Abstract
Grapevines (Vitis vinifera) are colonized by ubiquitous microorganisms known as endophytes, which may have advantageous or neutral effects without causing disease symptoms. Certain endophytes are uncultivable, so culture-independent approaches such as next generation sequencing (NGS) can help for a better understanding of their ecology and distribution. To date, there are no studies which directly link NGS results with taxa derived from a culturing approach, integrating morphological and multi-gene phylogenetic analysis of endophytes. In this study, a culture-dependent and high-resolution culture-independent approach (next generation sequencing) were used to identify endophytes in grapevine stems. In the culture-dependent approach, a total of 94 isolates were recovered from 84 of 144 healthy grapevine stem fragments (colonization rate = 58.3%). The study is unique as we used subsets of combined multi-gene regions to identify the endophytes to species level. Based on each multi-gene phylogenetic analysis, 28 species belong to 19 genera (Acremonium, Alternaria, Arthrinium, Ascorhizoctonia, Aspergillus, Aureobasidium, Bipolaris, Botryosphaeria, Botrytis, Chaetomium, Cladosporium, Curvularia, Hypoxylon, Lasiodiplodia, Mycosphaerella, Nigrospora, Penicillium, Phoma, Scopulariopsis) were identified. A higher number of culturable fungi were obtained from 13 year-old vines, followed by eight and three year-old vines. In the culture-independent approach, a fungal richness of 59 operational taxonomic units (OTU) was detected, being highest in 13 year-old grapevines, followed by eight and three years. Even though the cultivation approach detected lower fungal richness, the results related to stem are consistent for fungal community composition and richness. Comparison of the fungal taxa identified by the two approaches resulted in an overlap of 53% of the fungal genera. Due to interspecific variability of the sequences from NGS, in many cases the OTUs (even with the highly abundant ones) were only assignable to order, family or genus level. Incorporating multi-gene phylogenies we successfully identified many of the NGS derived OTUs with poor taxonomic information in reference databases to the genus or species levels. Hence, this study signifies the importance of applying both culture-dependent and culture-independent approaches to study the fungal endophytic community composition in Vitis vinifera. This principle could also be applied to other host species and ecosystem level studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Endophytes are microorganisms that reside asymptomatically within interior tissues of living plants for all or part of their life cycle without damaging the host plant (Navarro-Meléndez and Heil 2014; Zhou et al. 2015). Fungal endophytes have been found in all plant species studied in the plant kingdom (Hyde and Soytong 2008; Sánchez et al. 2010; Tejesvi et al. 2010; García et al. 2013; Bonfim et al. 2016; Busby et al. 2016), and have been associated with lichens (Li et al. 2007; Chagnon et al. 2016; Muggia et al. 2017) and sea grasses (Supaphon et al. 2017). The biodiversity of endophytes in a plant can be significant; in certain species, more than 100 endophytic taxa have been discovered (Tan and Zou 2001; Stone et al. 2004). Over the last three decades, endophytic fungi have fascinated taxonomists, mycologists, ecologists (Promputtha et al. 2007, 2010; Purahong and Hyde 2011), chemists and evolutionary biologists (Garoé et al. 2012).
Vitis vinifera L. is an extensively grown, highly important crop and naturally hosts a reservoir of microorganisms. Therefore, a complete survey of the grapevine endophytes under natural conditions is of upmost importance, as grape production and quality can be affected by the vineyard’s active microbial community (Pinto et al. 2014; Busby et al. 2016). Recent studies have shed some light upon the bacterial endophytic communities in grapevines (Bulgari et al. 2011; Compant et al. 2011; Andreolli et al. 2016; Lòpez-Fernàndez et al. 2017), while investigations on fungal endophytic communities have been rare and often limited to culture-dependent approach (Compant et al. 2011; González and Tello 2011; Campisano 2012; Morgan et al. 2017). However, knowledge of the diversity, distribution or influence of endophytic fungi in the development or prevention of certain fungal diseases is still incomplete and the majority of studies have involved European grapevines (Bruez et al. 2016; Rondot and Reineke 2016; Varanda et al. 2016).
Although fungal endophyte research has received considerable attention, their ecology and community composition are poorly characterized, due to methodological limitations. Endophytic fungi have traditionally been studied and described based on culture-dependent approach and characterization of morphological characters in culture (Hyde and Soytong 2008; González and Tello 2011; Busby et al. 2016). This approach is still used worldwide (Ghimire et al. 2011; González and Tello 2011; Ko et al. 2011; Rocha et al. 2011; Heinonsalo et al. 2016; Bhattacharyya et al. 2017; Mahmoud et al. 2017; Morgan et al. 2017), but the results must be considered with some caution, since it is a selective method and subject to factors including surface sterilization techniques, culturing media, incubation conditions, and ability of some fungi to sporulate in culture (Clay et al. 2016; Steinrucken et al. 2016). Many factors such as sampling site, tissue specificity, plant age, physiology or associated vegetation can influence the composition of endophytic communities (Martín-García et al. 2011; Vivas et al. 2015; Donayre et al. 2014; Wicaksono et al. 2015; Christian et al. 2016; Yadav et al. 2016; Dastogeer et al. 2017). The fungal endophyte colonization frequency vary with the age of the host (Arnold and Lutzoni 2007; Goveas et al. 2011; Park et al. 2012; Gupta and Chaturvedi 2017). Nascimento et al. (2015) reported that the rates of endophyte colonization varies with the plant age/development. Several other studies also found that endophyte colonization varied with the plant age (Osono and Mori 2005; Olejniczak and Lembicz 2007; Gupta and Chaturvedi 2017; Fuchs et al. 2017; Liu et al. 2017b).
Endophyte research could benefit from advances in molecular techniques that infer the genetic structure of cultures and the taxonomic composition of endophyte communities. In this sense, multi-locus genetic analyses of isolates are much-needed to obtain consistent information from evolutionary and ecologically determinant loci (Cai et al. 2009; Hyde et al. 2014, 2016; Ariyawansa et al. 2015; Liu et al. 2015). Even though a culture-dependent approach may contain biases, it provides reliable morphological and molecular taxonomic information of fungal endophytes (Ko et al. 2011).
Recently, meta-barcoding approaches have become important tools for assessment of the mycobiomes (Setati et al. 2015; Deagle et al. 2017; Lobo et al. 2017). The use of whole plant tissues for DNA extraction and molecular analysis of the fungal barcode is an alternative tool for the study of endophytic fungi (Duong et al. 2006; McKinnon 2016; Tejesvi et al. 2016; Liu et al. 2017a; Ruiz-Pérez and Zambrano 2017). These culture-independent approaches have been used to investigate the genetic diversity and population structure of endophytes, especially for those taxa that do not grow on standard media (Bullington and Larkin 2015; Ting et al. 2015; David et al. 2017; Purahong et al. 2018). Lücking and Moncada (2017) suggested that bulk of new fungal taxa is revealed through environmental high throughput sequencing with an astounding extent of information. However, these techniques have serious limitations in identifying the majority of unknown taxa into species level and obtaining correct names, since many sequences deposited in GenBank are associated with erroneous taxon names and many species groups cannot be discriminated by using ITS or other portions of the rDNA, in particular in the Ascomycota. Another fact is that many fungi have not been sequenced (Cai et al. 2009; Crouch et al. 2009; Nilsson et al. 2012, 2015).
NGS are mainly based on ITS regions, the fungal DNA barcode (Schoch et al. 2012), but using short fragments such as the ITS2 fragment. Such short sequences or even the whole sequences of ITS do not give the reliable sequence alignments to derive a phylogenetic tree at the species level. Furthermore due to their high inter- and intra-specific variability the taxonomic assignments at the generally agreed threshold of 97% similarity are not consistent for species level identification (Nilsson et al. 2008). Thus, the fungal taxonomic results derived from NGS are probably reasonable only to the genus level. Thus, they are usually reported at the genus level or even higher taxonomic levels such as family or order (Ovaskainen et al. 2010; Purahong et al. 2017a). To date there are no studies that directly link the NGS based fungal OTUs to the culture-dependent morphologically and molecularly (incorporating phylogenetic analysis using multiple genes) identified fungal species that are derived from the same sample.
The present study focused on (i) the comparison of a culture-dependent approach (culturing applying identification using morphological and phylogenetic analysis of multiple genes based identification), versus a culture-independent approach (meta-barcoding of the fungal ITS rDNA barcode) for characterization of diversity and community composition of fungal endophytes associated with stems of grapevines (Vitis vinifera cv. Summerblack) with different ages (3, 8 and 13 years old), (ii) assessing the shared community between two approaches detected from the same grapevine stem and (iii) revealing the potential functions of the endophytic fungal communities inhabiting the grapevine stem. For a better comparison, the same plant organ (stems of grapevines) located in the same vineyard was investigated. We hypothesized that (i) diverse fungal endophytes inhabit stems of grapevines and that the different approaches will reveal different fungal communities implying a higher diversity from the culture-independent approach as compared to culture-dependent approach, (ii) fungal endophytic communities are influenced by the age of grapevine plants and (iii) frequent taxa should be detected with both culture-dependent and culture-independent approaches.
Material and methods
Site description and sampling strategy
Samples were collected during summer of 2015 from a vineyard in Beijing, which comprised three age levels of grapevines (3, 8 and 13 years old Vitis vinifera cv. Midnight beauty). Growers spray a fungicide with pyraclostrobin and lime sulphur for four-to-five times a year. The cultivation style is ‘rain-shelter cultivation’. This region has a temperate and continental monsoon climate, with a mean annual temperature of 26 °C. Mean annual precipitation ranges from 550 to 960 mm of which more than 45% usually falls in August (China Agriculture Yearbook 2014). Asymptomatic grape samples from four stems (or trunks; two inner and two outer parts) were collected from one grapevine. Three healthy grapevines (without any disease symptom) for each age level (3, 8 and 13 years) were selected as replicates and processed within 24 h for fungal endophyte isolation. The remaining samples from these three grapevines per age were subsampled, pooled and homogenized for culture-independent mycobiome analysis through paired-end Illumina sequencing.
Culture-dependent approach
Isolation and identification of endophytic fungi
Following pilot tests, the optimum conditions for surface sterilization were established (Kaewkla and Franco 2016). Samples were cut into 0.5 × 0.5 cm2 sections. Under sterile conditions, tissue segments were surface disinfected in 70% ethanol for 1 min, 1.5% sodium hypochlorite solution for 1 min and three times in sterile-distilled water. To test the efficacy of this method, random surface-disinfected samples were repeatedly imprinted on PDA petri dishes, followed by incubation for two weeks at 20 °C to confirm the absence of epiphytes. After disinfection, samples were placed on PDA with the vascular vessels facing the medium. The plates were incubated for 7–15 days at 20 °C, and all morphologically different colonies were isolated. Fungal isolates were selected and grouped together as morphotypes (Lacap et al. 2003), according to the morphological characters such as the spore production, spore length and morphology, aerial mycelium colour, texture and form, exudates and growth rate.
DNA isolation and PCR
Fungal material for DNA extraction was harvested from 1 to 2 weeks-old cultures grown on potato dextrose agar (PDA) by scraping the mycelium. Specific gene regions were amplified with particular primers, i.e. ITS1 and ITS4 to amplify the internal transcribed spacers (ITS) (White et al. 1990), LROR and LR5 to amplify the large subunit rDNA (LSU) (Vilgalys and Hester 1990), NS1 and NS4 to amplify region of nuclear small subunit rDNA (SSU) (White et al. 1990), a fragment of translation elongation factor 1-α (TEF) was amplified using EF-728F and EF-986R (Carbone and Kohn 1999), GPD1 and GPD2 to amplify glyceraldehyde 3-phosphate dehydrogenase (GPDH) (Berbee et al. 1999), RPB2–5F and RPB2–7cR to amplify RNA polymerase second largest subunit (RPB2) (Sung et al. 2007), HSP60for+ and HSP60rev+ to amplify heat shock protein (HSP60) (Staats et al. 2005), BT2A and BT2B to amplify β- tubulin (TUB) (Glass and Donaldson 1995), ACT-512F and ACT783R to amplify partial actin gene (ACT) (Carbone and Kohn 1999). The amplification reactions were performed in 25 μl final volumes and consisted of TaKaRa Ex-Taq DNA polymerase 0.3 μl, 12.5 μl of 2× PCR buffer with 2.5 μl of dNTPs, 1 μl of each primer, 9.2 μl of double-distilled water and 100–500 ng of DNA template. PCR products were checked on 1% agarose electrophoresis gels stained with ethidium bromide. PCR products were Sanger sequenced by Sunbiotech Company, Beijing, China.
Sequence alignment and phylogenetic analyses
A BLAST search with the ITS sequence data was used to reveal the closest matching taxa of endophytes. After they were identified in to genus level, other necessary gene regions were sequenced for particular genera. The sequences obtained in this study were aligned with sequences retrieved from GenBank using MAFFT (http://www.ebi.ac.uk/Tools/msa/mafft/) (Katoh and Toh 2010) and were manually optimized with BioEdit (Hall 2006). All available type sequences of each genus were included in a preliminary phylogenetic analysis and phylogenetically closely related species were selected for further analysis of the combined gene regions. Maximum parsimony analysis (MP) was performed using phylogenetic analysis using PAUP (v. 4.0b10) (Swofford 2003). Ambiguously aligned regions were excluded from all analyses and gaps were treated as missing data. Trees were inferred using the heuristic search option with TBR branch swapping and 1000 random sequence additions. Branches of zero length were collapsed, and all equally most parsimonious trees were saved. The trees were visualized with TreeView v. 1.6.6 (Page 1996).
For the Bayesian analyses, the models of evolution were estimated using MrModeltest v. 2.3 (Nylander 2004). Posterior probabilities (PP) were determined by Bayesian Markov Chain Monte Carlo (BMCMC) sampling in MrBayes 3.0b4 (Ronquist and Huelsenbeck 2003), using the estimated model of evolution. Six simultaneous Markov chains were run for 1,000,000 generations, and trees were sampled every 100th generation (resulting in 10,000 total trees). The first 2000 trees, which represented the burn-in phase of the analyses, were discarded and the remaining 8000 trees were used to calculate PP in the majority-rule consensus tree. The sequences used for phylogenetic analysis were deposited in GenBank and provided in the Supporting Information (Table S1).
Culture-independent approach
Endophytic mycobiome analysis: paired-end sequencing
Total genomic DNA from homogenized stem samples was extracted using the CTAB/SDS method. DNA concentration and purity was monitored on 1% agarose gels. Accordingly, the DNA was diluted to 1 ng/μL using sterile water and used as PCR template. Nuclear ribosomal internal transcribed spacer (ITS1) region was amplified using specific primers (ITS5-1737F and ITS2-2043R) (Huang et al. 2016) with sample specific barcodes. All PCR reactions were carried out with Phusion® High-Fidelity PCR Master Mix (New England Biolabs). Thirty-five cycles (95 °C for 45 s, 56 °C for 45 s, and 72 °C for 60 s) were performed with a final extension at 72 °C for 7 min. Samples with amplified products of 400–450 bp were chosen for further analysis. These PCR products were quantified using SYB green and all products were mixed in equimolar ratios. The PCR product mix was then purified with Qiagen Gel Extraction Kit (Qiagen, Germany). Sequencing libraries were generated using TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, USA) following the manufacturer’s recommendations and index codes were added. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. At last, the library was sequenced on an Illumina HiSeq2500 platform and 250 bp paired-end reads were generated.
Endophytic mycobiome analysis: bioinformatic analysis
Paired-end reads were assigned to samples based on their unique barcode and truncated by cutting off the barcode and primer sequence. The unique barcode sequence for each sample is provided in the Supporting Information (Table S2). Paired-end reads were merged using FLASH (V1.2.7, http://ccb.jhu.edu/software/FLASH/) (Magoč and Salzberg 2011), and the splicing sequences were called raw tags as described in Bokulich et al. (2013). Quality filtering on the raw tags was performed under specific filtering conditions to obtain the high-quality clean tags (Bokulich et al. 2013) according to the QIIME (V1.7.0, http://qiime.org/index.html) (Caporaso et al. 2010) quality controlled process. The tags were compared with the reference database (Unite Database, https://unite.ut.ee/) using UCHIME algorithm (UCHIME Algorithm, http://www.drive5.com/usearch/manual/uchime_algo.html) (Edgar et al. 2011) to detect chimera sequences, and then the chimera sequences were removed (Haas et al. 2011). Sequences analyses of the clean tags were performed using the Uparse software (Uparse v7.0.1001, http://drive5.com/uparse/) (Edgar 2013). Sequences with ≥ 97% similarity were assigned to the same OTUs. Representative sequences of each OTU were screened for further annotation. Taxonomic assignment of the representative sequences was done against the Unite Database (https://unite.ut.ee/) (Kõljalg et al. 2013) using the Blast algorithm. OTUs abundance was normalized to the sample with the least sequences (55, 822). Singletons were removed from the dataset. All subsequent analyses were performed based on this normalized dataset. Raw Illumina reads were submitted to the Sequence Read Archive (SRA) of National Center for Biotechnology Information (NCBI) under the BioProject number PRJNA433252.
Comparison of the NGS and culture based endophytes
Similarity of the endophyte community derived from the NGS analysis with that of the culture-based approach was done using the CD-HIT-EST-2D algorithm (http://weizhong-lab.ucsd.edu/cd-hit/) to compare ITS2 sequence similarity between two datasets using a 90% similarity to see the genus level similarity of the two databases followed by a manual BLAST based identification of the respective OTUs. Functional group assignment of each OTU was done using the FUNGuild data base to (Nguyen et al. 2016; https://github.com/UMNFuN/FUNGuild).
Statistical analysis
All statistical analyses were performed using PAST (Hammer et al. 2001). To visualize the endophytic community compositions among different age levels of grape plants derived from culture-dependent and culture-independent approaches, we used non-metric multidimensional scaling (NMDS) analysis based on Jaccard distance measure (presence-absence OTU matrix). The stress values from NMDS were zero in both cases. To test for the difference in endophytic community compositions among different age levels of grape plants we used cluster analysis based on Jaccard (presence-absence OTU matrix) distance measure. To assess the coverage of the sequencing depth in mycobiome analysis, individual rarefaction analysis (with 95% confidence) was performed for each sample using the “diversity” function.
Results
Culture-dependent approach: low fungal diversity and strong effect of grapevine age
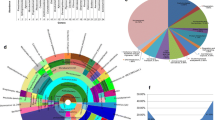
In total, 94 isolates were recovered from 84 of the 144 stem samples analyzed (colonization rate = 58.3%); thus, 60 stem fragments did not yield any endophytic fungi. All of the culturable endophytic fungi recovered were ascomycetes, and were distributed in five classes (Dothideomycetes, Eurotiomycetes, Leotiomycetes, Pezizomycetes and Sordariomycetes). As shown in Fig. 1, 71.2% of the isolates were assigned to Dothideomycetes, of which Pleosporales and Capnodiales were dominant, accounting for 55.2 and 31.3% of this group. The remaining Dothideomycetes isolates belonged to Botryosphaeriales and Dothideales, accounting for 8.9 and 4.4%, respectively. There were 12.7% of isolates in Sordariomycetes. Of the Sordariomycetes group, Sordariales (27.2%) and Xylariales (27.2%) were prominent, while 18.1, 18.1, 9.0% respectively were in Hypocreales, Trichosphaeriales and Microascales. A considerable fraction of isolates (11.7%) were Eurotiomycetes with all belonging to Eurotiales. In addition, 2.1 and 1.1% of isolates belong to Leotiomycetes and Pezizomycetes correspondingly.
Based on morphology and phylogenetic analysis of subsets of combined ITS, LSU, SSU, TEF, GPDH, RPB2, HSP60, TUB and ACT sequence data, the isolates obtained from the culture-dependent method were identified as 28 species (Table 1), 9 of which were observed only once. Alternaria alternata was the most abundant (relative abundance = 28.7%), followed by Cladosporium (21.1%), Aspergillus (10.6%), Botryosphaeria dothidea (5.3%), Aureobasidium pullulans (3.2%), Bipolaris sorokiniana (3.2%), Chaetomium globosum (3.2%) and Phoma herbarum (3.2%). The relative abundance of the other species were between 2.1 and 1.1% (Table 1). Fungal morphology and phylogenetic analysis of most frequent taxa are presented in Figs. 2, 3, 4 and 5 and the other prominent taxa are presented in Figs. 6, 7 and 8. Fungal richness of the culture-dependent approach ranged from 23 isolates (three year-old grapevines) to 32 isolates (13 year-old grapevines). Non-metric multidimensional scaling (NMDS) ordination and cluster analysis showed different fungal endophytic community composition among different ages of grapevine (Fig. 10). In three year-old stems, we detected 13 species, in eight year-old, 16 species and in 13 year-old, 17 species. The fungal endophytic community comprised several taxa, known as plant pathogens (Table 1). As an example, Botryosphaeria dothidea (associated with Botryosphaeria dieback in grapevine) could be considered as abundant, representing 5.3% of the total strains characterized (Table 1; Fig. 4).
Alternaria alternata. a, b Colonies on PDA (14 days old) from surface and reverse, c Conidia attached to conidiophore, d–i conidia. Scale bars: c–i = 20 µm. Phylogenetic tree inferred from maximum likelihood (ML) and Bayesian inference (BI) using combined ITS, LSU, GPDH, EF and RPB2 sequence data of the genus Alternaria. Only the topology generated from the ML analysis is shown. ML values greater than the 90% are indicated. Bayesian Posterior Probability greater than 0.90 are indicated with thick branch. Taxa isolated in this study are in blue
Aspergillus pseudoglaucus (a–e), Aspergillus japonicas (f–k), Aspergillus niger (l–p). a, b Colonies on PDA (14 days old) from surface and reverse, c–f conidia. g, h Colonies on PDA (14 days old) from surface and reverse, i–m conidia. n, o Colonies on PDA (14 days old) from surface and reverse, p–s conidia. Scale bars: c–f = 20 µm, i–m = 20 µm, p–s = 20 µm. Phylogenetic tree inferred from maximum likelihood (ML) and Bayesian inference (BI) using ITS sequence data of the genus Aspergillus. Only the topology generated from the ML analysis is shown. ML values greater than the 90% are indicated. Bayesian Posterior Probability greater than 0.90 are indicated with thick branch. Taxa isolated in this study are in blue
Botryosphaeria dothidea. a, b Colonies on PDA (14 days old) from surface and reverse, c–e conidia. Scale bars: c–e = 20 µm. Phylogenetic tree inferred from maximum likelihood (ML) and Bayesian inference (BI) using combined ITS and TEF sequence data of the genus Botryosphaeria. Only the topology generated from the ML analysis is shown. ML values greater than the 90% are indicated. Bayesian Posterior Probability greater than 0.90 are indicated with thick branch. Taxa isolated in this study are in blue
Cladosporium cladosporioides (a–d), Cladosporium ramotenellum (e–h), Cladosporium silenes (i–m), Cladosporium sphaerospermum (n–r), Cladosporium tenellum (s–v), Cladosporium tenuissimum (w–z) a, e, i, n, s, w Colonies on PDA (14 days old) from surface and reverse, b–d, f–h, j–m, o–r, t–v, x–z Conidia. Scale bars: b–d, f–h, j–m, o–r, t–v, x–z = 20 µm. Phylogenetic tree inferred from maximum likelihood (ML) and Bayesian inference (BI) using combined ITS, TEF and ACT sequence data of the genus Cladosporium. Only the topology generated from the ML analysis is shown. ML values greater than the 90% are indicated. Bayesian Posterior Probability greater than 0.90 are indicated with thick branch. Taxa isolated in this study are in blue
Phylogenetic trees inferred from maximum likelihood (ML) and Bayesian inference (BI). a Combined LSU, SSU and ITS sequence data of the genus Aureobasidium, b combined ITS, GDPH, TEF and LSU sequence data of the genus Bipolaris, c combined ITS, GDPH, TEF and LSU sequence data of the genus Curvularia, d combined LSU, ITS, BT and RPB2 sequence data of the genus Phoma. Only the topology generated from the ML analysis is shown. ML values greater than the 90% are indicated near the node. BI greater than 0.90 are indicated with thick branch. Taxa isolated in this study are in blue
Phylogenetic trees inferred from maximum likelihood (ML) and Bayesian inference (BI). a ITS sequence data of the genus Acremonium. b ITS sequence data of the genus Arthrinium. c Combined G3PDH, HSP60 and RPB2 sequence data of the genus Botrytis. d Combined ITS, TUB2, RPB2 and LSU sequence data of the genus Chaetomium. Only the topology generated from the ML analysis is shown. ML values greater than the 90% are indicated near the node. BI greater than 0.90 are indicated with thick branch. Taxa isolated in this study are in blue
Phylogenetic trees inferred from maximum likelihood (ML) and Bayesian inference (BI). a ITS sequence data of the genus Ascorhizoctonia, b Combined ITS and EF sequence data of the genus Lasiodiplodia, c ITS sequence data of the genus Nigrospora. d Combined ITS and BT sequence data of the genus Hypoxylon. Only the topology generated from the ML analysis is shown. ML values greater than the 90% are indicated near the node. BI greater than 0.90 are indicated with thick branch. Taxa isolated in this study are in blue
Culture-independent approach: mycobiome with low diversity and strong effect of grapevine age
Despite the high number of sequences per sample (55, 822 reads), low fungal richness was detected (59 OTUs in total, Table 2) ranging from 23 (three year-old samples) to 43 (13 year-old samples) OTUs (Fig. 9). The richness of endophytic fungi from different age levels were significantly different (P < 0.05), being highest at 13 years followed by eight and three years (Fig. 9). The majority of fungal OTUs were rare: 51 out of 59 had relative abundance lower than 0.1% (Table 2). Fungi identified by the culture-independent approach belonged to three phyla, Ascomycota (93.6%), Basidiomycota (4.2%) and Zygomycota (2.1%). Ascomycetous taxa were distributed among five classes: Dothideomycetes (34%), Eurotiomycetes (40.9%), Leotiomycetes (4.5%), Pezizomycetes (4.5%) and Sordariomycetes (15.9%). The frequently detected OTUs were CladosporiumOTU_1 (39.03%), PleosporaceaeOTU_2 (33.53%), CladosporiumOTU_4 (12.54%), AscomycotaOTU_15 (11.60%), CadophoraOTU_5 (1.60%), CladosporiumOTU_6 (0.56%), BotryosphaeriaOTU_7 (0.45%) and AscomycotaOTU_3 (0.29%). Non-metric multidimensional scaling (NMDS) ordination and cluster analysis showed different fungal endophytic community composition among different ages of grapevine (Fig. 10). In three year-old stems, we frequently detected four OTUs: PleosporaceaeOTU_2 (56.30%), AscomycotaOTU_15 (24.75%), CladosporiumOTU_1 (13.05%) and CadophoraOTU_5 (4.74%). In eight year-old stems, PleosporaceaeOTU_2 (43.66%), CladosporiumOTU_1 (31.14%), CladosporiumOTU_4 (14.76%) and AscomycotaOTU_15 (9.50%) were often detected. CladosporiumOTU_1 (72.92), CladosporiumOTU_4 (22.77%), BotryosphaeriaOTU_7 (1.32%) and CladosporiumOTU_6 (1.02%) were commonly associated with 13 year-old grapevines (Table 2).
Non-metric multidimensional scaling (NMDS) ordinations and cluster analysis of fungal community composition derived from culture-dependent (a, b) and high resolution culture-independent (c, d) approaches. Similarity from cluster analysis is ranged from 0 (completely different) to 1 (completely overlap)
Comparison between culture-dependent and culture-independent approaches
The results regarding the influence of age of grapevine plants on endophytic fungal communities were similar (Fig. 10). Detected all fungal genera were Acremonium, Alternaria, Arthrinium, Ascorhizoctonia, Aspergillus, Aureobasidium, Bipolaris, Botryosphaeria, Botrytis, Cadophora, Chaetomium, Chaetothyriales, Cladosporium, Cryptococcus, Curvularia, Eupenicillium, Exophiala, Hypoxylon, Kernia, Lasiodiplodia, Lophiostoma, Mortierella, Mycosphaerella, Nigrospora, Oidiodendron, Penicillium, Phialosimplex, Phoma, Pyrenochaeta, Scopulariopsis, Tomentella and Toxicocladosporium (Tables 1, 2). However, only Acremonium, Aspergillus, Botryosphaeria, Botrytis, Cladosporium, Lasiodiplodia and Phoma were detected in both approaches. The results from both approaches showed that members of ascomycetes were dominant in the endophytic fungal community inhabiting grapevine stems. However, we identified members of Basidiomycetes (TomentellaOTU_29, CryptococcusOTU_41 and AgaricomycetesOTU_46) in the culture-independent approach. Direct matching of the ITS sequences of fungal endophytes detected from these two approaches confirmed that the results obtained in most cases are consistent, except for Curvularia, where the sequence similarity was lower than 90% and did not cluster together and for Penicillium digitatum that clustered with its sexual morph state Eupenicillium. In total, direct matching of ITS sequences of fungal endophytes detected by the two approaches matched 13/28 fungal species from the culture-dependent approach and 16/59 fungal OTUs from the culture-independent approach at genera (90% similarity) or species (97% similarity or higher) levels (Table 3). We were able to assign 9/16 and 7/16 OTUs from NGS at genus and species levels, respectively (Table 3). Five fungal OTUs that were identified only at the phylum, order or family level in the culture-independent approach were identified at genus level by direct matching (91–95% similarity). These include AscomycotaOTU_3 (overall relative abundance = 0.29) and AscomycotaOTU_15 (overall relative abundance = 11.60; Cladosporium), AscomycotaOTU_22 (overall relative abundance = 0.01%; Chaetomium), EurotialesOTU_25 (overall relative abundance = 0.01%; Aspergillus) and PleosporaceaeOTU_2 (overall relative abundance = 33.53%; Alternaria) (Table 3). All frequently detected genera in the culture-dependent approach (i.e. with relative abundance higher than 5%; Alternaria, Aspergillus, Botryosphaeria and Cladosporium) were also detected in culture-independent approach. Alternaria (detected as PleosporaceaeOTU_2 in culture-independent approach), Cladosporium sp. and Botryosphaeria were the frequently detected endophytes in both approaches. Aspergillus sp. was frequently detected in the culture-dependent approach, but exhibited low relative abundances in the culture-independent approach. On the other hand, one highly detected fungal taxon from culture-independent approach (Cadophora sp.) was not isolated in the culture-dependent approach.
Fungal guilds and endophytic fungal functional groups
Fungal guild analysis showed that endophytes from the culture-independent approach potentially comprised various functional groups. This study indicates that the symptomless endophytes living in grapevine stems, can also exist in numerous life modes, such as being saprotrophs, pathogens or symbionts. Cadophora and Cladosporium sp. were frequently detected endophytes. The prevailing pathogen was Botryosphaeria dothidea. All saprotrophs were detected at low relative abundances (less than 0.1%) (Table 2). A list of all fungal endophytes detected in this study, with their possible functions, are listed in Table 2. The culture-independent approach revealed that fungal endophytes comprised endophytes, saprotrophs and pathogens.
Discussion
Comparison of culture-dependent versus culture-independent approaches
This is the first study conducted to compare the diversity and community composition of fungal endophytes in stems of grapevine (Vitis vinifera) using a culture-dependent approach, incorporating multigene phylogenetic analysis and a culture-independent approach using meta-barcoding and paired-end Illumina sequencing. The traditional approach for studying the diversity of endophytic fungi is the culture-dependent approach. Many studies have used sequence data from the ITS region to identify and evaluate endophytic fungi (Guo et al. 2000, 2001, 2003; Arnold 2002; Lacap et al. 2003; Promputtha et al. 2005, 2007, 2010; Tejesvi et al. 2011; Jeewon et al. 2003; Haghighi and Shahdoust 2015). In the present study we used subgroups of combined ITS, LSU, SSU, TEF, GPDH, RPB2, HSP60, TUB and ACT sequence data to identify the endophytes obtained from grapevine stems. Two isolates of Acremonium belong to A. alternatum were identified using analysis of ITS sequence data (Fig. 7a). Twenty-seven Alternaria isolates were subjected to combined analysis of ITS, LSU, GPDH, TEF and RPB2 sequence data and all Alternaria isolates were identified as A. alternata sensu stricto (Fig. 2). Two Arthrinium isolates were identified as A. rasikravindrii by using ITS sequence data (Fig. 7b). Analysis of ITS sequence data, showed that one isolate was Ascorhizoctonia sp. (Fig. 8a) and confirmed the identification of four Aspergillus species as A. japonicas, A. niger, A. pseudodeflectus and A. pseudoglaucus (Fig. 3). Analysis of combined LSU, SSU and ITS sequence data identified three Aureobasidium taxa to be A. pullulans (Fig. 6a). The phylogeny inferred from combined ITS, GDPH, TEF and LSU sequence data resolved three Bipolaris isolates as B. sorokiniana (Fig. 6b). Combined ITS and TEF sequence data identified five Botryosphaeria taxa to be B. dothidea (Fig. 4) and resolved one isolate as Lasiodiplodia theobromae (Fig. 8d). Two Botrytis isolates were subjected to combined G3PDH, HSP60 and RPB2 sequence data and were identified as B. cinerea (Fig. 7c). Analysis of combined ITS, TUB2, RPB2 and LSU sequence data, resolved three isolates as Chaetomium globosum (Fig. 7d). The phylogenetic tree inferred from combined ITS, TEF and ACT sequence data resolved 20 Cladosporium strains as C. cladosporioides, C. ramotenellum, C. silenes, C. sphaerospermum, C. tenellum and C. tenuissimum (Fig. 5). Analysis of combined ITS, GDPH, TEF and LSU sequence data from four Curvularia isolates resolved C. americana (Fig. 6c). One isolate of Hypoxylon lateripigmentum was identified by combined ITS and BT sequence data (Fig. 8c). An isolate of Mycosphaerella was identified as M. graminicola by the combined analysis of ITS, ACT and TEF sequence data. Two Nigrospora isolates were identified as N. oryzae and N. sphaerica using ITS sequence data (Fig. 8b). Analysis of combined ITS, LSU and SSU sequence data resolved the two Penicillium isolates as P. digitatum. A phylogeny inferred from combined LSU, ITS, BT and RPB2 sequence data resolved the three Phoma isolates as P. herbarum (Fig. 6d). An isolate of Scopulariopsis was analysed with combined ITS, LSU and SSU sequence data and was identified as Scopulariopsis brevicaulis. The current study identified all isolated taxa from culture-dependent method to species level with strong support in multi-gene phylogenetic analysis, setting a robust goal for future fungal community studies. Some endophytes such as Acremonium, Alternaria, Aureobasidium, Penicillium and Phoma have been used as biocontrol agents against pathogens in grapevines (Tables 1, 4).
Traditional taxonomy and nomenclature is unable to accurately document the vast number of unrecognized taxa, regardless of any doubts one might have to formally describe fungi based on DNA sequence data only (Lücking and Moncada 2017). The aim of environmental sequence nomenclature is to place names on hundreds of thousands of species of fungi that would otherwise be left undescribed (Lücking and Moncada 2017). Environmental high throughput sequencing reads almost 1000 times more than Sanger sequences for the fungal barcoding marker (Jayawardena et al. 2018). A recent study conducted by Lücking and Moncada (2017) showed that a formally recognized unnamed lichenicolous basidiomycete (in Agonimia and Normandina thalli) is a new genus, with seven new species, although no physical type specimens could be preserved. Lücking and Moncada (2017) suggested that this opens the door to the formal recognition of thousands of species of voucher less taxa detected through environmental sequencing techniques.
The main limitation of the culture-dependent approach is that unculturable species and some slow growing or weakly competitive species may not be isolated. To overcome the shortcomings of the culture-dependent approach, culture-independent approaches have been suggested as an alternative (Peršoh 2015; Hoppe et al. 2016; Gomez et al. 2017; Zapka et al. 2017). However, culture-dependent approaches do not seem to resolve species accurately, since taxa are not resolved to species level. Some studies have shown that endophytic fungi recovered by culture-dependent approaches are different from those detected by culture-independent approaches, and some isolated strains were never found in the culture-independent methods (Campisano 2012; Kraková et al. 2017; Mendoza et al. 2017). This fact was also experienced in the current study as we obtained 9 fungal genera from the culture-dependent approach, which were absent in culture-independent approach. Nevertheless, our study demonstrates that all frequently detected fungal genera from the culture-dependent approach can be detected in culture-independent approach when we used a high resolution technique with a high quality dataset. In this study, we used paired-end Illumina sequencing that provided a minimum of 55, 822 high quality sequences per sample (saturated rarefaction curves for all samples) and revealed all frequently detected fungal genera from culture-dependent approach that have relative abundance higher than 5%. The most frequently detected genera in culture-dependent and culture-independent approaches are also mostly consistent, except some OTUs, such as Aspergillus (frequently detected in culture-dependent approach, but become less frequent in culture-independent approach) and Cadophora sp. (commonly detected fungal taxon in the culture-independent approach, but not detected in the culture-dependent approach). One reason for this observation might be that Aspergillus sp. have a fast growing ability, but possibly occurring in low amounts in the tissue samples. Thus, in the culture-dependent approach, they could grow fast and result in a high number of isolates, whereas they could not be detected in next generation sequencing. Cadophora sp. are classified as endophytes in this study based on FUNGUILD, but some members of this genus can be plant pathogens that may not be able to grow quickly on artificial media without their host (Travadon et al. 2015).
Although there are some differences in the fungal taxa and richness of the two approaches (culture-dependent < culture-independent approaches), the results regarding the effect of stem ages on fungal community composition and richness are consistent. Studies on the community of endophytes have often ignored the impact of plant age (Fuchs et al. 2017). To our knowledge, this is the first study showing endophytic fungal composition in grapevine in three different age levels. However, our results based on both culture-dependent and culture-independent approaches indicate that endophytic fungal community and richness is maximum at 13 years. Further studies are needed to confirm whether there is any correlation with age level and the number of endophytic taxa in various hosts. Despite the highest per sample sequence read coverage we detected low diversity of fungal endophytes in grapevine stem samples. The same outcome was perceived in culture-dependent approach, which comprises only 28 species from 94 isolates. The colonization rate was also very low, with only 58.3% of the studied stem fragments yielding endophytes, signifying that nearly half of the fragments may not support culturable endophytes.
Why was there no species overlap between the two approaches in the present study?
The current study indicates that NGS data are only accurate at the genus or family levels. Seven fungal genera obtained by the culture-dependent approach (Acremonium, Aspergillus, Botryosphaeria, Botrytis, Cladosporium, Lasiodiplodia and Phoma) overlapped with those of the culture-independent approach (Table 3). NGS for fungal community analysis mostly acquires short sequence fragments of ITS (full, ITS1 or ITS2) that eventually is not adequate for species level identification at the currently agreed 97% sequence similarity for all fungi (Nilsson et al. 2008; Garnica et al. 2016). The ITS rDNA marker is not consistent due to their high variability, therefore not reliable for species level identification (Nilsson et al. 2008). In our study, we experienced that using NGS data to identify the taxa in a community is not accurate at the species level, as compared to the analyses using multigene sequence data using cultures from the culture-dependent method. ITS sequence data can be regarded as taxonomically less-informative for most of the fungal taxa belonging to Dothideomycetes and/or Sordariomycetes, which were identified through the culture-dependent approach which might account to the variable inter- and intra-specific variation of the ITS fragment.
Matching of endophytic fungi detected from the two approaches: opportunity to assign more correct taxonomic information to NGS datasets and reference databases
In this study, we were able to match sequencing and taxonomic data from culture-dependent techniques to assign taxonomic information at genera and species level to 9 and 7 fungal OTUs from the culture-independent approach. This matching allows us to assign better correct taxonomic information and functions to the fungal OTUs detected in culture-independent approach. Next generation sequencing often results in sequences that are associated with taxa which have not been reported in previous studies, as well as sequences that are not linked with any fungal sequences in GenBank (Tejesvi et al. 2010; Ko et al. 2011; Taylor et al. 2016). One of the major reasons for this might be the insufficient number of cultures based on reference sequences in GenBank or other databases, since early fungal identifications did not provide genetic data. In this study, we demonstrate that some commonly detected endophytes from the culture-independent approach can only be identified to phylum, order or family levels. These include PleosporaceaeOTU_2 (overall relative abundance 33.53%), which was later found to match the most frequently isolated endophytic species Alternaria alternata. NGS technologies together with general culturing methods allow synchronized exploration of a more complete picture of endophytic communities in host plants (Hardoim et al. 2015). This concept applies when fungal genera are identified by sequencing the pure cultures obtained in culture-dependent method and then relate to OTUs obtained from the culture-independent method with a defined similarity threshold based on the intra- and inter-specific variation of the isolate.
Potential functions of the endophytic fungal communities inhabiting the grapevine stem
Taxonomy based functional assignment of fungi has been used to study potential roles of endophytes in plant community structure and ecosystem functioning (Green et al. 2008; Roe et al. 2010). In the present study, culture-dependent and culture-independent approach allows the identification of potential roles of identified fungal taxa as endophytes, saprotrophs, pathogens or symbionts in the grapevine fungal community. Although the grapevines look healthy, they were already colonized by pathogens that may be inactive until suitable conditions arise to cause diseases. In this study, we identified two important fungal pathogens (Botryosphaeria dothidea and Botrytis cinerea) from both culture-dependent and independent approaches. A number of endophytes revealed by the culture-independent approach are classified as potential saprotrophs. This fact has been confirmed by earlier studies implying endophytes can change their lifestyle to become saprotrophs (Purahong and Hyde 2011; Fesel and Zuccaro 2016; Szink et al. 2016) and they may play important role in plant litter decomposition, especially at early decomposition stage (Purahong et al. 2016).
In general this study has shown the potential of using both approaches in a given study to link the NGS datasets with culture-based fungal isolates that are morphologically and phylogenetically identified. Such a complementary approach enables to properly identify and give correct taxonomic information to the fungal endophytes identified with the NGS approach. NGS studies without reference cultures may be misleading and many of the data generated in the past must be seen as very critical.
References
Andreolli M, Lampis S, Zapparoli G, Angelini E, Vallini G (2016) Diversity of bacterial endophytes in 3 and 15 year-old grapevines of Vitis vinifera cv. Corvina and their potential for plant growth promotion and phytopathogen control. Microbiol Res 183:42–52
Ariyawansa HA, Hyde KD, Jayasiri SC, Buyck B, Chethana KWT, Dai DQ, Dai YC, Daranagama DA, Jayawardena RS, Lücking R, Ghobad-Nejhad M, Niskanen T, Thambugala KM, Voigt K, Zhao RL, Li GJ, Doilom M, Boonmee S, Yang ZL, Cai Q, Cui YY, Bahkali AH, Chen J, Cui BK, Chen JJ, Dayarathne MC, Dissanayake AJ, Ekanayaka AH, Hashimoto A, Hongsanan S, Jones EBG, Larsson E, Li WJ, Li QR, Liu JK, Luo ZL, Maharachchikumbura SSN, Mapook A, McKenzie EHC, Norphanphoun C, Konta S, Pang KL, Perera RH, Phookamsak R, Phukhamsakda C, Pinruan U, Randrianjohany E, Singtripop C, Tanaka K, Tian CM, Tibpromma S, Abdel-Wahab MA, Wanasinghe DN, Wijayawardene NN, Zhang JF, Zhang H, Abdel-Aziz FA, Wedin M, Westberg M, Ammirati JF, Bulgakov TS, Lima DX, Callaghan TM, Callac P, Chang CH, Coca LF, Dal-Forno M, Dollhofer V, Fliegerová K, Greiner K, Griffith GW, Ho HM, Hofstetter V, Jeewon R, Kang JC, Wen TC, Kirk PM, Kytovuori I, Lawrey JD, Xing J, Li H, Liu ZY, Liu XZ, Liimatainen K, Thorsten Lumbsch H, Matsumura M, Moncada B, Nuankaew S, Parnmen S, Santiago ALCMDA, Sommai S, Song Y, de Souza CAF, de Souza- Motta CM, Su HY, Suetrong S, Wang Y, FongWS Yuan HS, Zhou LW, Réblová M, Fournier J, Camporesi E, Luangsa-ard JJ, Tasanathai K, Khonsanit A, Thanakitpipattana D, Somrithipol S, Diederich P, Millanes AM, Common RS, Stadler M, Yan JY, Li XH, Lee HW, Nguyen TTT, Lee HB, Battistin E, Marsico O, Vizzini A, Vila J, Ercole E, Eberhardt U, Simonini G, Wen HA, Chen XH, Miettinen O, Spirin V, Hernawati NV (2015) Fungal diversity notes 111–252 taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 75:27–274
Arnold A (2002) Neotropical fungal endophytes: diversity and ecology. PhD thesis, University of Arizona, Tucson
Arnold AE, Lutzoni F (2007) Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots? Ecology 88:541–549
Assante G, Dallavalle S, Malpezzi L, Nasini G, Burruano S, Torta L (2005) Acremines A-F, novel secondary metabolites produced by a strain of an endophytic Acremonium, isolated from sporangiophores of Plasmopara viticola in grapevine leaves. Tetrahedron 61:7686–7692
Berbee ML, Pirseyedi M, Hubbard S (1999) Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 91:964–977
Bhattacharyya LH, Borah G, Parkash V, Bhattacharyya PN (2017) Fungal endophytes associated with the ethnomedicinal plant Meyna Spinosa Roxb. Curr Life Sci 3:1–5
Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59
Bonfim JA, Vasconcellos RLF, Baldesin LF, Sieber TN, Cardoso EJBN (2016) Dark septate endophytic fungi of native plants along an altitudinal gradient in the Brazilian Atlantic forest. Fungal Ecol 20:202–210
Bruez E, Baumgartner K, Bastien S, Travadon R, Guerin-Dubrana L, Rey P (2016) Various fungal communities colonise the functional wood tissues of old grapevines externally free from grapevine trunk disease symptoms. Aust J Grape Wine Res 22:288–295
Brum MCP, Araújo WL, Maki CS, Azevedo JL (2012) Endophytic fungi from Vitis labrusca L. (‘Niagara Rosada’) and its potential for the biological control of Fusarium oxysporum. Genet Mol Res 11:4187–4197
Bulgari D, Casati P, Crepaldi P, Daffonchio D, Quaglino F (2011) Restructuring of endophytic bacterial communities in grapevine yellows-diseased and recovered Vitis vinifera L. plants. Appl Environ Microbiol 77:5018–5022
Bullington L, Larkin BG (2015) Using direct amplification and next-generation sequencing technology to explore foliar endophyte communities in experimentally inoculated western white pines. Fungal Ecol 17:170–178
Burruano S, Alfonzo A, Piccolo SL, Conigliaro G, Mondello V, Torta L, Moretti M, Assante G (2008) Interaction between Acremonium byssoides and Plasmopara viticola in Vitis vinifera. Phytopathol Mediter 47:122–131
Busby PE, Ridout M, Newcombe G (2016) Fungal endophytes: modifiers of plant disease. Plant Mol Biol 90:645–655
Cai L, Hyde KD, Taylor PWJ, Weir BS, Waller J, Abang MM, Zhang JZ, Yang YL, Phoulivong S, Liu ZY, Prihastuti H, Shivas RG, McKenzie EHC, Johnston PR (2009) A polyphasic approach for studying Colletotrichum. Fungal Divers 39:183–204
Campisano A (2012) Profiling of grapevine fungal endophytic community using Automated Ribosomal Intergenic Spacer Analysis (ARISA). Istituto Agario Di San Michele All Adige
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, González Peña A, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Carbone I, Kohn LM (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91:553–556
Chagnon PL, U’Ren JM, Miadlikowska J, Lutzoni F, Arnold AE (2016) Interaction type influences ecological network structure more than local abiotic conditions: evidence from endophytic and endolichenic fungi at a continental scale. Oecologia 180:181–191
China Agriculture Yearbook (2014) Ministry of Agriculture, PRC. China Agriculture Press. People’s Republic of China, Beijing
Christian NS, Sullivan C, Visser N, Clay K (2016) Plant host and geographic location drive endophyte community composition in the face of perturbation. Microb Ecol. https://doi.org/10.1007/s00248-016-0804-y
Clay K, Shearin ZRC, Bourke KA, Bickford WA, Kowalski KP (2016) Diversity of fungal endophytes in non-native Phragmites australis in the Great Lakes. Biol Invasions 18:2703–2716
Compant S, Birgit M, Colli-Mull J, Gangl H, Sessitsch A (2011) Endophytes of grapevine flowers, berries, and seeds: identification of cultivable bacteria, comparison with other plant parts, and visualization of niches of colonization. Microb Ecol 62:188–197
Crouch JA, Clarke BB, Hillman BI (2009) What is the value of ITS sequence data in Colletotrichum systematics and species diagnosis? A case study using the falcate-spored graminicolous Colletotrichum group. Mycologia 101:648–656
Dastogeer KMG, Li H, Sivasithamparam K, Jones MGK, Wylie SJ (2017) Host specificity of endophytic mycobiota of wild Nicotiana plants from arid regions of Northern Australia. Microb Ecol 75:74–87
David AS, Seabloom EW, May G (2017) Disentangling environmental and host sources of fungal endophyte communities in an experimental beachgrass study. Mol Ecol 26:21. https://doi.org/10.1111/mec.14354
de Felice DV, Solfrizzo M, De Curtis F, Lima G, Visconti A, Castoria R (2008) Strains of Aureobasidium pullulans can lower ochratoxin A contamination in wine grapes. Phytopathology 98:1261–1270
Deagle BE, Clarke LJ, Kitchener JA, Polanowski AM, Davidson AT (2017) Genetic monitoring of open ocean biodiversity: an evaluation of DNA metabarcoding for processing continuous plankton recorder samples. Mol Ecol Resour. https://doi.org/10.1111/1755-0998.12740
Donayre DKM, Dalisay TU, Bayot RG, Baltazar AM (2014) Diversity and tissue specificity of endophytic fungi in barnyard grass (Echinochloa glabrescens Munro ex Hook. f.). Asia Life Sci 23:725–741
Duong LM, Jeewon R, Lumyong S, Hyde KD (2006) DGGE coupled with ribosomal DNA genes phylogenies reveal uncharacterized fungal endophytes. Fungal Divers 23:121–138
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Elmer PAG, Reglinski T (2006) Bio-suppression of Botrytis cinerea in grapes. Plant Pathol 55:155–177
Fesel PH, Zuccaro A (2016) Dissecting endophytic lifestyle along the parasitism/mutualism continuum in Arabidopsis. Curr Opin Microbiol 32:103–112
Fuchs B, Krischke M, Mueller MJ, Krauss J (2017) Plant age and seasonal timing determine endophyte growth and alkaloid biosynthesis. Fungal Ecol 29:52–58
García E, Alonso A, Platas G, Sacristán S (2013) The endophytic mycobiota of Arabidopsis thaliana. Fungal Divers 60:71–89
Garnica S, Schön ME, Abarenkov K, Riess K, Liimatainen K, Niskanen T, Dima B, Soop K, Frøslev TG, Jeppesen TS, Peintner U, Kuhnert-Finkernagel R, Brandrud TE, Saar G, Oertel B, Ammirati JF (2016) Determining threshold values for barcoding fungi: lessons from Cortinarius (Basidiomycota), a highly diverse and widespread ectomycorrhizal genus. FEMS Microbiol Ecol. https://doi.org/10.1093/femsec/fiw045
Garoé NT, Cabrera R, Burgos-Reyes RL, Da Silva E, Giménez C, Cosoveanu A, Brito N (2012) Endophytic fungi from Vitis vinifera L. isolated in Canary Islands and Azores as potential biocontrol agents of Botrytis cinerea Pers.:Fr. J Horticult For Biotechnol 16:1–6
Ghimire SR, Charlton ND, Bell JD, Krishnamurthy YL, Craven KD (2011) Biodiversity of fungal endophyte communities inhabiting switchgrass (Panicum virgatum L.) growing in the native tallgrass prairie of northern Oklahoma. Fungal Divers 47:19–27
Glass NL, Donaldson GC (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 61:1323–1330
Gomez CA, Budvytiene I, Zemek AJ, Banaei N (2017) Performance of targeted fungal sequencing for culture-independent diagnosis of invasive fungal disease. Clin Infect Dis. https://doi.org/10.1093/cid/cix728
González V, Tello ML (2011) The endophytic mycota associated with Vitis vinifera in central Spain. Fungal Divers 47:29–42
Goveas SW, Madtha R, Nivas SK, D’Souza L (2011) Isolation of endophytic fungi from Coscinium fenestratum—a red listed endangered medicinal plant. Eurasia J Biosci 5:48–53
Green LE, Porras-Alfaro A, Sinsabaugh RL (2008) Translocation of nitrogen and carbon integrates biotic crust and grass production in desert grassland. J Ecol 96:1076–1085
Guo LD, Hyde KD, Liew ECY (2000) Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytol 147:617–630
Guo LD, Hyde KD, Liew ECY (2001) Detection and taxonomic placement of endophytic fungi within frond tissues of Livistona chinensis based on rDNA sequences. Mol Phylogenet Evol 20:1–13
Guo LD, Huang GR, Wang Y, He WH, Zheng WH, Hyde KD (2003) Molecular identification of white morphotype strains of endophytic fungi from Pinus tabulaeformis. Mycol Res 107:680–688
Gupta S, Chaturvedi P (2017) Foliar endophytic diversity of Centella asiatica (L.) urban in relation to different seasons and leaf age. Int J Curr Microbiol Appl Sci 6:468–477
Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, Petrosino JF, Knight R, Birren BW (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504
Haghighi MT, Shahdoust E (2015) Molecular detection of endophytic, Myrothecium spp. by ITS sequencing technique. Int J Res Stud Biosci 3:60–66
Hall T (2006) Bioedit. Department of Microbiology, North Carolina State University. http://www.mbioncsuedu/BioEdit/Bioedithtml
Hammer Q, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron 4:1–9
Hardoim PR, van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, Döring M, Sessitsch A (2015) The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev 79:293–320
Heinonsalo J, Buée M, Vaario LM (2016) Root-endophytic fungi cause morphological and functional differences in Scots pine roots in contrast to ectomycorrhizal fungi. Can J Bot 95:203–210
Hoppe B, Purahong W, Wubet T, Kahl T, Bauhus J, Arnstadt T, Hofrichter M, Buscot F, Krüger D (2016) Linking molecular deadwood-inhabiting fungal diversity and community dynamics to ecosystem functions and processes in Central European forests. Fungal Divers 77:367–379
Huang Y, Kuang Z, Wang W, Cao L (2016) Exploring potential bacterial and fungal biocontrol agents transmitted from seeds to sprouts of wheat. Biol Control 98:27–33
Hyde KD, Soytong K (2008) The fungal endophyte dilemma. Fungal Divers 33:163–173
Hyde KD, Nilsson RH, Alias SA, Ariyawansa HA, Blair JE, Cai L, de Cock WAM, Dissanayake AJ, Glocking SL, Goonasekara ID, Gorczak M, Hahn M, Jayawardena RS, van Kan JAL, Laurence H, Lévesque A, Li XH, Liu JK, Maharachchikumbura SSN, Manamgoda DS, Martin FN, McKenzie EHC, McTaggart AR, Mortimer PE, Nair PVR, Pawlowska J, Rintoul TL, Shivas RG, Spies ARCFJ, Summerell BA, Taylor PWJ, Terhem RB, Udayanga D, Vaghefi N, Walthe G, Wilk M, Wrzosek M, Xu JC, Yan J, Zhou N (2014) One stop shop: backbone trees for important phytopathogenic genera: I. Fungal Divers 67:21–125
Hyde KD, Hongsanan S, Jeewon R, Bhat DJ, McKenzie EHC, Jones EBG, Phookamsak R, Ariyawansa HA, Boonmee S, Zhao Q, Awad Abdel-Aziz FA, Abdel-Wahab MA, Banma S, Chomnunti P, Cui B, Daranagama DA, Das K, Dayarathn MC, de Silva NI, Dissanayake AJ, Doilom M, Ekanayaka AH, Gibertoni TB, Neto AG, Huang SK, Jayasiri SC, Jayawardena RS, Konta S, Lee HB, Li W, Lin C, Liu JK, Lu YZ, Luo ZL, Manawasinghe IS, Manimohan P, Mapook A, Niskanen T, Norphanphoun C, Papizadeh M, Perera RH, Phukhamsakda C, Richter C, Santiago ALCMA, Santos ERD, Senanayake IC, Tanaka K, Tennakoon TMDS, Thambugala KM, Tian Q, Tibpromma S, Thongbai B, Vizzini A, Wanasinghe DN, Wijayawardene NN, Wu H, Yang J, Zeng X, Zhang H, Zhang JF, Bulgakov TS, Camporesi E, Bahkali AH, Amoozegar MA, Neta LSA, Ammirati JF, Baghela A, Bhatt RP, Bojantchev D, Buyck B, Silva GA, Lima CLF, Oliveira RJV, Souza CAF, Dai YC, Dima B, Duong TT, Ercole E, Mafalda-Freire F, Ghosh A, Hashimoto A, Kamolhan S, Kang JC, Karunarathna SC, Kirk PM, Kytovuori I, Lantieri A, Liimatainen K, Liu ZY, Liu XH, Lucking R, Medardi G, Mortimer PE, Nguyen TTT, Promputtha I, Raj KNA, Reck MA, Lumyong S, Fazeli SAS, Stadler M, Soudi MR, Su HY, Takahashi T, Tangthirasunun N, Uniya P, Wang Y, Wen TC, Xu JC, Zhang Z, Zhao Y, Zhou JL, Zhu L (2016) Fungal diversity notes 367–491: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 80:1–270
Jayawardena RS, Purahong W, Zhang W, Wubet T, Li XH, Liu M, Zhao W, Hyde KD, Liu JH, Yan JY (2018) Biodiversity of fungi on Vitis vinifera L. revealed by traditional and high-resolution culture-independent approaches. Fungal Divers. https://doi.org/10.1007/s13225-018-0398-4
Jeewon R, Liew ECY, Simpson JA, Hodgkiss IJ, Hyde KD (2003) Phylogenetic significance of morphological characters in the taxonomy of Pestalotiopsis species. Mol Phylogenet Evol 27:372–383
Kaewkla O, Franco CMM (2016) Kribbella pittospori sp. nov., an endophytic actinobacterium isolated from the surface-sterilized stem of an Australian native apricot tree, Pittosporum angustifolium. Int J Syst Evol Microbiol 66:2284–2290
Katoh K, Toh H (2010) Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform 9:286–298
Ko TWK, Stephenson SL, Bahkali AH, Hyde KD (2011) From morphology to molecular biology: can we use sequence data to identify fungal endophytes? Fungal Divers 50:113–120
Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson KH (2013) Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5277
Kortekamp A (1997) Epicoccum nigrum link: a biological control, agent of Plasmopara viticola (Berk. et Curt.) Berl. et De Toni? Vitis 36:215–216
Kraková L, Šoltys Otlewska A, Pietrzak K, Purkrtová S, Savická D, Puškárová A, Bučková M, Szemes T, Budiš J, Demnerová K, Gutarowska B, Pangallo D (2017) Comparison of methods for identification of microbial communities in book collections: Culture-dependent (sequencing and MALDI-TOF MS) and culture-independent (Illumina MiSeq). Int Biodeterior Biodegrad. https://doi.org/10.1016/j.ibiod.2017.02.015
Lacap DC, Hyde KD, Liew ECY (2003) An evaluation of the fungal ‘morphotype’ concept based on ribosomal DNA sequences. Fungal Divers 12:53–66
Li WC, Zhou J, Guo SY, Guo LD (2007) Endophytic fungi associated with lichens in Baihua mountain of Beijing, China. Fungal Divers 25:69–80
Liu JK, Hyde KD, Jones EBG, Ariyawansa HA, Bhat DJ, Boonmee S, Maharachchikumbura SSN, McKenzie EHC, Phookamsak R, Phukhamsakda C, Shenoy BD, Abdel-Wahab MA, Buyck B, Chen J, Chethana KWT, Singtripop C, Dai DQ, Dai YC, Daranagama DA, Dissanayake AJ, Doilom M, Dsouza MJ, Fan XL, Goonasekara ID, Hirayama K, Hongsanan S, Jayasiri SC, Jayawardena RS, Karunarathna SC, Li WJ, Mapook A, Norphanphoun C, Pang KL, Perera RH, Persoh D, Pinruan U, Senanayake IC, Somrithipol S, Suetrong S, Tanaka K, Thambugala KM, Tian Q, Tibpromma S, Udayanga D, Wijayawardene NN, Wanasinghe DN, Wisitrassameewong K, Zeng XY, Abdel-Aziz FA, Adamcık S, Bahkali AH, Boonyuen N, Bulgakov T, Callac P, Chomnunti P, Greiner K, Hashimoto A, Hofstetter V, Kang JC, Lewis D, Li XH, Liu XZ, Liu ZY, Matsumura M, Mortimer PE, Rambold G, Randrianjohany E, Sato G, Sri-Indrasutdhi V, Tian CM, Verbeken A, von Brackel W, Wang Y, Wen TC, Xu JC, Yan JY, Zhao RL, Camporesi E (2015) Fungal Diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal Divers 72:1–197
Liu W, Zhou Z, Liu Y, Hu X, Guo Y, Li J (2017a) Application of high-throughput internal transcribed spacer rRNA metagenomics analysis in deciphering endophytic fungi diversity of Dendrobium officinale. J Biobased Mater Bioenergy 11:106–118
Liu T, Greenslade A, Yang S (2017b) Levels of rhizome endophytic fungi fluctuate in Paris polyphylla var. yunnanensis as plants age. Plant Divers 39:60–64
Lobo J, Shokralla S, Costa MH, Hajibabaei M, Costa FO (2017) DNA metabarcoding for high-throughput monitoring of estuarine macrobenthic communities. Sci Rep 7:15618
Lòpez-Fernàndez S, Mazzoni V, Pedrazzoli F, Pertot I, Campisano A (2017) A phloem-feeding insect transfers bacterial endophytic communities between grapevine plants. Front Microbiol 8:834
Lücking R, Moncada B (2017) Dismantling Marchandiomphalina into Agonimia (Verrucariaceae) and Lawreymyces gen. nov. (Corticiaceae): setting a precedent to the formal recognition of thousands of voucherless fungi based on type sequences. Fungal Divers 84:119–138
Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963
Mahmoud FM, Krimi Z, Maciá-Vicente JG, Errahmani MB, Lopez-Llorca LV (2017) Endophytic fungi associated with roots of date palm (Phoenix dactylifera) in coastal dunes. Revista Iberoamericana de Micología 34:116–120
Martín-García J, Espiga E, Pando V, Diez JJ (2011) Factors influencing endophytic communities in poplar plantations. Silva Fennica 45:169–180
Martini M, Musetti R, Grisan S, Polizzotto R (2009) DNA-dependent detection of the grapevine fungal endophytes Aureobasidium pullulans and Epicoccum nigrum. Plant Dis 93:993–998
McKinnon A (2016) Plant tissue preparation for the detection of an endophytic fungus in planta. Microbial Based Biopestic 1477:167–173
Mendoza LM, Neef A, Vignolo G, Belloch C (2017) Yeast diversity during the fermentation of Andean chicha: a comparison of high-throughput sequencing and culture-dependent approaches. Food Microbiol 67:1–10
Morgan HH, Toit M, Setati ME (2017) The grapevine and wine microbiome: insights from high-throughput amplicon sequencing. Front Microbiol 8:820
Muggia L, Kopun T, Grube M (2017) Effects of growth media on the diversity of culturable fungi from lichens. Molecules 22:824–846
Musetti R, Vecchione A, Stringher L, Borselli S, Zulini L, Marzani C, D’Ambrosio M, Sanità di Toppi L, Pertot I (2006) Inhibition of sporulation and ultrastructural alterations of grapevine downy mildew by the endophytic fungus Alternaria alternata. Phytopathology 96:689–698
Nascimento TL, Oki Y, Lima DMM, Almeida-Cortez JS, Fernandes GW, Souza-Motta CM (2015) Biodiversity of endophytic fungi in different leaf ages of Calotropis procera and their antimicrobial activity. Fungal Ecol 14:79–86
Navarro-Meléndez AL, Heil M (2014) Symptomless endophytic fungi suppress endogenous levels of salicylic acid and interact with the jasmonate-dependent indirect defense traits of their host, lima bean (Phaseolus lunatus). J Chem Ecol 40:816–825
Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG (2016) FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20:241–248
Nilsson RH, Kristiansson E, Ryberg M, Hallenberg N, Larsson K (2008) Intraspecific ITS variability in the kingdom fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol Bioinform 4:193–201
Nilsson RH, Tedersoo L, Abarenkov K, Ryberg M, Kristiansson E, Hartmann M, Schoch CL, Nylander JAA, Bergsten J, Porter TM, Jumpponen A, Vaishampayan P, Ovaskainen O, Hallenberg N, Bengtsson-Palme J, Eriksson KM, Larsson KH, Larsson E (2012) Five simple guidelines for establishing basic authenticity and reliability of newly generated fungal ITS sequences. MycoKeys 4:37–63
Nilsson RH, Tedersoo L, Ryberg M (2015) A comprehensive, automatically updated fungal ITS sequence dataset for reference-based chimera control in environmental sequencing efforts. Microbes Environ 30:145–150
Nylander JAA (2004) MrModeltest v2. Program distributed by the author Evolutionary Biology Centre. Uppsala University, Uppsala
Olejniczak P, Lembicz M (2007) Age-specific response of the grass Puccinellia distans to the presence of a fungal endophyte. Oecologia 152:485–494
Osono T, Mori A (2005) Seasonal and leaf age-dependent changes in occurrence of phyllosphere fungi in giant dogwood. Mycoscience 46:273–279
Ovaskainen O, Nokso-Koivisto J, Hottola J, Rajala T, Pennanen T, Ali-Kovero H, Miettinen O, Oinonen P, Auvinen P, Paulin L, Larsson KH, Mäkipää R (2010) Identifying wood-inhabiting fungi with 454 sequencing—what is the probability that BLAST gives the correct species? Fungal Ecol 3:274–283
Page RDM (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Park YH, Kim YC, Park SU, Lim HS, Kim JB, Cho BK, Bae H (2012) Age-dependent distribution of fungal endophytes in Panax ginseng roots cultivated in Korea. J Ginseng Res 36:327–333
Peršoh D (2015) Plant-associated fungal communities in the light of meta’omics. Fungal Divers 75:1–25
Pinto C, Pinho D, Sousa S, Pinheiro M, Egas C, Gomes AC (2014) Unravelling the diversity of grapevine microbiome. PLoS ONE 9:e85622
Promputtha I, Jeewon R, Lumyong S, McKenzie EHC, Hyde KD (2005) Ribosomal DNA fingerprinting in the identification of non sporulating endophytes from Magnolia liliifera (Magnoliaceae). Fungal Divers 20:167–186
Promputtha I, Lumyong S, Dhanasekaran V, McKenzie EHC, Hyde KD, Jeewon R (2007) A phylogenetic evaluation of whether endophytes become saprotrophs at host senescence. Microb Ecol 53:579–590
Promputtha I, Hyde KD, McKenzie EHC, Peberdy JF, Lumyong S (2010) Can leaf degrading enzymes provide evidence that endophytic fungi becoming saprobes? Fungal Divers 41:89–99
Purahong W, Hyde KD (2011) Effects of fungal endophytes on grass and non-grass litter decomposition rates. Fungal Divers 47:1–7
Purahong W, Wubet T, Lentendu G, Schloter M, Pecyna MJ, Kapturska D, Hofrichter M, Krüger D, Buscot F (2016) Life in leaf litter: novel insights into community dynamics of bacteria and fungi during litter decomposition. Mol Ecol 25:4059–4074
Purahong W, Pietsch KA, Lentendu G, Schöps R, Bruelheide H, Wirth C, Buscot F, Wubet T (2017a) Characterization of unexplored deadwood mycobiome in highly diverse subtropical forests using culture-independent molecular technique. Front Microbiol. https://doi.org/10.3389/fmicb.2017.00574
Purahong W, Wubet T, Krüger D, Buscot F (2018) Molecular evidence strongly supports deadwood inhabiting fungi exhibiting unexpected tree species preferences in temperate forests. ISME J 12:289–295
Rocha ACS, Garcia D, Uetanabaro APT, Carneiro RTO, Araujo IS, Mattos CRR, Goes-Neto A (2011) Foliar endophytic fungi from Hevea brasiliensis and their antagonism on Microcyclus ulei. Fungal Divers 47:75–84
Roe AD, Rice AV, Bromilow SE, Cooke JEK, Sperling FAH (2010) Multilocus species identification and fungal DNA barcoding: Insights from blue stain fungal symbionts of the mountain pine beetle. Mol Ecol Resour 10:946–959
Rondot Y, Reineke A (2016) Endophytic Beauveria bassiana in grapevine Vitis vinifera (L.) reduces infestation with piercing-sucking insects. Biol Control. https://doi.org/10.1016/j.biocontrol.2016
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Ruiz-Pérez CA, Zambrano MM (2017) Endophytic microbial community DNA extraction from the plant phyllosphere. Bio-protocol 7:1–5
Sánchez S, Bills GF, Domínguez Acuña L, Zabalgogeazcoa I (2010) Endophytic mycobiota of leaves and roots of the grass Holcus lanatus. Fungal Divers 41:115–123
Schena L, Ippolito A, Zahavi T, Cohen L, Nigro F, Droby S (1999) Genetic diversity and biocontrol activity of Aureobasidium pullulans isolates against postharvest rots. Postharvest Biol Technol 17:189–199
Schmid F, Moser G, Müller H, Berg G (2011) Functional and structural microbial diversity in organic and conventional viticulture: organic farming benefits natural biocontrol agents. Appl Environ Microbiol 77:2188–2191
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA 109:6241–6246
Setati ME, Jacobson D, Bauer FF (2015) Sequence-based analysis of the Vitis vinifera L. cv Cabernet Sauvignon grape must mycobiome in three South African vineyards employing distinct agronomic systems. Front Microbiol 6:1358
Staats M, van Baarlen P, van Kan JAL (2005) Molecular phylogeny of the plant pathogenic genus Botrytis and the evolution of host specificity. Mol Biol Evol 22:333–346
Steinrucken TV, Bissett A, Powell JR, Raghavendra AKH, van Klinken RD (2016) Endophyte community composition is associated with dieback occurrence in an invasive tree. Plant Soil 405:311–323
Stone JK, Polishook JD, White JF Jr (2004) Endophytic fungi. In: Mueller G, Foster M, Bills G (eds) Biodiversity of fungi. Inventory and monitoring methods. Academic, Amsterdam, p 728
Sung GH, Sung JM, Hywel-Jones NL, Spatafora JW (2007) A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): identification of localized incongruence using a combinational bootstrap approach. Mol Phylogenet Evol 44:1204–1223
Supaphon P, Phongpaichit S, Sakayaroj J, Rukachaisirikul V, Kobmoo N, Spatafora JW (2017) Phylogenetic community structure of fungal endophytes in seagrass species. Bot Mar. https://doi.org/10.1515/bot-2016-0089
Swofford DL (2003) PAUP*: phylogenetic analysis using parsimony and other methods, version 4. Sinauer Associates, Sunderland
Szink I, Davis EL, Ricks KD, Koide RT (2016) New evidence for broad trophic status of leaf endophytic fungi of Quercus gambelii. Fungal Ecol 22:2–9
Tan RX, Zou WX (2001) Endophytes: a rich source of functional metabolites. Nat Prod Rep 18:448–459
Taylor DL, Walters WA, Lennon NJ, Bochicchio J, Krohn A, Caporaso JG, Pennanen T (2016) Accurate estimation of fungal diversity and abundance through improved lineage-specific primers optimized for illumina amplicon sequencing. Appl Environ Microbiol 82:7217–7226
Tejesvi MV, Ruotsalainen AL, Markkola AM, Pirttilä AM (2010) Root endophytes along a primary succession gradient in northern Finland. Fungal Divers 41:125–134
Tejesvi MV, Kajula M, Mattila S, Pirttilä AM (2011) Bioactivity and genetic diversity of endophytic fungi in Rhododendron tomentosum Harmaja. Fungal Divers 47:97–107
Tejesvi MV, Picart P, Kajula M, Hautajärvi H, Ruddock L, Kristensen HH, Tossi A, Sahl HG, Ek S, Mattila S, Pirttilä AM (2016) Identification of antibacterial peptides from endophytic microbiome. Appl Microbiol Biotechnol 100:9283–9293
Ting AS, Akinsanya MA, Goh JK, Lim SP (2015) Metagenomics study of endophytic bacteria in Aloe vera using next-generation technology. Genomics Data 6:159–163
Travadon R, Lawrence DP, Rooney-Latham S, Gubler WD, Wilcox WF, Rolshausen PE, Baumgartner K (2015) Cadophora species associated with wood-decay of grapevine in North America. Fungal Biol 119:53–66
Varanda CMR, Oliveira M, Materatski P, Landum M, Clara MIE, Felix MR (2016) Fungal endophytic communities associated to the phyllosphere of grapevine cultivars under different types of management. Fungal Biol 120:1525–1536
Vilgalys R, Hester M (1990) Rapid genetic identification and maping of enzimatically amplified ribosomal DNA from several Crytococcus species. J Bacteriol 172:4238–4246
Vivas M, Kemler M, Slippers B (2015) Environmental maternal effects on the early phenotype and resistance Eucalyptus grandis and the structuring of fungal endophytic communities. In: 5th International Workshop on the Genetics of Host-Parasite Interactions in Forestry, At Orleans
White TJ, Bruns T, Lee J, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Wicaksono W, Jones EE, Monk J, Ridgway HJ (2015) Exploring factors that influence the composition of endophyte communities in Leptospermum scoparium (mānuka). In: 36th new phytologist symposium cell biology at the plant–microbe interface, At Munich, Germany
Yadav M, Yadav A, Kumar S, Yadav JP (2016) Spatial and seasonal influences on culturable endophytic mycobiota associated with different tissues of Eugenia jambolana Lam. and their antibacterial activity against MDR strains. BMC Microbiol 16:44–56
Zapka C, Leff J, Henley J, Tittl J, De Nardo E, Butler M, Griggs R, Fierer N, Edmonds-Wilson S (2017) Comparison of standard culture-based method to culture-independent method for evaluation of hygiene effects on the hand microbiome. mBio 8:e00093-17
Zhou SL, Yan SZ, Liu QS, Chen SL (2015) Diversity of endophytic fungi associated with the foliar tissue of a hemi-parasitic plant Macrosolen cochinchinensis. Curr Microbiol 70:58–66
Acknowledgements
This work was financially supported by Beijing Talent Program for Dr. Jiye Yan, CARS-29, Beijing science and technology project D17110001617002. We thank Dr. Heng Gui for his support to submit Raw Illumina reads to the Sequence Read Archive (SRA) of National Center for Biotechnology Information (NCBI).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dissanayake, A.J., Purahong, W., Wubet, T. et al. Direct comparison of culture-dependent and culture-independent molecular approaches reveal the diversity of fungal endophytic communities in stems of grapevine (Vitis vinifera). Fungal Diversity 90, 85–107 (2018). https://doi.org/10.1007/s13225-018-0399-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13225-018-0399-3