Abstract
High-mobility group A1 (HMGA1) has been suggested to play a significant role in tumor progression, but little is known about the accurate significance of HMGA1 in non-small cell lung cancer (NSCLC) patients. The aim of this study was to identify the role of HMGA1 in NSCLC. The expression status of HMGA1 was observed initially in NSCLC by Gene Expression Omnibus (GEO). The expression of HMGA1 messenger RNA (mRNA) and protein was examined in NSCLC and adjacent normal lung tissues through real-time PCR and immunohistochemistry. Meanwhile, the relationship of HMGA1 expression levels with clinical features and prognosis of NSCLC patients was analyzed. In our results, HMGA1 was overexpressed in NSCLC tissues compared with adjacent normal lung tissues in microarray data (GSE19804). HMGA1 mRNA and protein expressions were markedly higher in NSCLC tissues than in normal lung tissues (P < 0.001 and P = 0.010, respectively). Using immunohistochemistry, high levels of HMGA1 protein were positively correlated with the status of clinical stage (I–II vs. III–IV, P < 0.001), T classification (T1–T vs. T3–T4, P = 0.003), N classification (N0N1 vs. N2–N3, P < 0.001), M classification (M0 vs. M1, P = 0.002), and differentiated degree (high or middle vs. low or undifferentiated, P = 0.003) in NSCLC. Patients with higher HMGA1 expression had a significantly shorter overall survival time than did patients with low HMGA1 expression. Multivariate analysis indicated that the level of HMGA1 expression was an independent prognostic factor (P < 0.001) for the survival of patients with NSCLC. In conclusion, HMGA1 plays an important role on NSCLC progression and prognosis and may act as a convictive biomarker for prognostic prediction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer is the most common cause of cancer-related deaths worldwide [1]. Non-small cell lung cancer (NSCLC) is a frequent type of lung cancer, accounting for over 80 % of all lung cancer patients [2]. In spite of improved surgical techniques, advanced chemotherapeutic regimens, and novel targeted therapy, the clinical outcome of NSCLC patients remains unsatisfactory. The 5-year survival rate of patients with NSCLC is only 15 % [3]. Moreover, most patients with NSCLC are diagnosed at advanced stages because of the difficulty in making an early diagnosis. Thus, it is necessary to identify biomarkers which provide early diagnosis, accurate prognosis prediction.
The high-mobility group A (HMGA) proteins contain three AT hook DNA-binding motifs that mediate binding to the AT-rich regions in the minor groove of DNA. Two types of HMGA proteins, HMGA1 and HMGA2, with similar functions are encoded by two different genes at chromosomal loci 6p21.3 and 12q15, respectively [4]. HMGA plays an important role in several biological processes such as regulation of transcription, differentiation, and neoplastic transformation [5].
HMGA1 oncogene is widely expressed during embryonic development and is low or absent in most of the differentiated tissues in adults [6]. Recent study suggested that HMGA1 was induced by the Wnt/β-catenin pathway and maintains proliferation of gastric cancer cells [7]. Meanwhile, decreased expression of HMGA1 suppressed anchorage-independent cell proliferation, cellular motility, migration, and invasion in several cancer cell lines [8–11]. The high level of HMGA protein has been found in several malignant neoplasias, including colon cancer [12], breast cancer [13], prostatic cancer [14], pancreatic cancer [15], and ovarian cancer [16]. Furthermore, HMGA1 overexpression was associated with a highly malignant phenotype, also representing a poor prognostic factor since HMGA overexpression often correlated with the presence of metastasis, and with a short survival [17, 18]. Although the role of HMGA1 has been studied in many types of cancer, but little is known about the significance of HMGA1 in patients with NSCLC.
The aim of this study was to identify the pathological roles of HMGA1 in lung cancer. This study suggested that the messenger RNA (mRNA) and protein level of HMGA1 were increased in lung cancer tissue and positively associated with differentiated degree, clinical stage, and T/N/M classification. Furthermore, we found that overexpression of HMGA1 was a significant predictor of poor prognosis for NSCLC patients.
Materials and methods
Analysis of microarray data
Microarray data set (Gene Expression Omnibus (GEO) accession number: GSE19804) from 60 pairs of tumor and adjacent normal lung tissue specimens submitted by Lu was retrieved from the GEO database. Those differentially expressed genes were screened and identified by real-time PCR for the following study.
Samples collection
One hundred forty-five paraffin-embedded lung cancer tissues and 29 adjacent normal lung tissue samples were retrieved from the Affiliated Hospital of Jilin University and Occupational Disease Control and Prevention Hospital. Twenty pairs of fresh tumor and adjacent normal lung tissue samples were collected from the Affiliated Hospital of Jilin University and Occupational Disease Control and Prevention Hospital. All fresh samples were immediately preserved in liquid nitrogen. No patients had received any form of tumor-specific therapy before diagnosis. The histopathological diagnosis of all samples was respectively diagnosed by two pathologists. In these 145 NSCLC cases, there were 89 males and 56 females with age ranging from 23 to 76 years. The clinical follow-up time of patients ranged from 6 to 96 months. The clinical processes were approved by the Ethics Committees of the Affiliated Hospital of Jilin University and Occupational Disease Control and Prevention Hospital. Informed consent was obtained from all patients. The clinical stage and system treatment of all specimens were confirmed according to the seventh edition of the AJCC TNM system and NCCN guideline, respectively.
Real-time PCR
To investigate the mRNA level of HMGA1, total RNA of fresh tissues was reversely transcribed using PrimeScript® RT reagent Kit (TaKaRa). Quantitative real-time reverse transcription-PCR (qRT-PCR) was performed with SYBR® Premix Ex Taq™ II (TaKaRa) on a LightCycler (Roche), following the manufacturer’s instructions. The sequence-specific forward and reverse primers sequences for HMGA1 mRNA were 5′-TCCAGGAAGGAAACCAAGG-3′ and 5′-AGGACTCCTGCGAGATGC-3′, respectively. Forward and reverse primers sequences for GAPDH mRNA were 5′-GAGTCCACTGGCGTCTTC-3′ and 5′-GATGATCTTGAGGCTGTTGTC-3′, respectively. Relative quantification of mRNA expression was calculated by using the 2-△△Ct method. The raw data were presented as the relative quantity of HMGA1 mRNA, normalized with GAPDH, and relative to a calibrator sample. All qRT-PCR reactions were performed in triplicate.
Immunohistochemistry
To detect HMGA1 protein, immunohistochemistry experiments were performed. Paraffin-embedded sections were deparaffinized in xylene and rehydrated in a descending ethanol series (100, 90, 80, and 70 % ethanol) and double-distilled water according to standard protocols. High-pressure antigenic retrieval was performed in citrate buffer (pH 6.0) and boiled for 2 min. After antigen retrieval, sections were treated with 3 % hydrogen peroxide and 1 % bovine serum albumin to block the endogenous peroxidase activity and non-specific binding. The sections were incubated with HMGA1 antibody (Cell Signaling, #12094, dilution 1:250) overnight at 4 °C. The sections were washed three times and incubated with the biotinylated secondary antibody and streptavidin horseradish peroxidase complex for 25 min at room temperature, respectively. Sections were reacted with diaminobenzidine for 2 min, rinsed with tap water, and then counterstained with hematoxylin. In the end, sections were viewed under a light microscopy.
Evaluation of staining
Immunostaining results were reviewed and scored independently by two pathologists without knowledge of the clinical parameters. Any intensity of nuclear immunoreactivity was considered to present immunopositivity for HMGA1. A total of 1000 cells were counted at several high-power fields (×400) selected from different reactivity density regions including areas of high, moderate, low, and negative reactivity. The percentage of HMGA1-stained tumor cells was scored on a scale of 0–4 (0, no staining; 1, ≤5 %; 2, ≤30 %; 3, ≤50 %; 4, >50 %) [19]. For statistical analysis, a final staining scores of 0–2 and 3–4 were respectively considered to be low and high expression.
Statistical analysis
All data were analyzed by using SPSS 13.0 software. The unpaired T test was applied to test the differential mRNA expression of HMGA1 in lung cancer tissue compared to adjacent normal lung tissue. The chi-square test was used to analyze the correlation between HMGA1 expression and the clinicopathologic parameters of NSCLC patients. The Kaplan–Meier method and the log-rank test were used to the correlations between HMGA1 expression and the overall survival time of patients. The significance of survival variables was analyzed using the Cox multivariate proportional hazards model. Data was presented as mean ± SD. P values less than 0.05 were considered statistically significant.
Results
HMGA1 is highly expressed in NSCLC
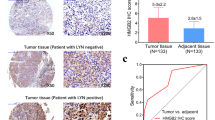
From our microarray data, HMGA1 was highly expressed in NSCLC tissues compared with adjacent normal lung tissues with an average of 1.87 folds (P < 0.001, Fig. 1). Using real-time PCR to measure the expression of HMGA1 transcripts, we found that the HMGA1 expression level was significantly increased with an average increase of 2.49-fold in NSCLC tissue in comparison to adjacent normal lung tissue (P < 0.001, Fig. 2). Furthermore, we measured subcellular localization and the expression levels of HMGA1 protein in 145 paraffin-embedded NSCLC samples and 29 paraffin-embedded normal lung tissues using immunohistochemical staining (Fig. 3). Specific HMGA1 protein staining was found in the nucleus. The expression of HMGA1 was significantly elevated in NSCLC compared with normal lung tissues (P = 0.010, Table. 1).
Immunohistochemical staining of HMGA1 in NSCLC tissues. a Negative expression of HMGA1 in normal lung tissues (original magnification ×400). b Negative expression of HMGA1 in lung cancer (original magnification ×400). c, d Weak expression of HMGA1 in lung cancer (original magnification ×400). e, f Strong expression of HMGA1 in lung cancer (original magnification ×400)
Relationship between clinicopathological characteristics and expression of HMGA1 in NSCLC patients
The relationship between clinicopathological characteristics and HMGA1 expression levels in patients with NSCLC were summarized in Table 2. We did not find any significant association of HMGA1 expression levels with patient’s gender (P = 0.227), age (P = 0.799), smoking (P = 0.077), and pathology classification (P = 0.363). However, HMGA1 was positively associated with clinical stage (I–II vs. III–IV, P < 0.001), T classification (T1–T vs. T3–T4, P = 0.003), N classification (N0–N1 vs. N2–N3, P < 0.001), M classification (M0 vs. M1, P = 0.002), and differentiated degree (high or middle vs. low or undifferentiated, P = 0.003) in NSCLC.
Survival analysis
To explore the prognostic role of HMGA1 expression for NSCLC, we analyzed the association between HMGA1 expression and patient survival using Kaplan–Meier analysis with the log-rank test. In 145 NSCLC cases with prognosis information, we found that the nuclear expression level of HMGA1 protein was significantly correlated with the overall survival of NSCLC patients. Patients with higher levels of HMGA1 expression had shorter survival rates than those with lower HMGA1 expression levels (P < 0.001, Fig. 4). Univariate analysis indicated that HMGA1 expression level, clinical stages, and T/N/M classifications were significantly correlated with patients’ survival (all P < 0.001, Table 3). To determine whether HMGA1 is an independent prognostic factor for NSCLC, we performed multivariate analyses using the Cox proportional hazards model. The results indicated that the level of high expression of HMGA1 was not an independent prognostic factor for NSCLC (P = 0.016, Table 3).
Discussion
HMGA1, which is a member of HMGA family, has been implicated in many biological processes [20]. HMGA1 proteins modulate gene expression by altering chromatin structure and orchestrating the assembly of transcription factor complexes to enhanceosomes within enhancer or promoter regions throughout the genome [20]. Recent studies indicated that the HMGA1 as a key factor enriched in undifferentiated tumors and embryonic stem cells [21], and is low or absent in most of the differentiated tissues [22], which suggested that HMGA1 plays an important role in cell proliferation and differentiation. In addition, present studies demonstrated that HMGA1 is also involved in processes of tumor invasion and metastasis [18, 23, 24]. In pancreatic cancer, HMGA1 promotes cell invasive and metastatic potential through PI3K/Akt/MMP-9 [25]. Furthermore, HMGA1 activity is not restricted to the regulation of cell proliferation and mobility and is necessary for several processes involved in induction of epithelial–mesenchymal transition and pluripotent stem cells [26–28].
HMGA1 is highly expressed in several types of human cancer, such as colon cancer [12], breast cancer [13], prostatic cancer [14], pancreatic cancer [15], and ovarian cancer [16], but little is known about HMGA1 in NSCLC patients. In a microarray analysis performed by Lu et al. (GSE19804), we found that HMGA1 level was higher in 60 NSCLC samples than in adjacent paired normal lung samples. Then, we present the evidence that mRNA and protein expressions of HMGA1 were increased in NSCLC through real-time PCR and immunohistochemistry, which were similar to the microarray data.
In order to further identify the role of HMGA1 in the development and progression of lung cancer. We analyzed the expression of HMGA1 in 145 NSCLC patients and found HMGA1 overexpression was significantly associated with clinical stage, T classification (tumor size), N classification (lymph node metastasis), M classification (distant metastasis), and differentiated degree. Overexpressed HMGA1 in NSCLC may accelerate tumor growth and enhance local cell invasion and metastasis. Our results may indicate that HMGA1 plays significant roles in NSCLC progression, including tumor proliferation, invasion, and metastasis. Similar to Wang et al.’s report, HMGA1 overexpression positively associated with clinical stage, lymph node metastasis, and histological grade in patients with laryngeal squamous cell carcinoma [29]. In human pituitary adenomas, HMGA1 expression also correlated with tumor size, tumor invasion, and histological grade [19]. Moreover, Takaha et al.’s study in vitro showed that HMGA1 knockdown markedly inhibited colony formation, significantly induced apoptosis, inhibited invasion potential, and induced anoikis in renal cell carcinoma [30]. These studies consistently suggest that overexpressed HMGA1 may play an unfavorable role in NSCLC pathogenesis. However, the correlation between HMGA1 expression and the survival of NSCLC patients has been seldom reported.
In the past few years, HMGA1 overexpression in tumor cells has been shown to be an independent poor prognostic factor in several types of tumors, such as lung cancer [31], colorectal cancer [17], liver cancer [18], pancreatic adenocarcinoma [32–34], and uveal melanomas [35]. In this study, we presented that HMGA1 expression in NSCLC was inversely correlated with patient’s overall survival in protein level. The patients with higher expression of HMGA1 protein had shorter survival time. According to multivariate analyses, overexpression of HMGA1 protein was a significant predictor of poor prognosis for NSCLC patients. These results were consistent with Sarhadi et al.’s report, which suggested that increased nuclear expression of HMGA1 correlated with poor survival of NSCLC patients [31]. Similarly, Takahashi et al. demonstrated that the expression of HMGA1 was significantly correlated with clinical stage and lymph node metastasis and as an independent biomarker for poor prognosis of colorectal cancer patients [17]. In pancreatic cancer, Liau et al. reported HMGA1 expression was predictive of poor patient survival and was an independent prognostic indicator through multivariate analysis [34].
In conclusion, this study indicated that the expression level of HMGA1 was significantly increased in NSCLC and correlated with the malignant status of NSCLC. Because of the limited sample size of patients in our study, further studies would be needed to verify the role of HMGA1 as a convictive clinical predictor for the survival of NSCLC patients.
References
Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: Cancer J Clin. 2014;64:9–29.
Rosell R, Karachaliou N. Lung cancer: maintenance therapy and precision medicine in NSCLC. Nat Rev Clin Oncol. 2013;10:549–50.
Spira A, Ettinger DS. Multidisciplinary management of lung cancer. N Engl J Med. 2004;350:379–92.
Johnson KR, Lehn DA, Reeves R. Alternative processing of mRNAs encoding mammalian chromosomal high-mobility-group proteins HMG-i and HMG-y. Mol Cell Biol. 1989;9:2114–23.
Reeves R, Nissen MS. The a.T-DNA-binding domain of mammalian high mobility group i chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem. 1990;265:8573–82.
Shah SN, Resar LM. High mobility group A1 and cancer: potential biomarker and therapeutic target. Histol Histopathol. 2012;27:567–79.
Xing J, Cao G, Fu C: HMGA1 interacts with beta-catenin to positively regulate wnt/beta-catenin signaling in colorectal cancer cells. Pathology Oncology Research: POR 2014.
Tesfaye A, Di Cello F, Hillion J, Ronnett BM, Elbahloul O, Ashfaq R, et al. The high-mobility group A1 gene upregulates cyclooxygenase 2 expression in uterine tumorigenesis. Cancer Res. 2007;67:3998–4004.
Di Cello F, Shin J, Harbom K, Brayton C. Knockdown of HMGA1 inhibits human breast cancer cell growth and metastasis in immunodeficient mice. Biochem Biophys Res Commun. 2013;434:70–4.
Xi Y, Li YS, Tang HB. High mobility group A1 protein acts as a new target of notch1 signaling and regulates cell proliferation in T leukemia cells. Mol Cell Biochem. 2013;374:173–80.
Larsson L, Jawert F, Magnusson B, Hasseus B, Kjeller G. Expression of high mobility group a proteins in oral leukoplakia. Anticancer Res. 2013;33:4261–6.
Chiappetta G, Manfioletti G, Pentimalli F, Abe N, Di Bonito M, Vento MT, et al. High mobility group HMGi(y) protein expression in human colorectal hyperplastic and neoplastic diseases. Int J Cancer J Int Cancer. 2001;91:147–51.
Chiappetta G, Botti G, Monaco M, Pasquinelli R, Pentimalli F, Di Bonito M, et al. HMGA1 protein overexpression in human breast carcinomas: correlation with ErbB2 expression. Clin Cancer Res: Off J Am Assoc Cancer Res. 2004;10:7637–44.
Tamimi Y, van der Poel HG, Denyn MM, Umbas R, Karthaus HF, Debruyne FM, et al. Increased expression of high mobility group protein i(y) in high grade prostatic cancer determined by in situ hybridization. Cancer Res. 1993;53:5512–6.
Abe N, Watanabe T, Masaki T, Mori T, Sugiyama M, Uchimura H, et al. Pancreatic duct cell carcinomas express high levels of high mobility group i(y) proteins. Cancer Res. 2000;60:3117–22.
Masciullo V, Baldassarre G, Pentimalli F, Berlingieri MT, Boccia A, Chiappetta G, et al. HMGA1 protein over-expression is a frequent feature of epithelial ovarian carcinomas. Carcinogenesis. 2003;24:1191–8.
Takahashi Y, Sawada G, Sato T, Kurashige J, Mima K, Matsumura T, et al. Microarray analysis reveals that high mobility group A1 is involved in colorectal cancer metastasis. Oncol Rep. 2013;30:1488–96.
Chang ZG, Yang LY, Wang W, Peng JX, Huang GW, Tao YM, et al. Determination of high mobility group A1 (HMGA1) expression in hepatocellular carcinoma: a potential prognostic marker. Dig Dis Sci. 2005;50:1764–70.
Wang EL, Qian ZR, Rahman MM, Yoshimoto K, Yamada S, Kudo E, et al. Increased expression of HMGA1 correlates with tumour invasiveness and proliferation in human pituitary adenomas. Histopathology. 2010;56:501–9.
Fedele M, Palmieri D, Fusco A. HMGA2: a pituitary tumour subtype-specific oncogene? Mol Cell Endocrinol. 2010;326:19–24.
Chiappetta G, Avantaggiato V, Visconti R, Fedele M, Battista S, Trapasso F, et al. High level expression of the HMGi(y) gene during embryonic development. Oncogene. 1996;13:2439–46.
Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507.
Abe N, Watanabe T, Izumisato Y, Suzuki Y, Masaki T, Mori T, et al. High mobility group A1 is expressed in metastatic adenocarcinoma to the liver and intrahepatic cholangiocarcinoma, but not in hepatocellular carcinoma: its potential use in the diagnosis of liver neoplasms. J Gastroenterol. 2003;38:1144–9.
Balcerczak M, Pasz-Walczak G, Balcerczak E, Wojtylak M, Kordek R, Mirowski M. HMGi(y) gene expression in colorectal cancer: comparison with some histological typing, grading, and clinical staging. Pathol Res Pract. 2003;199:641–6.
Liau SS, Jazag A, Whang EE. HMGA1 is a determinant of cellular invasiveness and in vivo metastatic potential in pancreatic adenocarcinoma. Cancer Res. 2006;66:11613–22.
Reeves R, Edberg DD, Li Y. Architectural transcription factor HMGi(y) promotes tumor progression and mesenchymal transition of human epithelial cells. Mol Cell Biol. 2001;21:575–94.
Shah SN, Kerr C, Cope L, Zambidis E, Liu C, Hillion J, et al. HMGA1 reprograms somatic cells into pluripotent stem cells by inducing stem cell transcriptional networks. PLoS One. 2012;7:e48533.
Pegoraro S, Ros G, Piazza S, Sommaggio R, Ciani Y, Rosato A, et al. HMGA1 promotes metastatic processes in basal-like breast cancer regulating emt and stemness. Oncotarget. 2013;4:1293–308.
Wang DS, Pan CC, Lai HC, Huang JM. Expression of HMGA1 and ezrin in laryngeal squamous cell carcinoma. Acta Oto-Laryngol. 2013;133:626–32.
Takaha N, Sowa Y, Takeuchi I, Hongo F, Kawauchi A, Miki T. Expression and role of HMGA1 in renal cell carcinoma. J Urol. 2012;187:2215–22.
Sarhadi VK, Wikman H, Salmenkivi K, Kuosma E, Sioris T, Salo J, et al. Increased expression of high mobility group a proteins in lung cancer. J Pathol. 2006;209:206–12.
van der Zee JA, ten Hagen TL, Hop WC, van Dekken H, Dicheva BM, Seynhaeve AL, et al. Differential expression and prognostic value of HMGA1 in pancreatic head and periampullary cancer. Eur J Cancer (Oxford, England : 1990). 2010;46:3393–9.
Hristov AC, Cope L, Di Cello F, Reyes MD, Singh M, Hillion JA, et al. HMGA1 correlates with advanced tumor grade and decreased survival in pancreatic ductal adenocarcinoma. Mod Pathol: Off J U S Can Acad Pathol Inc. 2010;23:98–104.
Liau SS, Whang E. High mobility group a: a novel biomarker and therapeutic target in pancreatic adenocarcinoma. Surg: J Royal Coll Surg Edinb Irel. 2009;7:297–306.
Qu Y, Wang Y, Ma J, Zhang Y, Meng N, Li H, et al. Overexpression of high mobility group A1 protein in human uveal melanomas: implication for prognosis. PLoS One. 2013;8:e68724.
Acknowledgements
This work was supported by the funding from the Rubber Manufacturing occupational hazard protection guidelines, the Ministry of Health project (2009_03-05), the Technology Development Foundation of Pudong District (PKJ2013-Y67), and the Experimental Animal Special Purpose Foundation of Science and Technology Commission of Shanghai Municipality (13140902901).
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Zhang, Z., Wang, Q., Chen, F. et al. Elevated expression of HMGA1 correlates with the malignant status and prognosis of non-small cell lung cancer. Tumor Biol. 36, 1213–1219 (2015). https://doi.org/10.1007/s13277-014-2749-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-2749-4