Abstract
The aim of the present study was to analyze the expression of sex-determining region Y-related high mobility group box 4 (SOX4) in non-small cell lung cancer (NSCLC) and its correlation with clinicopathologic characteristics, including the survival of NSCLC patients. To observe initially the expression status of SOX4 in lung squamous cell carcinoma and adenocarcinoma at gene expression omnibus. The expression of SOX4 mRNA and protein was examined in NSCLC tissues and normal lung tissues through real-time PCR and immunohistochemistry. Meanwhile, the relationship of SOX4 expression levels with clinical characteristics of 168 NSCLC patients was analyzed by immunohistochemistry. Univariate and multivariate analyses were performed to determine the association between SOX4 expression and prognosis of NSCLC patients. In our results, SOX4 expression was increased in NSCLC tissues compared with paired normal lung tissues in microarray data (GSE3268). SOX4 mRNA and protein expression were markedly higher in NSCLC tissues than in normal lung tissues (P = 0.001 and P = 0.001, respectively). Using immunohistochemistry, high levels of SOX4 protein were positively correlated with status of differentiated degree (high vs. middle, P = 0.004; high vs. low, P < 0.001), clinical stage (I–II vs. III–IV, P < 0.001), T classification (T1–T2 vs. T3–T4, P = 0.004), N classification (N0–N1 vs. N2–N3, P = 0.002), and M classification (M0 vs. M1, P = 0.011) in NSCLC. Moreover, the higher level of SOX4 expression was markedly correlated with poor overall survival in NSCLC patients (P < 0.001). Multivariate analysis suggested that increased SOX4 expression was a poor independent prognostic predictor for NSCLC patients (P = 0.002). In conclusion, SOX4 plays an important role on NSCLC progression and prognosis and may serve as a convictive prognostic biomarker for NSCLC patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer is the most common type of malignancy and is one of the leading causes of cancer-related deaths in the world [1, 2]. Non-small cell lung cancer (NSCLC) accounts for approximately 80 % of all lung cancers, with an annual increase in its incidence. Although advancements of diagnosis and treatment have improved the survival of lung cancer patients, the 5-year overall survival (OS) rate of NSCLC was 15 % [3]. Poor prognosis has been attributed to tumor invasion, metastasis, and recurrence. Therefore, it is urgent to identify biomarkers which provide early diagnosis, accurate prognosis prediction, and novel therapeutic target.
SOX4 is a member of the sex-determining region Y-related high mobility group box (SOX) transcription factor family, which consists of at least 20 highly conserved transcription factors in humans [4]. SOX4 gene is located on 6p22.3 and encodes a 47 kDa protein (474 amino acids) [5]. SOX4 protein is characterized by possession of the highly conserved high mobility group (HMG) DNA-binding domain, which mediates binding of SOX4 protein to a short target DNA sequence directly [5]. The 79-amino-acid HMG domains bind to the consensus target sequence (A/T)ACAA(T/A) in the minor grooves of DNA and modify the chromatin structure to generate a conformation that facilitates various DNA-dependent activities [5]. SOX4 involves in many developmental processes such as thymocyte development, nervous system development, embryonic cardiac development, and T cell differentiation pathway [6–8]. In addition, recent studies have suggested that SOX4 as one of the most frequently overexpressed protein in several types of human cancers mediated regulation of tumorigenesis and tumor progression such as hepatocellular carcinoma [9, 10], prostate cancer [11, 12], breast cancer [13, 14], colorectal cancer [15, 16], bladder cancer [17, 18], pineoblastoma [19], uterine leiomyoma [20], pancreatic cancer [21], pheochromocytoma [22], glioblastoma [23], endometrial cancer [24], medulloblastoma [25], and adenoid cystic carcinoma [26]. However, little is known about the significance of SOX4 in patients with NSCLC.
In lung cancer, SOX4 has been identified as a target of gene amplification at chromosome 6p [27]. Moreover, a microarray analysis indicated that SOX4 level significantly increased in small cell lung cancer (SCLC) compared to normal lung tissues [28]. So far, there is no study to evaluate the pathological role of SOX4 in NSCLC patients. The aim of this study is to estimate the association between SOX4 and clinical characteristics including prognosis in NSCLC patients and to explore the role of SOX4 in NSCLC development and progression.
Materials and methods
Analysis of microarray data
Microarray data sets (GEO accession number: GSE3268 and GSE19804) from five pairs of squamous cell carcinoma and adjacent normal lung tissue submitted by Wachi and sixty pairs of adenocarcinoma and adjacent normal lung tissue specimens submitted by Lu were retrieved from the GEO database. Those differentially expressed genes were screened and identified by Real-time PCR for the following study.
Sample collection
A total of 10 freshly frozen NSCLC samples and 10 normal lung samples were collected from Xianning Central Hospital. All fresh samples were immediately preserved in liquid nitrogen. One hundred and sixty-eight paraffin-embedded NSCLC specimens and forty-one normal lung specimens were retrieved from Xianning Central Hospital. No patients had received any form of tumor-specific therapy before diagnosis. Before the use of these clinical samples, prior consents from the patients and approval from the Institutional Ethics Committee of Xianning Central Hospital were obtained. The histopathological diagnosis of all samples was, respectively, diagnosed by two pathologists. The clinical staging was based on the 7th edition of the AJCC Cancer Staging Manual. In the 168 NSCLC cases, there were 98 males and 70 females with age ranging from 21 to 73 years (median 58.19 years). The clinical follow-up time of patients ranged from 6 to 60 months. OS was defined as the interval from the date of diagnosis to NSCLC-related death.
Real-time PCR
Total RNA was extracted using Trizol (Invitrogen) according to the manufacturer’s protocol. After purification, complementary DNA (cDNA) was synthesized from 10-μg total RNA using the Prime Script RT Master Mix (Takara). The primers (Invitrogen) were designed as follows: for human SOX4, the forward primer was 5′-CTTGACATGATTAGCTGGCATGATT-3′; and the reverse primer was 5′-CCTGTGCAATATGCCGTGTAGA-3′. For human GAPDH, the forward primer was 5′-CCCACTCCTCCACCTTTGAC-3′; and the reverse primer was 5′-ATGAGGTCCACCACCCTGTT-3′. The RT-PCR was conducted by SYBR Premix Ex TaqTM II (Takara) on LightCycler (Roche). Relative quantification of RNA expression was calculated using the 2−△△Ct method. The raw data were presented as the relative quantity of target mRNA, normalized with GAPDH, and relative to a calibrator sample. Each sample was examined in triplicate.
Western blot
Western blot was carried out as described [29] with anti-SOX4 (1:1,000; Abcam, ab80261). HRP-conjugated anti-rabbit IgG antibody was used as the secondary antibody (1:2,000; Cell signaling technology). Signals were detected using enhanced chemiluminescence reagents (Pierce).
Immunohistochemistry
Paraffin sections from NSCLC and normal lung specimens were deparaffinized in xylene and rehydrated in a descending ethanol series (100, 95, 90, 80, 70 % ethanol) and double-distilled water according to standard protocols. Heat-induced antigen retrieval was performed in citrate buffer and boiled for 10 min. After antigen retrieval, sections were treated with 3 % hydrogen peroxide and 1 % bovine serum albumin to block the endogenous peroxidase activity and non-specific binding. The sections were incubated with SOX4 antibody (Abcam, ab80261, dilution 1:100) overnight at 4 °C. After phosphate buffered saline washing, the tissue sections were incubated with the biotinylated secondary antibody and streptavidin–horseradish peroxidase complex, each for 20 min at room temperature. Diaminobenzidine was used as the chromogen, and tissue sections were counterstained with haematoxylin and then viewed under a bright-field microscope.
Evaluation of staining
The tissue sections stained immunohistochemically for SOX4 were reviewed, and scored separately by two pathologists blinded to the clinical parameters. Any disagreements were arbitrated by the third pathologists. Staining intensity was graded (0, negative; 1, weak; 2, moderate; 3, strong), and percentage of positive-staining cells was counted (0, 10; 1, 11–50; 2, 51–75; 3, >76 %). The final score was determined by the combined staining score and proportion score (intensity score × proportion score) [29]. The final staining score ranged from 0 to 9. For statistical analysis, final staining scores of 0–4 and 6–9 were, respectively, considered to be low and high expression.
Statistical analysis
All statistical analyses were performed using SPSS version 13.0 and GraphPad 5.0 software. The unpaired t test was applied to test the differential mRNA expression of SOX4 in NSCLC tissues compared to normal lung tissues. The Chi-square test was used to examine the differences of SOX4 protein expression between NSCLC tissues and normal lung tissues. The Chi-square test was applied to the examination of relationship between SOX4 expression levels and clinicopathologic characteristics. Survival curves were plotted using the Kaplan–Meier method and compared using the log-rank test. The significance of survival variables was analyzed using the Cox multivariate proportional hazards model. A P value of less than 0.05 was considered statistically significant.
Results
SOX4 mRNA and protein were overexpressed in NSCLC tissues
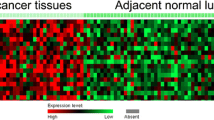
From our microarray data, SOX4 was highly expressed in lung squamous cell carcinoma and adenocarcinoma tissues compared with paired adjacent normal lung tissues (Fig. 1).
Furthermore, we performed real-time PCR to verify the expression of SOX4 mRNA transcripts in ten fresh NSCLC tissues and ten fresh normal lung tissues. Compared with normal lung tissues, NSCLC tissues showed higher expression levels of SOX4 mRNA (P = 0.001, Fig. 2).
We measured the expression levels of SOX4 protein in 168 archived paraffin-embedded NSCLC samples and 41 normal lung samples using immunohistochemical staining (Fig. 3a–f). Specific SOX4 protein staining was found in nucleus. Furthermore, we observed that SOX4 protein was overexpressed in 61.3 % (103/168) of NSCLC samples. In comparison, only 31.7 % of normal lung samples had highly expressed SOX4 protein, significantly lower than that in the NSCLC samples (P = 0.001, Table 1). Furthermore, we measured the expression levels of SOX4 protein in NSCLC tissues and adjacent normal lung tissues using Western blot. SOX4 protein was significantly overexpressed in NSCLC tissues compared with adjacent normal lung tissues (Fig. 4).
Association between clinicopathological characteristics and expression of SOX4 in NSCLC patients
The association between clinicopathological characteristics and SOX4 expression levels in patients with NSCLC was summarized in Table 2. We did not find any significant association of SOX4 expression levels with patient’s gender (P = 0.538), age (P = 0.618), smoking (P = 0.417), and pathology classification (P = 0.809). However, SOX4 was positively associated with differentiated degree (high vs. middle, P = 0.004; high vs. low, P < 0.001), clinical stage (I–II vs. III–IV, P < 0.001), T classification (T1–T2 vs. T3–T4, P = 0.004), N classification (N0–N1 vs. N2–N3, P = 0.002), and M classification (M0 vs. M1, P = 0.011) in NSCLC.
Survival analysis
To explore the prognostic value of SOX4 expression in NSCLC, we measured the association between the levels of SOX4 expression and patients’ survival using Kaplan–Meier analysis with the log-rank test. In 168 NSCLC patients with prognosis information, we found that the level of SOX4 protein expression was significantly associated with the OS of NSCLC patients, as patients with lower levels of SOX4 expression had better survival than those with higher levels of SOX4 expression (P < 0.001, Fig. 5). Furthermore, we also found that increased expression of SOX4 showed poor prognosis in NSCLC patients, regardless of clinical stage, T classification, N classification, and M classification. Multivariate analysis showed that decreased SOX4 expression was an independent poor prognostic factor for NSCLC patients (P = 0.002, Table 3).
Discussion
SOX4 is a member of the group C subfamily of the SOX transcription factors and plays a significant role during embryogenesis, where its expression is widespread and regulates the development of numerous tissues [30]. In many cancers, deregulated expression of SOX4 has been correlated with increased cancer cell growth, cell survival, suppression of apoptosis, and tumor progression through the induction of an epithelial-to-mesenchymal transition (EMT) and metastasis [30]. In adenoid cystic carcinoma, knockdown of SOX4 results in decreased viability and increased apoptosis, and contributes to malignant phenotype [31]. Moreover, Vervoort et al. reported that TGF-beta-mediated increased expression of SOX4 is required for the induction of a mesenchymal phenotype during EMT in human mammary epithelial cells [32]. Similar to Zhang et al’s study in breast cancer, they found that SOX4 expression induces EMT and promotes metastasis in vitro and in xenograft models in vivo [13].
SOX4 expression is increased in a wide variety of tumors, including leukemia, colorectal cancer, prostate cancer, and breast cancer, suggesting a fundamental role in the development of these malignancies [30]. However¸ the role of SOX4 in NSCLC is still unclear. In microarray analysis performed by Wachi et al. (GSE3268) and Lu et al. (GSE19804), we found SOX4 was higher level in lung squamous cell carcinoma and adenocarcinoma samples than in adjacent paired normal lung samples. Then, we performed real-time PCR to verify the expression of SOX4 mRNA and found SOX mRNA expression was also elevated in NSCLC samples. Furthermore, we present the evidence that protein expression of SOX4 was increased in NSCLC through immunohistochemistry, which was consistent with the microarray data and real-time PCR result.
In order to further explore the role of SOX4 in the development and progression of NSCLC, we first analyzed the expression of SOX4 in 168 NSCLC patients and found SOX4 overexpression was significantly associated with differentiated degree, clinical stage, tumor size (T classification), lymph node metastasis (N classification), and distant metastasis (M classification). SOX4 overexpressed in NSCLC may accelerate tumor growth and enhance local cell invasion and metastasis. Our results implicate that SOX4 may involve in NSCLC progression. Similarly, increased levels of SOX4 expression correlated with more aggressive tumors and poor prognosis in prostate cancer [11, 33]. In addition, Vervoort et al. demonstrated that SOX4 expression was elevated in breast cancer compared with normal mammary tissue and positively correlated with histologic grade and status of estrogen receptor, estrogen receptor and HER2 [13]. However, the correlation between SOX4 expression and the survival of NSCLC patients has been seldom reported.
In the past few years, SOX4 overexpression in tumor cells has been shown to be an independent prognostic factor in several types of tumors, which has a favorable or unfavorable prognostic significance according to tumor types. In primary gallbladder carcinoma, SOX4 overexpression was significantly related to better OS and disease-free survival, and SOX4 expression was an independent risk factor for both OS and disease-free survival through multivariate analyses [34]. Moreover, Jafarnejad et al. reported that SOX4 expression was remarkably reduced in metastatic melanoma compared with primary melanoma, and reduced SOX4 expression correlated with a poorer disease-specific survival of melanoma patients and was an independent prognostic factor [35]. On the contrary, there was more evidence indicating that overexpression of SOX4 was unfavorable prognosis factor in gastric cancer [36], acute lymphoblastic leukemia (ALL) [37], colorectal cancer [16], etc. In gastric cancer patients, overexpression of SOX4 was significantly correlated with metastasis, poor differentiation, and unfavorable prognosis [36]. Similarly, Ramezani-Rad et al.’s study showed high levels of SOX4 expression in ALL cells at the time of diagnosis predicted poor outcome, and identified SOX4 as a critical activator of PI3K/AKT and MAPK signaling in ALL cells [37]. The discrepancy of SOX4 expression in different types of human cancer may be attributed to tumor heterogeneity. In the present study, we first presented the evidence that SOX4 protein expression in NSCLC was inversely correlated with patient’s OS. The patients with higher expression of SOX4 protein had shorter survival time. According to multivariate analyses, increased expression of SOX4 protein was a significant predictor of unfavorable prognosis for NSCLC patients.
In summary, our study showed that mRNA and protein expression level of SOX4 were significantly increased in NSCLC tissues and associated with the malignant status of NSCLC. Moreover, our results indicated that SOX4 was a significant prognostic factor for NSCLC patients. Because of the limited patient sample size in our study, further studies are needed to strengthen these findings and verify the role of SOX4 as a reliable clinical predictor of outcome for NSCLC patients.
References
Siegel R, Ma J, Zou Z, Jemal A (2014) Cancer statistics, 2014. CA: Cancer J Clin 64(1):9–29. doi:10.3322/caac.21208
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA: Cancer J Clin 61(2):69–90. doi:10.3322/caac.20107
Rosell R, Karachaliou N (2013) Lung cancer: Maintenance therapy and precision medicine in NSCLC. Nat Rev Clin Oncol 10(10):549–550. doi:10.1038/nrclinonc.2013.152
Penzo-Mendez AI (2010) Critical roles for SoxC transcription factors in development and cancer. Int J Biochem Cell Biol 42(3):425–428. doi:10.1016/j.biocel.2009.07.018
Jafarnejad SM, Ardekani GS, Ghaffari M, Li G (2013) Pleiotropic function of SRY-related HMG box transcription factor 4 in regulation of tumorigenesis. Cell Mol Life Sci 70(15):2677–2696. doi:10.1007/s00018-012-1187-y
Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PA, van de Wetering M, Verbeek S, Lamers WH, Kruisbeek AM, Cumano A, Clevers H (1996) Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature 380(6576):711–714. doi:10.1038/380711a0
Schilham MW, Moerer P, Cumano A, Clevers HC (1997) Sox-4 facilitates thymocyte differentiation. Eur J Immunol 27(5):1292–1295. doi:10.1002/eji.1830270534
Cheung M, Abu-Elmagd M, Clevers H, Scotting PJ (2000) Roles of Sox4 in central nervous system development. Brain Res Mol Brain Res 79(1–2):180–191
Liao YL, Sun YM, Chau GY, Chau YP, Lai TC, Wang JL, Horng JT, Hsiao M, Tsou AP (2008) Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene 27(42):5578–5589. doi:10.1038/onc.2008.168
Hur W, Rhim H, Jung CK, Kim JD, Bae SH, Jang JW, Yang JM, Oh ST, Kim DG, Wang HJ, Lee SB, Yoon SK (2010) SOX4 overexpression regulates the p53-mediated apoptosis in hepatocellular carcinoma: clinical implication and functional analysis in vitro. Carcinogenesis 31(7):1298–1307. doi:10.1093/carcin/bgq072
Liu P, Ramachandran S, Ali Seyed M, Scharer CD, Laycock N, Dalton WB, Williams H, Karanam S, Datta MW, Jaye DL, Moreno CS (2006) Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res 66(8):4011–4019. doi:10.1158/0008-5472.can-05-3055
Moreno CS (2010) The Sex-determining region Y-box 4 and homeobox C6 transcriptional networks in prostate cancer progression: crosstalk with the Wnt, Notch, and PI3K pathways. Am J Pathol 176(2):518–527. doi:10.2353/ajpath.2010.090657
Zhang J, Liang Q, Lei Y, Yao M, Li L, Gao X, Feng J, Zhang Y, Gao H, Liu DX, Lu J, Huang B (2012) SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res 72(17):4597–4608. doi:10.1158/0008-5472.can-12-1045
Graham JD, Hunt SM, Tran N, Clarke CL (1999) Regulation of the expression and activity by progestins of a member of the SOX gene family of transcriptional modulators. J Mol Endocrinol 22(3):295–304
Sinner D, Kordich JJ, Spence JR, Opoka R, Rankin S, Lin SC, Jonatan D, Zorn AM, Wells JM (2007) Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol 27(22):7802–7815. doi:10.1128/mcb.02179-06
Andersen CL, Christensen LL, Thorsen K, Schepeler T, Sorensen FB, Verspaget HW, Simon R, Kruhoffer M, Aaltonen LA, Laurberg S, Orntoft TF (2009) Dysregulation of the transcription factors SOX4, CBFB and SMARCC1 correlates with outcome of colorectal cancer. Br J Cancer 100(3):511–523. doi:10.1038/sj.bjc.6604884
Aaboe M, Birkenkamp-Demtroder K, Wiuf C, Sorensen FB, Thykjaer T, Sauter G, Jensen KM, Dyrskjot L, Orntoft T (2006) SOX4 expression in bladder carcinoma: clinical aspects and in vitro functional characterization. Cancer Res 66(7):3434–3442. doi:10.1158/0008-5472.can-05-3456
Gunes S, Yegin Z, Sullu Y, Buyukalpelli R, Bagci H (2011) SOX4 expression levels in urothelial bladder carcinoma. Pathol Res Pract 207(7):423–427. doi:10.1016/j.prp.2011.05.005
Fevre-Montange M, Champier J, Szathmari A, Wierinckx A, Mottolese C, Guyotat J, Figarella-Branger D, Jouvet A, Lachuer J (2006) Microarray analysis reveals differential gene expression patterns in tumors of the pineal region. J Neuropathol Exp Neurol 65(7):675–684. doi:10.1097/01.jnen.0000225907.90052.e3
Litovkin KV, Ivanova OV, Bauer A, Hoheisel JD, Bubnov VV, Zaporozhan VN (2008) Microarray study of gene expression in uterine leiomyoma. Exp Oncol 30(2):106–111
Cao D, Hustinx SR, Sui G, Bala P, Sato N, Martin S, Maitra A, Murphy KM, Cameron JL, Yeo CJ, Kern SE, Goggins M, Pandey A, Hruban RH (2004) Identification of novel highly expressed genes in pancreatic ductal adenocarcinomas through a bioinformatics analysis of expressed sequence tags. Cancer Biol Ther 3(11):1081–1089 discussion 1090–1081
Molatore S, Liyanarachchi S, Irmler M, Perren A, Mannelli M, Ercolino T, Beuschlein F, Jarzab B, Wloch J, Ziaja J, Zoubaa S, Neff F, Beckers J, Hofler H, Atkinson MJ, Pellegata NS (2010) Pheochromocytoma in rats with multiple endocrine neoplasia (MENX) shares gene expression patterns with human pheochromocytoma. Proc Natl Acad Sci USA 107(43):18493–18498. doi:10.1073/pnas.1003956107
Lin B, Madan A, Yoon JG, Fang X, Yan X, Kim TK, Hwang D, Hood L, Foltz G (2010) Massively parallel signature sequencing and bioinformatics analysis identifies up-regulation of TGFBI and SOX4 in human glioblastoma. PLoS One 5(4):e10210. doi:10.1371/journal.pone.0010210
Huang YW, Liu JC, Deatherage DE, Luo J, Mutch DG, Goodfellow PJ, Miller DS, Huang TH (2009) Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res 69(23):9038–9046. doi:10.1158/0008-5472.can-09-1499
Lee CJ, Appleby VJ, Orme AT, Chan WI, Scotting PJ (2002) Differential expression of SOX4 and SOX11 in medulloblastoma. J Neurooncol 57(3):201–214
Frierson HF Jr, El-Naggar AK, Welsh JB, Sapinoso LM, Su AI, Cheng J, Saku T, Moskaluk CA, Hampton GM (2002) Large scale molecular analysis identifies genes with altered expression in salivary adenoid cystic carcinoma. Am J Pathol 161(4):1315–1323. doi:10.1016/s0002-9440(10)64408-2
Medina PP, Castillo SD, Blanco S, Sanz-Garcia M, Largo C, Alvarez S, Yokota J, Gonzalez-Neira A, Benitez J, Clevers HC, Cigudosa JC, Lazo PA, Sanchez-Cespedes M (2009) The SRY-HMG box gene, SOX4, is a target of gene amplification at chromosome 6p in lung cancer. Hum Mol Genet 18(7):1343–1352. doi:10.1093/hmg/ddp034
Bangur CS, Switzer A, Fan L, Marton MJ, Meyer MR, Wang T (2002) Identification of genes over-expressed in small cell lung carcinoma using suppression subtractive hybridization and cDNA microarray expression analysis. Oncogene 21(23):3814–3825. doi:10.1038/sj.onc.1205480
Li J, Guan HY, Gong LY, Song LB, Zhang N, Wu J, Yuan J, Zheng YJ, Huang ZS, Li M (2008) Clinical significance of sphingosine kinase-1 expression in human astrocytomas progression and overall patient survival. Clin Cancer Res 14(21):6996–7003. doi:10.1158/1078-0432.ccr-08-0754
Vervoort SJ, van Boxtel R, Coffer PJ (2013) The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe? Oncogene 32(29):3397–3409. doi:10.1038/onc.2012.506
Pramoonjago P, Baras AS, Moskaluk CA (2006) Knockdown of Sox4 expression by RNAi induces apoptosis in ACC3 cells. Oncogene 25(41):5626–5639. doi:10.1038/sj.onc.1209566
Vervoort SJ, Lourenco AR, van Boxtel R, Coffer PJ (2013) SOX4 mediates TGF-beta-induced expression of mesenchymal markers during mammary cell epithelial to mesenchymal transition. PLoS One 8(1):e53238. doi:10.1371/journal.pone.0053238
Haram KM, Peltier HJ, Lu B, Bhasin M, Otu HH, Choy B, Regan M, Libermann TA, Latham GJ, Sanda MG, Arredouani MS (2008) Gene expression profile of mouse prostate tumors reveals dysregulations in major biological processes and identifies potential murine targets for preclinical development of human prostate cancer therapy. Prostate 68(14):1517–1530. doi:10.1002/pros.20803
Wang C, Zhao H, Lu J, Yin J, Zang L, Song N, Dong R, Wu T, Du X (2012) Clinicopathological significance of SOX4 expression in primary gallbladder carcinoma. Diagn Pathol 7:41. doi:10.1186/1746-1596-7-41
Jafarnejad SM, Wani AA, Martinka M, Li G (2010) Prognostic significance of Sox4 expression in human cutaneous melanoma and its role in cell migration and invasion. Am J Pathol 177(6):2741–2752. doi:10.2353/ajpath.2010.100377
Shen R, Pan S, Qi S, Lin X, Cheng S (2010) Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 in gastric cancer. Biochem Biophys Res Commun 394(4):1047–1052. doi:10.1016/j.bbrc.2010.03.121
Ramezani-Rad P, Geng H, Hurtz C, Chan LN, Chen Z, Jumaa H, Melnick A, Paietta E, Carroll WL, Willman CL, Lefebvre V, Muschen M (2013) SOX4 enables oncogenic survival signals in acute lymphoblastic leukemia. Blood 121(1):148–155. doi:10.1182/blood-2012-05-428938
Acknowledgments
None.
Conflicts of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, D., Hao, T., Pan, Y. et al. Increased expression of SOX4 is a biomarker for malignant status and poor prognosis in patients with non-small cell lung cancer. Mol Cell Biochem 402, 75–82 (2015). https://doi.org/10.1007/s11010-014-2315-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-014-2315-9