of combined CT and TT genotype has been reported with reduced breast cancer risk in Spanish population [57].
We observed a strong linkage between VEGF −2549I/D and −2578C/A polymorphisms (\( \overset{^{\prime }}{D} \) = 0.99; r 2 = 0.97), −2549I/D and −460T/C (\( \overset{^{\prime }}{D} \) = 0.94; r 2 = 0.84) and −2578C/A and −460T/C polymorphisms (\( \overset{^{\prime }}{D} \) = 0.93; r 2 = 0.83). Linkage disequilibrium between promoter polymorphisms −2549I/D and −2578C/A of VEGF has also been reported in Polish, German, Swedish breast cancer patients [26] and Swedish colorectal cancer patients [70]. In the present study, we observed no significant correlation of VEGF polymorphisms with the advancing stage of breast cancer. Association of AA genotype of −2578C/A polymorphism has been reported with aggressiveness of tumour in gastric [79] and hepatocellular cancers [29].
The functional property of −2578C/A polymorphism has been shown to affect mRNA levels [86]. In metastatic breast cancer, comparison of the effect of paclitaxel with paclitaxel and bevacizumab combination showed that patients with −2578AA genotype had longer median overall survival as compared to −2578CA + CC genotype in the paclitaxel and bevacizumab combination [27]. Therefore, in the present study, we concluded that VEGF −2549I/D, −2578C/A and −460T/C polymorphisms are associated with risk to breast cancer in Punjab, North India. SNPs in the VEGF polymorphisms might influence the delivery of chemotherapy to the cancer cells and might consequently hold predictive information in relation to response [7, 87, 88]. A follow-up investigation of the subjects of the present study is ongoing and will provide further information on role of VEGF polymorphisms on metastasis risk and survival of breast cancer patients. VEGF is an important target in anti-cancer therapy, and findings about SNPs influencing VEGF-targeted therapies will help physicians to tailor therapy in individual manner.
Abbreviations
- VEGF :
-
Vascular endothelial growth factor
- SNP:
-
Single-nucleotide polymorphism
- UTR:
-
Untranslated region
- HWE:
-
Hardy-Weinberg equilibrium
- PCR:
-
Polymerase chain reaction
- RFLP:
-
Restriction fragment length polymorphism
- ARMS:
-
Amplification refractory mutation system
- OR:
-
Odds ratio
- CI:
-
Confidence interval
References
Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–74.
Schoppmann SF, Horvat R, Birner P. Lymphatic vessels and lymphangiogenesis in female cancer: mechanisms, clinical impact and possible implications for anti-lymphangiogenic therapies. Oncol Rep. 2002;9(3):455–60.
Hanahan D, Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86(3):353–64.
Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407(6801):249–57.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74.
Carmeliet P. Angiogenesis in health and disease. Nat Med. 2003;9(6):653–60.
Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. J Nat Med. 2003;9(6):669–76.
Alon T, Hemo I, Itin A, Pe’er J, Stone J, Keshet E. Vascular endothelial growth factor acts as a survival factor for newly formed retinal vessels and has implications for retinopathy of prematurity. Nat Med. 1995;1(10):1024–8.
Benjamin LE, Keshet E. Conditional switching of vascular endothelial growth factor (VEGF) expression in tumors: induction of endothelial cell shedding and regression of hemangioblastoma-like vessels by VEGF withdrawal. Proc Natl Acad Sci U S A. 1997;94(16):8761–6.
Vincenti V, Cassano C, Rocchi M, Persico G. Assignment of the vascular endothelial growth factor gene to human chromosome 6p21.3. Circulation. 1996;93(8):1493–5.
Brogan IJ, Khan N, Isaac K, Hutchinson JA, Pravica V, Hutchinson IV. Novel polymorphisms in the promoter and 5′ UTR regions of the human vascular endothelial growth factor gene. Hum Immunol. 1999;60(12):1245–9.
Watson CJ, Webb NJ, Bottomley MJ, Brenchley PE. Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: correlation with variation in VEGF protein production. Cytokine. 2000;12(8):1232–5.
Akiri G, Nahari D, Finkelstein Y, Le SY, Elroy-Stein O, Levi BZ. Regulation of vascular endothelial growth factor (VEGF) expression is mediated by internal initiation of translation and alternative initiation of transcription. Oncogene. 1998;17(2):227–36.
Huez I, Creancier L, Audigier S, Gensac MC, Prats AC, Prats H. Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol Cell Biol. 1998;18(11):6178–90.
Renner W, Kotschan S, Hoffmann C, Obermayer-Pietsch B, Pilger E. A common 936 C/T mutation in the gene for vascular endothelial growth factor is associated with vascular endothelial growth factor plasma levels. J Vasc Res. 2000;37(6):443–8.
Awata T, Inoue K, Kurihara S, Ohkubo T, Watanabe M, Inukai K, et al. A common polymorphism in the 5′-untranslated region of the VEGF gene is associated with diabetic retinopathy in type 2 diabetes. Diabetes. 2002;51(5):1635–9.
Mohammadi M, Ollier WE, Hutchinson IV. A functional association study of VEGF gene promoter polymorphisms with VEGF expression by stimulated pbm cells. Hum Immunol. 2003;64:S125.
Stevens A, Soden J, Brenchley PE, Ralph S, Ray DW. Haplotype analysis of the polymorphic human vascular endothelial growth factor gene promoter. Cancer Res. 2003;63(4):812–6.
Toi M, Inada K, Hoshina S, Suzuki H, Kondo S, Tominaga T. Vascular endothelial growth factor and platelet-derived endothelial cell growth factor are frequently coexpressed in highly vascularized human breast cancer. Clin Cancer Res. 1995;1(9):961–4.
Jubb AM, Pham TQ, Hanby AM, Frantz GD, Peale FV, Wu TD, et al. Expression of vascular endothelial growth factor, hypoxia inducible factor 1alpha, and carbonic anhydrase IX in human tumours. J Clin Pathol. 2004;57(5):504–12.
Gasparini G, Toi M, Gion M, Verderio P, Dittadi R, Hanatani M, et al. Prognostic significance of vascular endothelial growth factor protein in node-negative breast carcinoma. J Natl Cancer Inst. 1997;89(2):139–47.
Linderholm B, Lindh B, Tavelin B, Grankvist K, Henriksson R. p53 and vascular-endothelial-growth-factor (VEGF) expression predicts outcome in 833 patients with primary breast carcinoma. Int J Cancer. 2000;89(1):51–62.
Linderholm BK, Lindahl T, Holmberg L, Klaar S, Lennerstrand J, Henriksson R, et al. The expression of vascular endothelial growth factor correlates with mutant p53 and poor prognosis in human breast cancer. Cancer Res. 2001;61(5):2256–60.
Kushlinskii NE, Gershtein ES. Role of vascular endothelial growth factor during breast cancer. Bull Exp Biol Med. 2002;133(6):521–8.
Reich SJ, Fosnot J, Kuroki A, Tang W, Yang X, Maguire AM, et al. Small interfering RNA (siRNA) targeting VEGF effectively inhibits ocular neovascularization in a mouse model. Mol Vis. 2003;9:210–6.
Jin Q, Hemminki K, Enquist K, Lenner P, Grzybowska E, Klaes R, et al. Vascular endothelial growth factor polymorphisms in relation to breast cancer development and prognosis. Clin Cancer Res. 2005;11(10):3647–53.
Schneider BP, Wang M, Radovich M, Sledge GW, Badve S, Thor A, et al. ECOG 2100. Association of vascular endothelial growth factor and vascular endothelial growth factor receptor-2 genetic polymorphisms with outcome in a trial of paclitaxel compared with paclitaxel plus bevacizumab in advanced breast cancer: ECOG 2100. J Clin Oncol. 2008;26(28):4672–8.
Kidd LR, Brock GN, VanCleave TT, Benford ML, Lavender NA, Kruer TL, et al. Angiogenesis-associated sequence variants relative to breast cancer recurrence and survival. Cancer Causes Control. 2010;21(10):1545–57.
Kong SY, Park JW, Lee JA, Park JE, Park KW, Hong EK, et al. Association between vascular endothelial growth factor gene polymorphisms and survival in hepatocellular carcinoma patients. Hepatology. 2007;46(2):446–55.
Nasr HB, Chahed K, Bouaouina N, Chouchane L. Functional vascular endothelial growth factor -2578 C/A polymorphism in relation to nasopharyngeal carcinoma risk and tumor progression. Clin Chim Acta. 2008;395(1–2):124–9.
Dassoulas K, Gazouli M, Rizos S, Theodoropoulos G, Christoni Z, Nikiteas N, et al. Common polymorphisms in the vascular endothelial growth factor gene and colorectal cancer development, prognosis, and survival. Mol Carcinog. 2009;48(6):563–9.
Kjaer-Frifeldt S, Fredslund R, Lindebjerg J, Hansen TF, Spindler KL, Jakobsen A. Danish Colorectal Cancer Group. Prognostic importance of VEGF-A haplotype combinations in a stage II colon cancer population. Pharmacogenomics. 2012;13(7):763–70.
Jaiswal PK, Tripathi N, Shukla A, Mittal RD. Association of single nucleotide polymorphisms in vascular endothelial growth factor gene with bladder cancer risk. Med Oncol. 2013;30(2):509.
Bruyere F, Hovens CM, Marson MN, D’Arcier BF, Costello AJ, Watier H, et al. VEGF polymorphisms are associated with an increasing risk of developing renal cell carcinoma. J Urol. 2010;184(4):1273–8.
Yang B, Cross DF, Ollerenshaw M, Millward BA, Demaine AG. Polymorphisms of the vascular endothelial growth factor and susceptibility to diabetic microvascular complications in patients with type 1 diabetes mellitus. J Diabetes Complications. 2003;17(1):1–6.
Buraczynska M, Ksiazek P, Baranowicz-Gaszczyk I, Jozwiak L. Association of the VEGF gene polymorphism with diabetic retinopathy in type 2 diabetes patients. Nephrol Dial Transplant. 2007;22(3):827–32.
Salvarani C, Boiardi L, Casali B, Olivieri I, Cantini F, Salvi F, et al. Vascular endothelial growth factor gene polymorphisms in Behçet’s disease. J Rheumatol. 2004;31(9):1785–9.
Kamoun M, Houman MH, Hamzaoui A, Hamzaoui K. Vascular endothelial growth factor gene polymorphisms and serum levels in Behçet’s disease. Tissue Antigens. 2008;72(6):581–7.
Boiardi L, Casali B, Nicoli D, Farnetti E, Chen Q, Macchioni P, et al. Vascular endothelial growth factor gene polymorphisms in giant cell arteritis. J Rheumatol. 2003;30(10):2160–4.
Breunis WB, Biezeveld MH, Geissler J, Ottenkamp J, Kuipers IM, Lam J, et al. Vascular endothelial growth factor gene haplotypes in Kawasaki disease. Arthritis Rheum. 2006;54(5):1588–94.
George GP, Mittal RD. Association of polymorphic variants of Vascular Endothelial Growth Factor (VEGF) gene in relation to risk and androgen therapy response in Prostate cancer patients of North India. 2011. http://www.ichg2011.org/cgi-bin/showdetail.pl?absno=10216. Accessed 16 May 2014.
Prakash S, Prasad N, Sharma RK, Faridi RM, Agrawal S. Vascular endothelial growth factor gene polymorphisms in North Indian patients with end stage renal disease. Cytokine. 2012;58(2):261–6.
Lee SJ, Lee SY, Jeon HS, Park SH, Jang JS, Lee GY, et al. Vascular endothelial growth factor gene polymorphisms and risk of primary lung cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(3):571–5.
Kataoka N, Cai Q, Wen W, Shu XO, Jin F, Gao YT, et al. Population-based case–control study of VEGF gene polymorphisms and breast cancer risk among Chinese women. Cancer Epidemiol Biomarkers Prev. 2006;15(6):1148–52.
Cacev T, Loncar B, Seiwerth S, Spaventi S, Kapitanovic S. Vascular endothelial growth factor polymorphisms -1154G/A and -460 C/T are not associated with VEGF mRNA expression and susceptibility to sporadic colon cancer. DNA Cell Biol. 2008;27(10):569–74.
Langsenlehner U, Wolf G, Langsenlehner T, Gerger A, Hofmann G, Clar H, et al. Genetic polymorphisms in the vascular endothelial growth factor gene and breast cancer risk. The Austrian “tumor of breast tissue: incidence, genetics, and environmental risk factors” study. Breast Cancer Res Treat. 2008;109(2):297–304.
Langsenlehner T, Langsenlehner U, Renner W, Krippl P, Mayer R, Wascher TC, et al. Single nucleotide polymorphisms and haplotypes in the gene for vascular endothelial growth factor and risk of prostate cancer. Eur J Cancer. 2008;44:1572–6.
Onen IH, Konac E, Eroglu M, Guneri C, Biri H, Ekmekci A. No association between polymorphism in the vascular endothelial growth factor gene at position -460 and sporadic prostate cancer in the Turkish population. Mol Biol Rep. 2008;35(1):17–22.
Zhai R, Liu G, Asomaning K, Su L, Kulke MH, Heist RS, et al. Genetic polymorphisms of VEGF, interactions with cigarette smoking exposure and esophageal adenocarcinoma risk. Carcinogenesis. 2008;29(12):2330–4.
Al-Moundhri MS, Al-Nabhani M, Burney IA, Al-Farsi AA, Al-Bahrani B. Gastric cancer risk predisposition and prognostic significance of vascular endothelial growth factor (VEGF) gene polymorphisms—a case–control study in an Omani population. Mol Carcinog. 2009;48(12):1170–6.
Maltese P, Canestrari E, Ruzzo A, Graziano F, Falcone A, Loupakis F, et al. VEGF gene polymorphisms and susceptibility to colorectal cancer disease in Italian population. Int J Colorectal Dis. 2009;24(2):165–70.
Talar-Wojnarowska R, Gasiorowska A, Olakowski M, Lekstan A, Lampe P, Smolarz B, et al. Vascular endothelial growth factor (VEGF) genotype and serum concentration in patients with pancreatic adenocarcinoma and chronic pancreatitis. J Physiol Pharmacol. 2010;61(6):711–6.
Wall SJ, Jiang Y, Muschel RJ, DeClerck YA. Meeting report: proteases, extracellular matrix, and cancer: an AACR Special Conference in Cancer Research. Cancer Res. 2003;63(15):4750–5.
Murakami M, Nguyen LT, Hatanaka K, Schachterle W, Chen PY, Zhuang ZW, et al. FGF-dependent regulation of VEGF receptor 2 expression in mice. J Clin Invest. 2011;121(7):2668–78.
Menendez D, Krysiak O, Inga A, Krysiak B, Resnick MA, Schonfelder G. A SNP in the flt-1 promoter integrates the VEGF system into the p53 transcriptional network. Proc Natl Acad Sci U S A. 2006;103(5):1406–11.
Menendez D, Inga A, Snipe J, Krysiak O, Schonfelder G, Resnick MA. A single-nucleotide polymorphism in a half-binding site creates p53 and estrogen receptor control of vascular endothelial growth factor receptor 1. Mol Cell Biol. 2007;27(7):2590–600.
Rodrigues P, Furriol J, Tormo E, Ballester S, Lluch A, Eroles P. The single-nucleotide polymorphisms +936 C/T VEGF and -710 C/T VEGFR1 are associated with breast cancer protection in a Spanish population. Breast Cancer Res Treat. 2012;133(2):769–78.
Cancer incidence in state not so alarming, claims survey. Door-to-door study puts Punjab’s figures below the national average. In: The Tribune, Chandigarh, India. 2013. http://www.tribuneindia.com/2013/20130121/punjab.htm#3. Accessed 15 May 2014.
Singletary SE, Allred C, Ashley P, Lawrence W. Bassett LW, Berry D, et al. (2002) Revision of the American Joint Committee on Cancer staging system for breast cancer. J Clin Oncol. 2002;20(17):3628–36
Adeli K, Ogbonna G. Rapid purification of human DNA from whole blood for potential application in clinical chemistry laboratories. Clin Chem. 1990;36:261–4.
Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5.
Hicklin DJ, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol. 2005;23(5):1011–27.
Salven P, Perhoniemi V, Tykka H, Maenpaa H, Joensuu H. Serum VEGF levels in women with a benign breast tumor or breast cancer. Breast Cancer Res Treat. 1999;53(2):161–6.
Heer K, Kumar H, Read JR, Fox JN, Monson JR, Kerin MJ. Serum vascular endothelial growth factor in breast cancer: its relation with cancer type and estrogen receptor status. Clin Cancer Res. 2001;7(11):3491–4.
Pande D, Negi R, Khanna S, Khanna R, Khanna HD. Vascular endothelial growth factor levels in relation to oxidative damage and antioxidant status in patients with breast cancer. J Breast Cancer. 2011;14(3):181–4.
Kapahi R, Manjari M, Uppal MS, Singh NR, Sambyal V, Guleria K. Association of -2549 insertion/deletion polymorphism of vascular endothelial growth factor with breast cancer in North Indian patients. Genet Test Mol Biomarkers. 2013;17(3):242–8.
Shahbazi M, Fryer AA, Pravica V, Brogan IJ, Ramsay HM, Hutchinson IV, et al. Vascular endothelial growth factor gene polymorphisms are associated with acute renal allograft rejection. J Am Soc Nephrol. 2002;13(1):260–4.
Krippl P, Langsenlehner U, Renner W, Yazdani-Biuki B, Wolf G, Wascher TC, et al. A common 936 C/T gene polymorphism of vascular endothelial growth factor is associated with decreased breast cancer risk. Int J Cancer. 2003;106(4):468–71.
He Y, Ni J, Chen S, Jiang Y, Jia S, Gao Y. The vascular endothelial growth factor-2549 insertion/deletion polymorphism is not associated with susceptibility to hepatocellular carcinoma in Chinese. DNA Cell Biol. 2010;29(7):393–6.
Ungerback J, Elander N, Dimberg J, Soderkvist P. Analysis of VEGF polymorphisms, tumor expression of VEGF mRNA and colorectal cancer susceptibility in a Swedish population. Mol Med Rep. 2009;2(3):435–9.
Schneider BP, Radovich M, Sledge GW, Robarge JD, Li L, Storniolo AM, et al. Association of polymorphisms of angiogenesis genes with breast cancer. Breast Cancer Res Treat. 2008;111(1):157–63.
Zhang L, Zhang G, Wang P, Gong J, Cao Y, Tang L. Association of vascular endothelial growth factor -2578C/A gene polymorphism in Chinese patients with colon cancer. Genet Test Mol Biomarkers. 2011;15(3):117–21.
Li Y, Liang J, Liu X, Liu H, Yin B, Xiao J, et al. Correlation of polymorphisms of the vascular endothelial growth factor gene and the risk of lung cancer in an ethnic Han group of North China. Exp Ther Med. 2012;3(4):673–6.
Koukourakis MI, Papazoglou D, Giatromanolaki A, Bougioukas G, Maltezos E, Sivridis E. VEGF gene sequence variation defines VEGF gene expression status and angiogenic activity in non-small cell lung cancer. Lung Cancer. 2004;46(3):293–8.
Wang T, Hu K, Ren J, Zhu Q, Wu G, Peng G. Polymorphism of VEGF-2578C/A associated with the risk and aggressiveness of nasopharyngeal carcinoma in a Chinese population. Mol Biol Rep. 2010;37(1):59–65.
Hsiao PJ, Lu MY, Chiang FY, Shin SJ, Tai YD, Juo SH. Vascular endothelial growth factor gene polymorphisms in thyroid cancer. J Endocrinol. 2007;195(2):265–70.
Jacobs EJ, Feigelson HS, Bain EB, Brady KA, Rodriguez C, Stevens VL, et al. Polymorphisms in the vascular endothelial growth factor gene and breast cancer in the Cancer Prevention Study II cohort. Breast Cancer Res. 2006;8(2):R22.
Howell WM, Bateman AC, Turner SJ, Collins A, Theaker JM. Influence of vascular endothelial growth factor single nucleotide polymorphisms on tumour development in cutaneous malignant melanoma. Genes Immun. 2002;3(4):229–32.
Tzanakis N, Gazouli M, Rallis G, Giannopoulos G, Papaconstantinou I, Theodoropoulos G, et al. Vascular endothelial growth factor polymorphisms in gastric cancer development, prognosis, and survival. J Surg Oncol. 2006;94(7):624–30.
Ke Q, Liang J, Wang LN, Hu ZB, Jin GF, Zhou Y, et al. Potentially functional polymorphisms of the vascular endothelial growth factor gene and risk of gastric cancer. Mol Carcinog. 2008;47(8):647–51.
Hofmann G, Langsenlehner U, Renner W, Langsenlehner T, Yazdani-Biuki B, Clar H, et al. Common single nucleotide polymorphisms in the vascular endothelial growth factor gene and colorectal cancer risk. J Cancer Res Clin Oncol. 2008;134(5):591–5.
Li Y, Wang Y, Kang S, Wang N, Zhou RM, Duan YN, et al. Association of vascular endothelial growth factor gene polymorphisms with susceptibility to epithelial ovarian cancer. Int J Gynecol Cancer. 2010;20(5):717–23.
Saenz-Lopez P, Vazquez F, Cozar JM, Carretero R, Garrido F, Ruiz-Cabello F. VEGF polymorphisms are not associated with an increased risk of developing renal cell carcinoma in Spanish population. Hum Immunol. 2013;74(1):98–103.
Konac E, Onen HI, Metindir J, Alp E, Biri AA, Ekmekci A. Lack of association between -460 C/T and 936 C/T of the vascular endothelial growth factor and angiopoietin-2 exon 4G/A polymorphisms and ovarian, cervical, and endometrial cancers. DNA Cell Biol. 2007;26(7):453–63.
Tavakkoly-Bazzaz J, Amoli MM, Pravica V, Chandrasecaran R, Boulton AJ, Larijani B, et al. VEGF gene polymorphism association with diabetic neuropathy. Mol Biol Rep. 2010;37(7):3625–30.
Szeto CC, Chow KM, Poon P, Szeto CY, Wong TY, Li PK. Genetic polymorphism of VEGF: impact on longitudinal change of peritoneal transport and survival of peritoneal dialysis patients. Kidney Int. 2004;65(5):1947–55.
Willett CG, Boucher Y, di Tomaso E, Duda DG, Munn LL, Tong RT, et al. Direct evidence that the VEGF-specific antibody bevacizumab has antivascular effects in human rectal cancer. Nat Med. 2004;10(2):145–7.
Jain RK. Antiangiogenic therapy for cancer: current and emerging concepts. Oncology (Williston Park). 2005;19(4 Suppl 3):7–16.
Suganthalakshmi B, Anand R, Kim R, Mahalakshmi R, Karthikprakash S, Namperumalsamy P, et al. Association of VEGF and eNOS gene polymorphisms in type 2 diabetic retinopathy. Mol Vis. 2006;12:336–41.
Acknowledgments
We would like to thank the patients and control individuals for taking part in this study. This study was supported by the DBT grant BT/PR 13252/GBD/27/236/2009 sanctioned to KG and VS. Research fellowship (No. 3/1/3/JRF-2012/HRD) to RK from ICMR is duly acknowledged. We are thankful to Dr. Geeta Sharma, Principal, Sri Guru Ram Das Institute of Medical Sciences and Research, Vallah, Amritsar, Punjab, for their help in providing access to patients and facilities for execution of research work.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kapahi, R., Guleria, K., Sambyal, V. et al. Association of VEGF and VEGFR1 polymorphisms with breast cancer risk in North Indians. Tumor Biol. 36, 4223–4234 (2015). https://doi.org/10.1007/s13277-015-3059-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3059-1