Abstract
Purpose
Vascular endothelial growth factor (VEGF) is a key regulator of tumor-induced angiogenesis and is required for growth of tumors. We tested the hypothesis that VEGF gene polymorphisms may be associated with breast cancer.
Experimental design
We performed a case–control study including 804 female incident breast cancer patients and 804 female age-matched healthy control subjects. We selected seven VEGF candidate polymorphisms and determined genotypes by 5′-nuclease (TaqMan) assays. Furthermore, VEGF plasma levels and genotypes were analyzed in a group of 81 healthy volunteers (64 men and 17 women).
Results
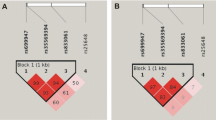
Haplotype analysis showed two separate blocks of high-linkage disequilibrium, formed by five polymorphisms upstream of the coding sequence (promoter and 5′ untranslated region) and two polymorphisms downstream of the coding sequence. None of the single polymorphisms or haplotypes was significantly associated with the presence of breast cancer. After Bonferroni correction for multiple testing, only one statistical signifcant association between VEGF genotypes and haplotypes and tumor characteristics was observed (-634C allele and small tumor size; p < 0.001). In a multivariate regression analysis including sex, age, VEGF genotypes, and haplotypes as covariates and VEGF plasma level as dependent variable, none of the VEGF polymorphism or haplotypes was a significant predictor of VEGF plasma levels.
Conclusions
Our findings do not support the hypothesis that VEGF polymorphisms are associated with breast cancer risk. The association of the VEGF -634C allele with small tumor size is in clear contrast to a previous publication and should be interpreted with caution until replicated by additional studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tumor growth and progression requires the formation of new blood vessels, a process called angiogenesis. Angiogenesis is a complex multifactorial process involving a variety of proangiogenic and proteolytic enzyme activators and inhibitors [1]. The most important regulator of angiogenesis is vascular endothelial growth factor (VEGF), which is overexpressed in several tumor tissues. VEGF is a disulfide-bonded dimeric glycoprotein, sharing close sequence homology with placenta growth factor, VEGF-B and VEGF-C, and lower sequence homology with platelet-derived growth factor (PDGF) [2]. VEGF plasma levels are highly predictive for tumor growth and survival rate of breast cancer patients [3, 4] and therapeutic strategies blocking VEGF action successfully inhibited tumor growth [4].
Several single nucleotide polymorphisms (SNPs) have been described in the VEGF gene, some of them have been associated with VEGF expression and/or clinical phenotypes. We have previously reported a significant association of one variant, VEGF 936C>T (rs3025039), with decreased risk for breast cancer [5]. This result was subsequently confirmed by one study [6], but refuted by two other studies [7, 8].
In order to replicate and expand previous data on the role of VEGF polymorphisms in breast cancer risk, we have determined seven SNPs and haplotypes of the VEGF gene in 804 incident breast cancer patients and 804 healthy age-matched population-based control subjects.
Materials and methods
Subjects
The Austrian “tumor of breast tissue: incidence, genetics, and environmental risk factors” (TIGER) study is an ongoing study investigating risk factors for breast cancer. TIGER consists of 804 consecutive female patients with histologically confirmed incident breast cancer without any other cancer diagnosis beside breast cancer. All patients were recruited between January 2000 and September 2004 from patients attending the Division of Oncology, Department of Internal Medicine, Medical University Graz, Austria. Patients were included in the aftercare measures program of the Division of Oncology Graz, providing follow-ups in regular intervals (3 months interval in years 1–3, 6 months interval in years 4–5, and 12 months interval in years 6–15 after diagnosis). Follow-up investigations included clinical check-up, laboratory (including CEA and CA15-3), radiological (bone scan, liver scan, chest X-ray, and mammograms), and gynecological analyses.
The majority of the participants of TIGER had not participated in any genetic association study before, a small fraction of TIGER participants (n = 18) were also included in a previous case–control study from the same Department [5].
For each patient of the TIGER study, one healthy female age-matched (±2 years) control subject was enrolled. Control subjects were recruited from local health screening studies, the presence of known current or previous malignant disease was excluded anamnestically.
The study was performed according to the Austrian Gene Technology Act and has been approved by the Ethical Committee of the Medical University Graz. Written informed consent was obtained from all participating subjects. All study participants (patients and controls) were Caucasians (Table 1).
Selection of VEGF polymorphisms
With the use of the public NCBI SNP database and available literature [9, 10–12], we selected VEGF candidate polymorphisms with a minor allele frequency of at least 0.10 and location in the promoter region, coding region or untranslated region of the VEGF gene. Using this approach, seven common VEGF polymorphisms were chosen for further analysis (Fig. 1).
Structure of the VEGF gene and position of candidate gene polymorphisms. Position of polymorphism are relative to the translation start, italic positions indicate alternative designations. Dashed lines indicate 13 kb region between upstream polymorphisms and downstream polymorphisms, containing the coding sequence (CDS) and seven introns
DNA isolation and genotyping assays
Genomic DNA was isolated by standard procedures. VEGF genotypes were determined between November 2005 and August 2006 using 5′-nuclease assays (TaqMan). Reaction conditions were as described previously [13]. Primers and probe sets are summarized in Table 2. The laboratory staff responsible for genotyping were blinded for case/control status.
Determination of VEGF plasma levels
The VEGF plasma levels were determined in 81 healthy volunteers (64 men and 17 women) using a commercially available enzyme immunoassay (human VEGF Quantikine, R&D Systems, Wiesbaden, Germany) as described previously [10]. All reactions were performed in duplicate. The assay was specific for VEGF165 and did not detect related molecules, e.g., PDGF or placental growth factor.
Construction of haplotypes and statistical analysis
Haplotypes and linkage disequilibrium were determined using the Haploview program (Version 2.05, http://www.broad.mit.edu/personal/jcbarret/haploview/). Assignment of individual haplotype pairs was performed by the PHASE Version 2.1 software [14]. Statistic analysis was done using SPSS 14.0 for Windows. Numeric values were analyzed by Student’s t-test, proportions of groups were compared by chi-squared test. Odds ratio (OR) and 95% confidence interval (95% CI) were calculated by logistic regression analysis. Threshold for significance was p < 0.05.
Due to the fact that precise frequencies of VEGF genotypes and haplotypes were not at the planning phase of the study, an a priori Power analysis was performed assuming a frequency of 0.1 for a hypothetical genetic risk marker. Using these condidtions, the present study had a Power of 0.99, 0.95 or 0.75 to detect or exclude an OR of 2.0, 1.7 or 1.5 for breast cancer. The statistical Power increased with higher frequencies and/or higher ORs, and decreased with lower frequencies and/or lower ORs of genetic markers.
Results
Tumor characteristics of TIGER participants are presented in Table 2. VEGF genotypes did not deviate from the Hardy Weinberg equilibrium in patients or controls. Haplotype analysis showed two separate blocks of high-linkage disequilibrium, formed by five polymorphisms upstream of the coding sequence (promoter and 5′ untranslated region) and two polymorphisms downstream of the coding sequence, respectively (Fig. 2). None of the single polymorphisms or haplotypes was significantly associated with the presence of breast cancer (Table 3).
Tumor characteristics of breast cancer patients stratified by VEGF genotypes and haplotypes are summarized in Tables 4 and 5. Five associations were below the significance treshold of 0.5. Applying Bonferroni correction for multiple testing, only the association of the -634G>C polymorphism with tumor size remained statistically significant. VEGF genotypes or haplotypes were furthermore not associated with HER2neu overexpression, estrogen receptor status or progesteron receptor status (data not shown).
The potential association of VEGF polymorphisms and haplotypes with VEGF plasma levels were determined in a multivariate linear regression analysis. Polymorphisms and haplotypes were entered assuming codominant effects (0 = polymorphism/haplotype not present; 1 = one copy present; 2 = two copies present). None of the VEGF polymorphism or haplotypes was a significant predictor of VEGF plasma levels. This did not change when sex and age were entered in the model as additional covariates (Table 6).
Discussion
Aim of the present study was to re-evaluate the association of VEGF gene polymorphisms and their haplotypes with breast cancer risk in a large case–control study including incident patients and population-based control subjects. No significant differences in allele, genotype, and haplotype distribution of the VEGF gene polymorphisms between breast cancer cases and controls were detected. Our study does not support the notion that VEGF polymorphisms do modify the risk of breast cancer.
Our data are in contrast to a case–control study we had performed previously [5]. In that study, including 500 Caucasian breast cancer cases and 500 controls, we observed a decreased risk for breast cancer in carriers of VEGF 936T allele. VEGF genotypes in that study were determined by a PCR-RFLP, which may be more error-prone than the TaqMan assay used in the present study. Furthermore, the present study included only incident breast cancer patients, whereas in the previous study incident and as well as prevalent patients had been included.
The fact that the significant association between the VEGF 936C>T polymorphism and breast cancer was not replicated in the present study remains puzzling. On the other hand, non-replication of significant primary genetic association results is a well-known phenomenon in the field of genetic epidemiology. Consequently, a number of methodical papers on genetic association studies have stressed the importance of studies confirming (or confuting) results of primary reports [15–17]. Replication of association studies is imperative to draw firm conclusions about the role of genetic risk factors.
Recently, Jin and co-workers investigated the association between four VEGF polymorphisms (-2578C>A, -1154G>A, -634G>C, and 936C>T) and breast cancer risk [8]. As main result of their study, no association between VEGF polymorphisms or haplotypes and the presence of breast cancer was observed. This is in line with our findings. However, Jin and co-workers reported an association of the VEGFR -634CC genotype and the -2578/-634 CC haplotype with high-tumor aggressiveness (large tumor and high-histologic grade). Interestingly, the same genotype was associated with smaller tumor size and had no effect on histologic grade in the present study. These opposing results underline again the utmost importance of replication of genetic studies.
In a study by Jacobs and co-workers, VEGF alleles -2578C and −1154G were associated with invasive, but not with in situ breast cancer. VEGF polymorphisms −634G>C and 936C>T were not related to breast cancer susceptibility [7]. Kataoka and co-workers reported that breast cancer risk was influenced by the VEGF 936C>T polymorphism, but not by the −1498T>C or the −634G>C polymorphism [6]. Taken together, on the basis of currently available data, a clear effect of VEGF genotypes on breast cancer risk is unlikely.
The hypothesis that polymorphisms of the VEGF might influence breast cancer risk has been built upon the notion that VEGF gene polymorphism are associated with altered VEGF gene expression. In the present study, we were unable to detect any clear effects of VEGF genotypes on VEGF plasma levels. This is in line with a recent publication from Berrahmoune and co-workers, who reported that plasma VEGF concentrations were under strong genetic control in healthy families, but not influenced by VEGF genotypes at positions −1498, −634 or 936 [18]. It is likely that substantial genetic determinants of vascular growth might be found in other candidates genes, such as those for hypoxia inducable factor (HIF1), VEGF receptor 1 (Kinase Insert Domain Receptor; KDR) or VEGF receptor 2 (FMS-Related Tyrosine Kinase 1; FLT [19]).
References
Carmeliet P (2000) Mechanisms of angiogenesis and arteriogenesis. Nat Med 6:389–395
Keck PJ, Hauser SD, Krivi G et al (1989) Vascular permeability factor, an endothelial cell mitogen related to PDGF. Science 246:1309–1312
Linderholm B, Lindh B, Tavelin B, Grankvist K, Henriksson R (2000) p53 and vascular-endothelial-growth-factor (VEGF) expression predicts outcome in 833 patients with primary breast carcinoma. Int J Cancer 89:51–62
Carmeliet P, Jain RK (2000) Angiogenesis in cancer and other diseases. Nature 407:249–257
Krippl P, Langsenlehner U, Renner W et al (2003) A common 936 C/T gene polymorphism of vascular endothelial growth factor is associated with decreased breast cancer risk. Int J Cancer 106:468–471
Kataoka N, Cai Q, Wen W, Shu XO, Jin F, Gao YT, Zheng W (2006) Population-based case-control study of VEGF gene polymorphisms and breast cancer risk among Chinese women. Cancer Epidemiol Biomarkers Prev 15:1148–1152
Jacobs EJ, Feigelson HS, Bain EB, Brady KA, Rodriguez C, Stevens VL, Patel AV, Thun MJ, Calle EE (2006) Polymorphisms in the vascular endothelial growth factor gene and breast cancer in the Cancer Prevention Study II cohort. Breast Cancer Res 8:R22
Jin Q, Hemminki K, Enquist K et al (2005) Vascular endothelial growth factor polymorphisms in relation to breast cancer development and prognosis. Clin Cancer Res 11:3647–3653
Awata T, Inoue K, Kurihara S, Ohkubo T, Watanabe M, Inukai K, Inoue I, Katayama S (2002) A common polymorphism in the 5’-untranslated region of the VEGF gene is associated with diabetic retinopathy in type 2 diabetes. Diabetes 51:1635–1639
Renner W, Kotschan S, Hoffmann C, Obermayer-Pietsch B, Pilger E (2000) A common 936 C/T mutation in the gene for vascular endothelial growth factor is associated with vascular endothelial growth factor plasma levels. J Vasc Res 37:443–448
Brogan IJ, Khan N, Isaac K, Hutchinson JA, Pravica V, Hutchinson IV (1999) Novel polymorphisms in the promoter and 5’ UTR regions of the human vascular endothelial growth factor gene. Hum Immunol 60:1245–1249
Watson CJ, Webb NJ, Bottomley MJ, Brenchley PE (2000) Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: correlation with variation in VEGF protein production. Cytokine 12:1232–1235
Langsenlehner U, Krippl P, Renner W et al (2005) Interleukin−10 promoter polymorphism is associated with decreased breast cancer risk. Breast Cancer Res Treat 90:113–115
Stephens M, Smith NJ, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989
Ioannidis JP, Gwinn M, Little J et al (2006) Human genome epidemiology network and the network of investigator networks. A road map for efficient and reliable human genome epidemiology. Nat Genet 38:3–5
Trikalinos TA, Ntzani EE, Contopoulos-Ioannidis DG, Ioannidis JP (2004) Establishment of genetic associations for complex diseases is independent of early study findings. Eur J Hum Genet 12:762–769
Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K (2002) A comprehensive review of genetic association studies. Genet Med 4:45–61
Berrahmoune H, Herbeth B, Lamont JV, Masson C, Fitzgerald PS, Visvikis-Siest S (2007) Heritability for plasma VEGF concentration in the Stanislas family study. Ann Hum Genet 71:54–63
Menendez D, Krysiak O, Inga A, Krysiak B, Resnick MA, Schonfelder G (2006) A SNP in the flt–1 promoter integrates the VEGF system into the p53 transcriptional network. Proc Natl Acad Sci USA 103:1406–1411
Acknowledgment
This study was supported by the Anniversary Fund of the Österreichische Nationalbank (Project Nr. 10609).
Author information
Authors and Affiliations
Corresponding author
Additional information
U. Langsenlehner and G. Wolf contributed equally to the study.
Rights and permissions
About this article
Cite this article
Langsenlehner, U., Wolf, G., Langsenlehner, T. et al. Genetic polymorphisms in the vascular endothelial growth factor gene and breast cancer risk. The Austrian “tumor of breast tissue: incidence, genetics, and environmental risk factors” study. Breast Cancer Res Treat 109, 297–304 (2008). https://doi.org/10.1007/s10549-007-9655-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-007-9655-z