Abstract
Background
Few studies have systematically explored a pathway approach: to test the association of multiple polymorphisms from multiple genes important to angiogenesis simultaneously with risk of breast cancer. We report our preliminary data evaluating the association of polymorphisms from seven genes known to influence angiogenesis with the likelihood of having breast cancer.
Methods
We recruited 715 controls and 520 subjects with breast cancer. Subjects provided a blood specimen and completed a questionnaire that included common breast cancer risk factors and breast cancer status. We evaluated candidate polymorphisms in the following genes: Hypoxia Inducible Factor-1 alpha (HIF1α), Vascular Endothelial Growth Factor (VEGF), VEGF Receptor 1 (VEGFR-1), VEGFR-2, endothelial Nitric Oxide Synthase (eNOS), Neuropilin-1 (NRP-1) and Neuropilin-2 (NRP-2). Testing for associations between each polymorphism and the presence or absence of breast cancer was performed.
Results
VEGF-2578 AA and -1498 CC genotypes were more common in cancer cases than controls (P = 0.06 and P = 0.04, respectively). These two genotypes remained significant predictors of breast cancer status after adjusting for non-genetic risk factors estimated by the Gail model (P = 0.03 and P = 0.03, respectively). When comparing women with invasive versus pre-invasive breast cancer, the eNOS-786 TT and eNOS 894 GG genotypes were associated with a greater likelihood of invasive disease and the eNOS 894 GG genotype was associated with a greater likelihood of having metastatic disease.
Conclusion
There is an association of the VEGF-2578A and -1498C alleles with increased breast cancer risk. This association remains significant when adjusted for Gail score-related risk factors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Angiogenesis plays a critical role in local growth of solid tumors and subsequently in the process of distant spread [1–6]. Extensive preclinical and clinical data in breast cancer support the important prognostic and therapeutic importance of angiogenesis [7, 8]. There is heterogeneity in degree of tumor angiogenesis and breast cancers with more robust angiogenic profiles having worse outcome. Fibrocystic lesions with the highest microvessel density (MVD) are associated with a greater risk of breast cancer [9]. MVD has been shown to be highest with histologically aggressive ductal carcinoma in situ (DCIS) lesions [10] and associated with increased VEGF expression [11]. High MVD in pre-malignant lesions has been associated with high risk of future breast cancer [9]. Also, high MVD in invasive disease has been correlated with a greater likelihood of metastatic disease [12] and a shorter relapse free and overall survival in patients with node-negative breast cancer [13].
As angiogenesis is not only a pathologic but also a normal physiologic process, it represents a truly host mediated process. Thus variability in genes that control this process may account for some of this heterogeneity and such variability may be the result of germline variability rather than somatic mutational events. Our prior work has compared genetic variability in breast tumors with that in germline DNA for several single nucleotide polymorphisms (SNPs) of genes known to modulate angiogenesis. In that study there was no difference detected between tumor and germline polymorphisms suggesting that this variability was an inherited event [14].
Specific SNPs in genes that modulate angiogenesis have previously been associated with increased risk of developing malignancy [15–20] and prognosis after developing malignancy [21–26]. The effect of this variability on incidence of breast cancer has been mixed. A polymorphism in Vascular Endothelial Growth Factor (VEGF) [17] has been shown to correlate in a positive fashion with the likelihood of having breast cancer, however, there are conflicting reports [27–29]. One limitation of many prior studies is that a limited number of polymorphisms or genes were evaluated. Furthermore, the combined effect of known, validated, non-genetic and inherited risk factors have rarely been accounted for.
Furthermore, it appears as if there is a significant difference in the angiogenic requirements of invasive breast cancer when compared to pre-invasive breast cancer (i.e. ductal carcinoma in situ). Thus, some women may be genetically predisposed to an invasive (versus pre-invasive) phenotype. A prior study by Jacobs et al. demonstrated an association between a specific VEGF polymorphism and invasive breast cancer which was not evident for ductal carcinoma in situ [27]. Other work has demonstrated that risk [30], vascular invasion [23], and outcome [31] in breast cancer may be influenced by genetic variability in endothelial Nitric Oxide Synthase (eNOS).
The goal of this study is to comprehensively evaluate the association between polymorphisms in genes that modulate the angiogenesis pathway with: (1) likelihood of breast cancer, (2) invasive versus pre-invasive disease, and (3) metastatic versus local/regional disease.
Materials and methods
Study population
This trial was approved by the IRB at Indiana University and all subjects consented to have blood collected and to provide information in the form of a detailed questionnaire. A total of 1,240 subjects were recruited within the “Friends for Life” project. The “Friends for Life” project was a recruitment effort performed at Indiana University with the goal of recruiting women with both current/prior breast cancer (both invasive disease and ductal carcinoma in situ) and healthy women with no history of breast cancer [32]. The majority of subjects were recruited in a single day recruitment drive focused around the “Susan G. Komen Race for the Cure” event in Indianapolis, IN, in April of 2005. The organization of this day involved the coordination of over 160 volunteers to carry out consent, phlebotomy and to assist subjects with the completion of a demographic/disease-specific questionnaire. This study was performed in the Indiana University Simon Cancer Center.
Five subjects were excluded because of inconsistencies in questionnaire data. Twenty-three additional patients did not have samples available for genotyping. This left 1,212 subjects for analysis. To have adequate power to detect genetic associations with the studied outcomes, analysis was performed in Caucasian subjects. Companion clinical data was obtained by questionnaire which was filled out by each subject. The subject demographics are outlined in Table 1.
Subject data
Subjects signed informed consent and donated up to 9 cm3 of whole blood which was subsequently extracted for genotypic analysis (see below). All pertinent demographic and medical information was obtained from a 5-page questionnaire that was completed by the subject. This included variables that would allow for the completion of the Gail score (i.e. age, age at first menses, age at first live birth, number of first degree relatives with breast cancer, and number of prior biopsies) as well as multiple others. Subjects also provided a thorough, current and complete medication list. For those who had been diagnosed with breast cancer, they also provided a description of interventions that they had incurred.
Non-genetic risk assessment
In order to accurately assess the genetic association between germline genetic variation and the likelihood of breast cancer, we chose to control for the degree of risk normally assessed in the routine clinical setting. The most well-validated and frequently used breast cancer risk assessment tool in this context is the Gail score [33, 34]. Six hundred and seventy-five women completed 100% of the questions that were necessary to complete a Gail score. Of these, 329 women (49%) indicated a current or prior disease diagnosis. A Gail score was not calculated for those who had not answered all of the necessary questions.
Candidate polymorphisms
The polymorphisms we tested for were selected via a bioinformatics approach with the goal of selecting genes known to modulate angiogenesis (see Table 2). We used the following criteria to select genes for study: (1) that the gene be part of a pathway for which there is a credible scientific basis to support its involvement in the angiogenesis pathway; (2) that the gene has an established, well-documented genetic polymorphism; (3) that the frequency of the polymorphism is high enough that its impact on cancer risk at a population level will be meaningful; and/or (4) that the polymorphism has some degree of likelihood to alter the function of the gene in a biologically relevant manner.
Genotyping
DNA was extracted from whole blood of study subjects utilizing Gentra Puregene extraction kit (Gentra Systems; Minneapolis, MN) per manufacturer’s instructions. Genotyping was performed using SYBR Green & Taqman-based Real Time PCR. Please see Supplement 1 for details.
Statistics
Minor allele frequencies and exact tests for conformity of genotype counts to Hardy–Weinberg proportions were calculated using Haploview version 3.32 [35]. Allele frequency differences between Caucasian and African American populations were compared using Pearson’s χ2 test. To control for potential confounding of association results due to population stratification, analysis was conducted within racial groups.
Association between marker genotypes and outcomes were tested using Pearson’s χ2 test or Fisher’s exact test where appropriate. Linear effects of marker genotypes on outcomes were tested with the Mantel–Haenszel (MH) χ2 test. Logistic regression models were fitted to assess the additive effect of marker genotypes in predicting breast cancer status after adjusting non-genetic risk factors estimated by the Gail model. Model fit between full and nested models was estimated with a likelihood ratio test.
For all statistical tests, α = 0.05 was defined as the level of statistical significance. All statistical analyses were conducted with SAS version 9.1 (SAS Institute, Inc., Cary, NC).
Results
Breast cancer risk
All of the selected SNPs were independently evaluated for their association with breast cancer status, invasive, and metastatic disease. One-hundred percent of subjects that had provided a blood specimen were successfully genotyped for all candidate SNPs. The allele frequencies are presented in Table 2. The VEGF-1498 C/T and the -2578 C/A alleles were associated with breast cancer risk in the Caucasian population (see Table 3). VEGF-2578 and -1498 are in high linkage disequilibrium in Caucasian (r 2 = 0.968), but less associated in African Americans (r 2 = 0.587) populations. The VEGF-1498 C/T and -2578 C/A genotypes had to be evaluated separately for Caucasian and African American women because there were significant differences in allele frequency (P = 2.2 × 10−10 and P = 7.3 × 10−5, respectively). The other selected SNPs from VEGF did not demonstrate an association with disease status. Also, when evaluating the selected SNPs of HIF1α, VEGFR-1, VEGFR-2, eNOS, NRP-1, & NRP-2 there were no significant associations identified with the likelihood of breast cancer.
Breast cancer risk considering Gail score (logistic regression)
Associations were also calculated with consideration of Gail score included. Six hundred and seventy-five of the 1,224 women had answered the necessary questions to successfully calculate a Gail score. 656 women had both Gail score and genotyping completed. When implementing the Gail score as a base model, only VEGF-2578 and -1498 added significantly to the logistic model with Gail score alone. Women with VEGF-2578 AA genotypes were observed to have twice the odds of having breast cancer than with those with AC or CC genotypes (P = 0.03; OR = 1.99, 95% CI = 1.06–3.74). Similarly, women with the VEGF-1498 CC genotype had significantly higher odds of disease than with those with CT or TT genotypes (P = 0.03; OR = 2.01, 95% CI = 1.08–3.76).
Invasive versus pre-invasive disease
Approximately 75% of the women with breast cancer reported that their diagnosis was invasive breast cancer versus 25% who reported DCIS. This fraction was constant whether the subject population was pre-menopausal or post-menopausal. eNOS-786T/C and eNOS 894 G/T alleles were both associated with a significantly higher prevalence of invasive (versus pre-invasive) disease. eNOS-786 TT genotype was significantly more common in subjects with invasive disease than in those with pre-invasive disease (P = 0.04). Although not significant at α = 5%, the eNOS 894 GG genotype was observed at a higher frequency in women with invasive disease as compared to pre-invasive disease (P = 0.08). When evaluating the pre-menopausal subgroup, the VEGFR-2 1416 AA genotype was more common in women with invasive breast cancer compared to DCIS (64% vs. 36%) (P = 0.09 Fisher’s exact test). The sample size of this latter cohort, however, was quite small (n = 61) and thus associations here are exploratory.
Local disease versus metastatic disease
Since angiogenesis is a hallmark that promulgates distant spread of disease, we also evaluated for an association between genotype and women with distant or local/regional disease. The eNOS 894 GG genotype was disproportionately seen in the metastatic setting (versus local/regional disease), with the proportion of subjects with metastatic disease changing linearly with genotype in the Caucasian cohort (P = 0.1 Fisher’s exact test and P = 0.04 Mantel–Haenszel test) and in the post-menopausal subgroup (P = 0.1 Fisher’s exact test and P = 0.03 Mantel–Haenszel test).
Recent versus remote date of diagnosis
To test for the possibility of survivor bias in our analysis, an unplanned analysis was performed comparing women with a relatively recent diagnosis (≤3 years) versus those who were diagnosed more remotely (>3 years). There was no significant difference in genotype frequency distribution for any marker studied in those women enrolled who were diagnosed recently versus remotely.
Discussion
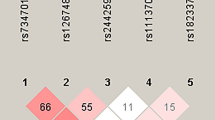
Angiogenesis clearly plays a role in the pathogenesis of breast cancer and this is supported with strong preclinical and clinical evidence [7]. Perhaps the most convincing evidence is the improvement in outcome for women with advanced breast cancer that receive anti-angiogenesis therapy [8]. A prior study has demonstrated a decreased risk of breast cancer for those who have the VEGF 936T allele [17]. Other studies, however, have refuted this finding [27–29]. Preclinical work has demonstrated that variability in the VEGF promoter does indeed affect expression [36]. The results here suggest that VEGF promoter polymorphisms do play a role in the risk of breast cancer, and that risk is present even when known clinical variables are considered (see Fig. 1 for established SNPs in VEGF). These data have important mechanistic implications and suggest that targets within the VEGF pathway are potentially valuable tools for breast cancer prevention.
Furthermore, previous work has demonstrated that variability in the eNOS gene has been associated with breast cancer risk [30] and has demonstrated a marginal association with the likelihood of having invasive disease (compared to pre-invasive disease) [23] and also a greater likelihood of having distant disease (compared with local/regional disease) [31]. Our data further support an association between eNOS genetic variability and likelihood of tumor invasiveness and spread. In total, these findings support the hypothesis that genetic variability in the angiogenesis pathway may contribute to heterogeneity in the pathogenesis of breast cancer.
There are several limitations to this study. First, there is an inherent survivor bias in the recruitment method used. While a portion of the subjects that had been recruited were recently diagnosed, some had been disease free for several years. Thus, it is possible that the breast cancer cohort was biased toward a more indolent disease phenotype as women who would go on to die of disease shortly after diagnosis would not be captured by this analysis. It is clearly possible that this more indolent phenotype may also have a different genetic makeup than a more aggressive disease type and that this may limit the generalizability of the results. Because of this concern, we performed an exploratory analysis of women who were diagnosed recently (defined as ≤3 years) versus those diagnosed remotely (defined as >3 years). Although the time point of 3 years was chosen arbitrarily, the median overall survival for women with advanced disease was taken into consideration to make this a conservative analysis. While this analysis is clearly exploratory, the lack of major genetic differences strongly suggests that there was no overt survivor bias in this study. Another limitation to this study is that the health history was self reported. While the likelihood of false reporting is low for certain demographic questions, the likelihood of false reporting for questions regarding type of breast cancer is most certainly higher.
One of the major limitations to prior studies has been that only one gene or one polymorphism has been evaluated in this complex pathway. Although our current study did not include all possible SNPs, it did encompass a large number of established candidates. A second major limitation of other prior studies is that few have considered non-genetic variables. Since there are well-established clinical variables that predict the likelihood of breast cancer an unintentional imbalance in these variables could significantly alter accurate determination of genotypic associations. We attempted to avoid this imbalance with the calculation of the Gail score when possible. This approach also allowed us to assess the iterative value of testing for these variants in the current clinical setting where the Gail model is widely used.
There are several well established and widely used variables that predict increased risk of breast cancer [37]. The elucidation of these variables has allowed the testing and FDA approval of a drug which actually decreases the future risk of disease (i.e. tamoxifen) in women at high risk [37]. Unfortunately, the reduction in risk to date has been confined to the ER+ subgroup of tumors. This is not surprising as the majority of agents tested to date have a primary mechanism of action that involves limiting estrogen effect. Thus, understanding non-hormonal pathways that increase the likelihood of breast cancer is important and may ultimately lead to therapeutic agents that decrease the incidence of either hormone receptor positive, or endocrine-unresponsive breast cancer.
In conclusion, it appears that associations of germline inherited variability in genes that control angiogenesis with the incidence of breast cancer are predominantly derived from the VEGF gene itself. This effect appears to have predictive power that adds to the Gail model for breast cancer risk. Also, the likelihood of having invasive and metastatic disease may also be due to variability in genes that modulate angiogenesis. It follows that genes involved in angiogenesis may be targets of future therapies designed to prevent breast cancer.
References
Folkman J (2001) Angiogenesis-dependent diseases. Semin Oncol 28:536–542
Folkman J (1985) Tumor angiogenesis. Adv Cancer Res 43:175–203
Folkman J (1995) Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat Med 1:27–31
Plate KH, Breier G, Risau W (1994) Molecular mechanisms of developmental and tumor angiogenesis. Brain Pathol 4:207–218
Weinstat-Saslow D, Steeg PS (1994) Angiogenesis and colonization in the tumor metastatic process: basic and applied advances. FASEB J 8:401–407
Parangi S, O’Reilly M, Christofori G et al (1996) Antiangiogenic therapy of transgenic mice impairs de novo tumor growth. Proc Natl Acad Sci U S A 93:2002–2007
Schneider BP, Miller KD (2005) Angiogenesis of breast cancer. J Clin Oncol 23:1782–1790
Schneider BP, Sledge GW Jr (2007) Drug insight: VEGF as a therapeutic target for breast cancer. Nat Clin Pract Oncol 4:181–189
Guinebretiere JM, Le Monique G, Gavoille A et al (1994) Angiogenesis and risk of breast cancer in women with fibrocystic disease. J Natl Cancer Inst 86:635–636
Guidi AJ, Fischer L, Harris JR et al (1994) Microvessel density and distribution in ductal carcinoma in situ of the breast. J Natl Cancer Inst 86:614–619
Guidi AJ, Schnitt SJ, Fischer L et al (1997) Vascular permeability factor (vascular endothelial growth factor) expression and angiogenesis in patients with ductal carcinoma in situ of the breast. Cancer 80:1945–1953
Weidner N, Semple JP, Welch WR et al (1991) Tumor angiogenesis and metastasis–correlation in invasive breast carcinoma. N Engl J Med 324:1–8
Weidner N, Folkman J, Pozza F et al (1992) Tumor angiogenesis: a new significant and independent prognostic indicator in early-stage breast carcinoma. J Natl Cancer Inst 84:1875–1887
Schneider BP, Skaar TC, Sledge GW et al (2006) Analysis of angiogenesis genes from paraffin-embedded breast tumor and lymph nodes. Breast Cancer Res Treat 96:209–215
Lin CC, Wu HC, Tsai FJ et al (2003) Vascular endothelial growth factor gene-460 C/T polymorphism is a biomarker for prostate cancer. Urology 62:374–377
McCarron SL, Edwards S, Evans PR et al (2002) Influence of cytokine gene polymorphisms on the development of prostate cancer. Cancer Res 62:3369–3372
Krippl P, Langsenlehner U, Renner W et al (2003) A common 936 C/T gene polymorphism of vascular endothelial growth factor is associated with decreased breast cancer risk. Int J Cancer 106:468–471
Cheon KT, Choi KH, Lee HB et al (2000) Gene polymorphisms of endothelial nitric oxide synthase and angiotensin-converting enzyme in patients with lung cancer. Lung 178:351–360
Medeiros R, Morais A, Vasconcelos A et al (2002) Endothelial nitric oxide synthase gene polymorphisms and genetic susceptibility to prostate cancer. Eur J Cancer Prev 11:343–350
Iughetti P, Suzuki O, Godoi PH et al (2001) A polymorphism in endostatin, an angiogenesis inhibitor, predisposes for the development of prostatic adenocarcinoma. Cancer Res 61:7375–7378
Howell WM, Bateman AC, Turner SJ et al (2002) Influence of vascular endothelial growth factor single nucleotide polymorphisms on tumour development in cutaneous malignant melanoma. Genes Immun 3:229–232
Balasubramanian SP, Brown NJ, Reed MW (2002) Role of genetic polymorphisms in tumour angiogenesis. Br J Cancer 87:1057–1065
Ghilardi G, Biondi ML, Cecchini F et al (2003) Vascular invasion in human breast cancer is correlated to T→786C polymorphism of NOS3 gene. Nitric Oxide 9:118–122
Hefler LA, Ludwig E, Lampe D et al (2002) Polymorphisms of the endothelial nitric oxide synthase gene in ovarian cancer. Gynecol Oncol 86:134–137
Medeiros RM, Morais A, Vasconcelos A et al (2002) Outcome in prostate cancer: association with endothelial nitric oxide synthase Glu-Asp298 polymorphism at exon 7. Clin Cancer Res 8:3433–3437
Tanimoto K, Yoshiga K, Eguchi H et al (2003) Hypoxia-inducible factor-1alpha polymorphisms associated with enhanced transactivation capacity, implying clinical significance. Carcinogenesis 24:1779–1783
Jacobs EJ, Feigelson HS, Bain EB et al (2006) Polymorphisms in the vascular endothelial growth factor gene and breast cancer in the cancer prevention study II cohort. Breast Cancer Res 8:R22
Jin Q, Hemminki K, Enquist K et al (2005) Vascular endothelial growth factor polymorphisms in relation to breast cancer development and prognosis. Clin Cancer Res 11:3647–3653
Langsenlehner U, Wolf G, Langsenlehner T et al (2007) Genetic polymorphisms in the vascular endothelial growth factor gene and breast cancer risk. The Austrian “tumor of breast tissue: incidence, genetics, and environmental risk factors” study. Breast Cancer Res Treat, doi:10.1007/s10549-007-9655-z
Lu J, Wei Q, Bondy ML et al (2006) Promoter polymorphism (-786t>C) in the endothelial nitric oxide synthase gene is associated with risk of sporadic breast cancer in non-Hispanic white women age younger than 55 years. Cancer 107:2245–2253
Choi JY, Lee KM, Noh DY et al (2006) Genetic polymorphisms of eNOS, hormone receptor status, and survival of breast cancer. Breast Cancer Res Treat 100:213–218
Lemler S, Rufenbarger C, Skaar T et al (2005) Creating a DNA bank for pharmacogenomic epidemiology: the Friends for Life study. Breast Cancer Res Treat 94:S153 (Abstract #3084)
Costantino JP, Gail MH, Pee D et al (1999) Validation studies for models projecting the risk of invasive and total breast cancer incidence. J Natl Cancer Inst 91:1541–1548
Gail MH, Brinton LA, Byar DP et al (1989) Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst 81:1879–1886
Barrett JC, Fry B, Maller J et al (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265
Stevens A, Soden J, Brenchley PE et al (2003) Haplotype analysis of the polymorphic human vascular endothelial growth factor gene promoter. Cancer Res 63:812–816
Cuzick J, Powles T, Veronesi U et al (2003) Overview of the main outcomes in breast-cancer prevention trials. Lancet 361:296–300
Acknowledgements
This work was supported by ASCO Career Development Award, Breast Cancer Research Foundation, Catherine Peachey Fund, GCRC CReFF Award & Indiana University Simon Cancer Center.
Author information
Authors and Affiliations
Corresponding author
Additional information
On Behalf of the “Friends for Life” Consortium.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Schneider, B.P., Radovich, M., Sledge, G.W. et al. Association of polymorphisms of angiogenesis genes with breast cancer. Breast Cancer Res Treat 111, 157–163 (2008). https://doi.org/10.1007/s10549-007-9755-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-007-9755-9