Abstract
Based on an unexpected result of obtaining molecular sequence data from tropical representatives of the genus Normandina, we revised the biological concept of the neotropical taxon Marchandiomphalina foliacea. The obtained data let us conclude that M. foliacea is not a basidiomycete, as originally proposed, but belongs in Verrucariaceae, in the genus Agonimia, including its perithecia which had been identified with the lichenicolous Norrlinia peltigericola. The ITS (and nuLSU) sequences previously obtained from M. foliacea, seemingly confirming its status as a basidiomycete, are from an unmanifested lichenicolous fungus, present also in numerous specimens of Normandina. ITS data suggest the presence of seven lineages that can be recognized at the species level, forming two clusters: one cluster of three lineages found in thalli of M. foliacea, and a second cluster of four lineages found in thalli of Normandina. This pattern is similar to what has recently been found in the basidiomycete genus Cyphobasidium occurring predominantly in Parmeliaceae lichens. We propose the combination of Omphalina foliacea into the genus Agonimia, as Agonimia foliacea (P.M. Jørg.) Lücking & Moncada, comb. nov., and place Marchandiomphalina in synonymy with Agonimia. To formally recognize the unnamed lichenicolous basidiomycete present in Agonimia and Normandina thalli, we take advantage of provision ICN Art. 40.5 in the Code and describe the unmanifested fungus as a new genus, with seven new species, even if no physical type specimens can be preserved (except for the corresponding host lichens which, however, do not show the features of the fungus): Lawreymyces Lücking & Moncada, gen. nov. (Type: L. palicei), with L. bogotensis Lücking & Moncada, sp. nov., L. columbiensis Lücking & Moncada, sp. nov., L. confusus Lücking & Moncada, sp. nov., L, foliaceae Lücking & Moncada, sp. nov., L. palicei Lücking & Moncada, sp. nov., L. pulchellae Lücking & Moncada, sp. nov., and L. spribillei Lücking & Moncada, sp. nov. This opens the door to the formal recognition of thousands of species of voucherless fungi detected through environmental sequencing techniques under the current Code.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nearly three decades ago, Jørgensen (1989) described the enigmatic lichen fungus Omphalina foliacea P.M. Jørg. from the northern Andes and Costa Rica. The sterile taxon was considered to represent a basidiolichen and provisionally placed in Omphalina. Simultaneously, Santesson (1989) reported three presumably lichenicolous fungi from this lichen, including the verrucarialean Norrlinia peltigericola (Nyl.) Theiss. & Syd. and two species in the genera Lichenopeltella (Trichothyriaceae) and Stigmidium (Mycosphaerellaceae). Based on ITS and nuLSU sequence data, Palice et al. (2005) concluded that O. foliacea was indeed a basidiomycete, but found it to be unrelated to other lichenized species of Omphalina, which in the meanwhile had been transferred to the genus Lichenomphalia (Redhead et al. 2002). Omphalina foliacea clustered instead within the so-called hymenochaetoid clade as defined by Binder et al. (2005). A broad phylogenetic analysis further resolved the phylogenetic position of this taxon in the order Corticiales, family Corticiaceae, related to genera such as the plant-parasitic, resupinate fungi Corticium and Erythricium and the lichenicolous Marchandiobasidium and Marchandiomyces (Lawrey et al. 2007, 2008). Consequently, a new genus, Marchandiomphalina, was erected for O. foliacea (Diederich and Lawrey 2007).
As part of an unrelated study about the performance of four ribosomal markers to reconstruct the phylogeny of the higher Ascomycota, Lumbsch et al. (2005) also analyzed sequences obtained from the presumed lichenicolous fungus Norrlinia peltigericola growing on Omphalina foliacea, from the same material from which the ITS and nuLSU sequences of the presumed host mycobiont had been obtained (Palice 4369 from Ecuador). They resolved N. peltigericola as closely related to Agonimia in the Verrucariaceae. The same taxon was again included in a study focusing on squamulose Verrucariaceae by Muggia et al. (2010), who showed the fungus to be nested within Agonimia, thus apparently representing the only lichenicolous taxon in a genus otherwise forming squamulose lichens.
In an attempt to assess species richness of lichenized fungi in the Colombian Andes using the ITS barcoding locus, we sampled material of numerous species, including Marchandiomphalina foliacea and its commonly encountered accompanying fungus, Norrlinia peltigericola. Our initial sequence data appeared to confirm previous findings by Palice et al. (2005) and Lumbsch et al. (2005), although morpho-anatomical comparison with the type of N. peltigericola from Peltigera canina in Finland suggested that the fungus growing on neotropical-montane thalli of M. foliacea was likely not conspecific with that species, something also observed by Santesson (1989). We were also puzzled by the striking similarities between the thallus of M. foliacea and that of various species of Agonimia, considering the Marchandiomphalina-Norrlinia association a prime example of a presumed lichenicolous fungus and its host mimicking a fertile lichen, similar to the case of Normandina and Lauderlindsaya (David and Hawksworth 1989; Diederich and Sérusiaux 1993; Mares et al. 1993; McCune 1997). In Normandina, it was eventually shown that the perithecia first attributed to a lichenicolous fungus actually belong to the lichen mycobiont (Aptroot 1991; Hoffmann and DePriest 2000), a fact now widely accepted (Muggia et al. 2010; Frisch and Ohmura 2015).
In addition to the ITS barcoding inventory in the Colombian Andes, in a separate project we started to assemble a broader data set for Normandina from a world-wide sample, initially with ITS and mtSSU sequence data from Colombia, Brazil, and Hawaii. To our surprise, while part of the samples produced the expected sequences, a larger number of samples from Colombia consistently yielded the correct mtSSU sequences matching Normandina, whereas the ITS sequences from the same samples fell within the Basidiomycota, in close vicinity of Marchandiomphalina foliacea. With this seemingly unexplainable result, we arrived again at the interface between two biological associations that seemed to have misguided lichenologists for a long time: the lichenized ascomycete Normandina, with its perithecia initially thought to be a lichenicolous fungus, and the lichenized basidiomycete Marchandiomphalina, apparently infected by an ascomycete in the genus Agonimia. With the above constellation and considering that Normandina and Marchandiomphalina are frequently found in the same andine habitats, this appeared to be a perfect setup for complications through mixups in molecular studies. However, we could eventually exclude any methodical error, and logical analysis of the data only allowed one conclusion: a surprising reinterpretation of the biological nature of Marchandiomphalina foliacea and its presumed verrucarialean companion which, after all, are one and the same species, forming a typical, squamulose, richly fruiting Agonimia, infected by an unmanifested basidiomycete that also associates with Normandina thalli and might be specific to Verrucariaceae lichens. In the light of recent findings by Spribille et al. (2016), who discovered basidiomycete yeasts in the genus Cyphobasidium to associate particularly with Parmeliaceae lichens, this might be another example of a highly specific basidiomycete-ascomycete association in lichens.
Materials and methods
A total of 31 new sequences were generated via Sanger sequencing, 12 for the mitochondrial small subunit rDNA (mtSSU) and 19 for the internal transcribed spacer (ITS), from two samples (four subsamples) of Marchandiomphalina foliacea and 13 samples (18 subsamples) of Normandina pulchella originating from Colombia, Brazil, and Hawaii. DNA extraction and sequencing was performed both in the Pritzker Laboratory for Molecular Systematics and Evolution at the Field Museum, Chicago, and in the Molecular Laboratory of the Botanical Garden and Botanical Museum (BGBM), Berlin (with sequencing at Macrogen, Inc., South Korea and Amsterdam). DNA was extracted using the QIAGEN DNeasy Plant Mini Kit and the SIGMA-ALDRICH REDExtract-N-Amp™ Tissue PCR Kit. Dilutions of 10:1 and 10:2 were used for PCR amplifications, with the primer pairs mrSSU1 and muSSU3R for the mtSSU (Zoller et al. 1999; Zhou and Stanosz 2001) for the mtSSU and ITS1F and ITS4 (Gardes and Bruns 1993; White et al. 1990) for the ITS. The 25 µl PCR reactions contained 2.5 µl buffer, 2.5 µl dNTP mix, 1 µl of each primer (10 µM), 5 µl BSA, 2 µl Taq, 2 µl genomic DNA extract and 9 µl distilled water. The thermal cycling parameters were set as follows: initial denaturation for 3–5 min at 95 °C, followed by 30–35 cycles of 1 min at 95 °C, 1 min at 52–54 °C, 1 min at 72 °C, and final elongation for 7 min at 72 °C. Amplification products were mounted on 1% agarose gels stained with ethidium bromide and either, after cutting of the target bands, purified using the QIAGEN QIAquick PCR Purification Kit (or the Nucleo Spin DNA Purification Kit; Macherey–Nagel), or the PCR product was directly cleaned with ExoSAP-IT PCR Product Cleanup (Affymetrix). For PCR done at the Field Museum, fragments were sequenced using the Big Dye Terminator reaction kit (ABI PRISM, Applied Biosystems). Sequencing and PCR amplifications were performed using the same sets of primers. Cycle sequencing was executed with the following setting: 25 cycles of 95 °C for 30 s, 48 °C for 15 s, 60 °C for 4 min. Sequenced products were precipitated with 10 µl of sterile dH2O, 2 µl of 3 M Napa, and 50 µl of 95% EtOH and subsequently loaded on an ABI 3100 (Applied Biosystems) automatic sequencer. For PCR done at the BGBM, sequences were generated by MACROGEN Inc. (Seoul, South Korea, and Amsterdam, The Netherlands). Sequences were checked with GenBank accessions using Nucleotide BLAST® (Altschul et al. 1990; Boratyn et al. 2013), assembled with DNASTAR SeqMan 4.03 (Swindell and Plasterer 1997) and GENEIOUS 8.1.0 (Kearse et al. 2012), manually inspected and adjusted and, after quality control within the context of multiple alignments for each locus, submitted to GenBank (Table 1).
The newly generated sequences were assembled in BIOEDIT 7.2.5 (Hall 1999, 2011) with mtSSU and ITS sequences downloaded GenBank and aligned with MAFFT 7.294 using the -auto option and the sort function (Katoh and Standley 2013). For the mtSSU from the Normandina samples, we initially downloaded all available sequences (298) of Verrucariaceae and, after sorting and inspection, narrowed down the sample to 33 sequences representing the genera Agonimia and Normandina, including one sequence from the perithecia on Marchandiomphalina identified as Norrlinia and another from a sterile thallus lobe of Marchandiomphalina (Table 1). After realignment using MAFFT, trimming, and manual inspection, the final alignment had a length of 850 bases (Suppl. File S1). For the ITS from the Normandina samples, we initially downloaded all available sequences (897) of Verrucariaceae and, after alignment, sorting, and inspection, narrowed down the data set to 191 sequences including all available genera and species within the family. The full alignment had a length of 764 bases (Suppl. File S2); because of alignment ambiguity of the ITS across all Verrucariaceae, we subjected the data set to analysis of ambiguously aligned regions using the GUIDANCE webserver (Penn et al. 2010a, b) and retained two subsets with columns having alignment confidence scores of 70% and higher (466 bases long; Suppl. File S3) and 90% and higher (350 bases long; Suppl. File S4). After phylogenetic analysis of the subsets, the seven Normandina ITS sequences were retained with Hydropunctaria maura as outgroup for a full alignment of 580 bases (Suppl. File S5). For the ITS sequences corresponding to Marchandiomphalina, we added two sequences from GenBank of that genus and three sequences of Marchandiobasidium and Erythricium as outgroup following Lawrey et al. (2007), for a data set of 17 sequences and an alignment length of 807 bases (Suppl. File S6).
All data sets were subjected to maximum likelihood search using RAxML 8.2.8 either locally or on the CIPRES Science Gateway server (Miller et al. 2010; Stamatakis 2015), with non-parametric bootstrapping using between 500 and 1000 replicates under the GTRGAMMA model.
Results and discussion
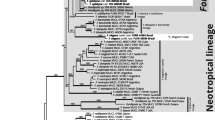
We obtained ITS and mtSSU sequence data from 13 specimens (18 subsamples) of Normandina from Colombia, Brazil, and Hawaii. All mtSSU sequences blasted with Verrucariaceae and previously deposited sequences of Normandina. Phylogenetic and phenotype analysis suggested these specimens to represent at least four different species, three of them undescribed (Fig. 1); these are being treated in a parallel paper (Moncada and Lücking, in prep.).
Best-scoring maximum likelihood tree of mtSSU sequence data for the genera Agonimia and Normandina in Verrucariaceae, including sterile thallus lobes of Marchandiomphalina foliacea (MON5417) and perithecia from thalli of Marchandiomphalina foliacea (AY300896) previously identified as Norrlinia peltigericola. Bootstrap support values of 70% and higher are indicated below the branches. Letters denote lineages corresponding to the same specimens used in Figs. 2 and 3; specimens from which disparate mtSSU (Verrucariaceae) and ITS sequences (Marchandiomphalina) were obtained are underlined
The ITS sequences obtained from the Normandina samples formed two distinct entities. One entity of seven sequences blasted consistently with ITS sequences of Verrucariaceae in GenBank; phylogenetic analysis indicated four distinct lineages (Suppl. Fig. S7, Fig. 2). The other entity of eight sequences blasted with Marchandiomphalina foliacea and in phylogenetic analysis formed a strongly supported cluster with the latter, sister to a clade formed by the genera Marchandiobasidium and Erythricium in Corticiaceae (Fig. 3; see also Lawrey et al. 2007). These ITS sequences exhibited highly structured variation, forming seven lineages in two unsupported clusters, one corresponding to M. foliacea samples (Fig. 3A–C) and the other to Normandina samples (Fig. 3D–G); there also was a strong correlation of the latter with the Normandina lineages obtained from mtSSU and ITS data (Figs. 1, 2, 3).
Best-scoring maximum likelihood tree of ITS sequence data for the genus Normandina. Bootstrap support values of 70% and higher are indicated below the branches. Letters denote lineages with specimens corresponding to the same specimens used in Fig. 1
Best-scoring maximum likelihood tree of ITS sequence data for Marchandiomphalina foliacea, including sequences obtained from several specimens of Normandina. Bootstrap support values of 70% and higher are indicated below the branches. Letters denote lineages with specimens corresponding to the same specimens used in Fig. 1; specimens from which disparate mtSSU (target ascomycete mycobiont) and ITS sequences (lichenicolous basidiomycete) were obtained are underlined
The mtSSU sequences from perithecia on the thallus of Marchandiomphalina collected in Ecuador, originally identified as Norrlinia peltigericola, and from sterile thallus lobes collected in Colombia, were identical and clustered within Agonimia (Fig. 1A; see also Muggia et al. 2010). In contrast, the ITS sequences from the thallus of the same samples (Ecuador: Palice 4369; Colombia: Lücking 33368c) were identified as Marchandiomphalina, although different from each other, and clustered with those from other thalli from the same areas (Ecuador: Palice 2509; Colombia: Lücking 33368a, b; Fig. 3: clades A, B), in a pattern very similar to what we found in a larger part of the Normandina samples. This is particularly striking considering that, in one sample of Marchandiomphalina and several of Normandina, the extracted DNA came from phenotypically homogeneous, sterile thallus lobes.
The finding that the same DNA extract amplified one marker correctly for the target mycobiont and another marker for a different fungus is not rare and rather commonly observed in lichens, the reason for this being differential affinities of the primers to the primer binding sites, either due to slight base variations or to the presence of introns at the binding sites. Often, a marker is amplified for more than one fungus and then generates mixed sequence signal. For instance, we obtained a distinctly mixed signal for a sample of ITS from a thallus of Marchandiomphalina foliacea (MON0522; Colombia, Moncada 4895) that was not used in our analysis due to signal ambiguity but nicely illustrates the presence of two distinct fungi, in which one dominated the signal and gave a non-ambiguous blast result (Fig. 4). Very likely, this effect depends on the primer pairs used, in this case with mrSSU1 and mrSSU3R for the mtSSU and ITS1F and ITS4 for the ITS. We did not test other primer pairs but we predict that with alternative or specifically designed primers, it should be possible to obtain specific results. Remarkably, in all specimens of Normandina studied, we obtained clean ITS, which either belonged to the Normandina ascomycete or the Marchandiomphalina basidiomycete. There was no case of a mixed signal as in the single case of Marchandiompalina illustrated above. Also, mtSSU consistently yielded an ascomycete in all samples.
Example of mixed ITS sequence data obtained from a sample of Marchandiomphalina foliacea not used in the present analysis (MON0522; Colombia, Moncada 4895), including unambiguous blast results with Omphalina foliacea ITS sequences accessioned in GenBank (corresponding to samples Palice 2509 and 4369)
We considered various hypotheses to explain this unexpected result. The idea of an unrecognized species of Marchandiomphalina resembling a Normandina could be immediately rejected, since in those specimens of Normandina in which the ITS clustered with Marchandiomphalina, the mtSSU consistently clustered with Normandina, demonstrating that we had genuine Normandina at hand; also, this hypothesis would not have explained the presence of Agonimia mtSSU in sterile thallus lobes of Marchandiomphalina. The only logical explanation was a contaminant situation. With the current biological concept of Marchandiomphalina and Normandina both being lichen formers, this would have implied that the Normandina samples yielding the aberrant ITS were contaminated with fragments of Marchandiomphalina thalli, and the sterile Marchandiomphalina lobes with non-discernable hyphae of Agonimia. Both Marchandiomphalina and Normandina form soredia or goniocysts, and therefore accidental cross-contamination upon study seemed not unlikely. However, three observations contradicted such an assumption. First, very limited, visibly homogeneous material for DNA extraction was taken from carefully treated samples, after cleaning and removing any alien material, if present, from the thalli; it was therefore unlikely that fragments of other lichens consistently contaminated the extracts. Second, accidental contamination with other lichen fungi should exhibit a stochastic pattern instead of resulting in sequences of the same genus of an alien lichen fungus in all contaminated samples, especially since the sampled biota are very diverse but no Marchandiomphalina thalli were present in any of the Normandina samples and viceversa (all collected Normandina specimens were epiphytic and those of Marchandiomphalina terrestrial). Third, although all aberrant ITS sequences in the Normandina samples formed a strongly supported clade with Marchandiomphalina, they formed distinct lineages strongly correlated with the Normandina lineages based on mtSSU sequences. Such a specific pattern would be impossible to obtain if the contaminant was a lichen former with no specific relationship to the contaminated samples.
Since undoubtedly there was a contaminant situation, but the contaminant could not have been accidental or a lichen former, the alternative explanation for the observed pattern was for the contaminant to be a specific, lichenicolous fungus. Following the idea that Marchandiomphalina foliacea is a lichen former, one could envision a scenario where one and the same genus, Marchandiomphalina, or even one and the same species, M. foliacea, was able to form either a lichen or alternatively invade other lichens as lichenicolous fungus. In various lineages, such as Arthoniales, Lecanorales, Ostropales, and Teloschistales, lichen formers and lichenicolous fungi are closely related and may occur in the same genus or even the same species (Rambold and Triebel 1992; Lawrey and Diederich 2003, 2016), such as in Arthonia, Carbonea, Gyalideopsis, Lecanora, Opegrapha, Rhizocarpon, Tephromela, and Toninia. Certain species grow as parasites on other lichens when young and later form autonomous thalli; the best-known example for this peculiar biology is Diploschistes muscorum (Skop.) R. Sant. (Graphidaceae), but this phenomenon is also found in the genera Placopyrenium (Verrucariaceae) and Protoparmelia (Parmeliaceae). Facultatively lichenicolous growth in fungi that are otherwise lichen formers has been reported from several Diploschistes species, as well as from Piccolia nannaria (Tuck.) Lendemer & Beeching (unknown affinity), Placopyrenium cinereoatratum (Degel.) Orange (Verrucariaceae), and Rinodina santorinensis J. Steiner (Physciaceae). However, in all these cases, the involved lichen fungi form crustose to at best microsquamulose thalli and are never foliose as in Marchandiomphalina. An interesting case is the family Parmeliaceae, which contains almost exclusively foliose to fruticose macrolichens, but also the three lichenicolous genera Nesolechia, Phacopsis, and Raesenenia (Crespo et al. 2010; Divakar et al. 2015). All three are closely related to macrolichen fungi, namely Punctelia, Relicina, and Protousnea, although they are not nested within those genera and there is no known case in Parmeliaceae where a macrolichen genus (or species) would include both lichenized and lichenicolous lineages or forms (the only such cases are found in the crustose genus Protoparmelia). A frequently reported case of a macrolichen presumably invading another lichen is that of Xanthoria parietina forming apothecia on thalli of Physcia tenella (Ott 1987), although similar cases were found to represent pigment-poor, grey Xanthoria polycarpa growing intermingled with Physcia spp. (Honegger et al. 1996); however, this is not considered a lichenicolous growth but accidental, mechanical hybridization (Kirk et al. 2008).
In cases of facultatively lichenicolous growth at genus or species level, the propagules causing the infection are invariably of fungal nature, usually ascospores. Marchandiomphalina foliacea, besides being a foliose lichen (Fig. 5A), is presumably not known to produce fungal spores but instead disperses with goniocysts (Fig. 5B); hence, a strategy to invade other lichen thalli to achieve a symbiotic association with the host photobiont would not make sense. On the other hand, since Normandina also produces similar vegetative propagules (soredia), the possibility that both lichen formers may form mechanical hybrids deserves attention. However, both lichens were reported to have different photobionts, Coccomyxa in Marchandiomphalina (Jørgensen 1989) and Diplosphaera in Normandina (Thüs et al. 2011), the latter originally identified as Nannochloris (Tscherrnak-Woess 1981). Different photobionts would render mechanical hybridization unlikely, although the photobiont in Marchandiomphalina has not been studied with molecular methods and hence its precise identify remains unresolved. Mechanical hybridization would also be expected to occur stochastically between species, but a stochastical pattern is contradicted by the highly structured variation found in the ITS data, with three lineages obtained from Marchandiomphalina samples and four from Normandina samples, forming two clades corresponding to either Marchandiomphalina or Normandina thalli. Such a pattern, on the other hand, is consistent with the hypothesis that the basidiomycetous ITS sequences represent lichenicolous fungi, in which case the Marchandiomphalina thallus would not be what it originally seemed based on anatomical and molecular analysis.
Comparative morphology of Marchandiomphalina (≡ Agonimia) foliacea and other species of Agonimia. A–J Marchandiomphalina (≡ Agonimia) foliacea; general habit (A, C, D), underside with goniocysts (B), close up of individual lobes (F–J), disposition of perithecia (C–I), and presence of the lichenicolous fungus Stigmidium joergensenii (I, J) (A Colombia, Lücking & Moncada 33368; B–F Colombia, Lücking & Moncada 34080; G–I Ecuador, Palice 4369; J Colombia, Cleef 9596). K A. globulifera, thallus with perithecia (France, Diederich 14478). L A. tristicula, thallus with perithecia (Greece, Sipman & Raus 55732). All photographs by the authors except K, kindly provided by P. Diederich
The rationale for considering Omphalina foliacea a basidiomycete were the supposedly thick hyphae associated with the algal cells forming the goniocysts on the underside, reported to be up to 7 µm thick (Jørgensen 1989). We examined several specimens of O. foliacea from Costa Rica and Colombia and found the hyphae to be 3–5 µm thick. We also studied material of the type species of Agonimia, A. tristicula (Nyl.) Zahlbr., for comparison. Both M. foliacea and A. tristicula have a similar body plan, with a squamulose thallus forming an upper, paraplectenchymatous cortex, a thick photobiont layer with several layers of algal cells, and a lower layer producing irregular to rounded goniocysts but lacking a cortex. Apart from the obvious differences in size, with 0.1–0.5 mm wide squamules in A. tristicula and 1–5 mm wide lobes in M. foliacea, anatomically there are indeed size differences in the cortical cells (7–15 µm diam. with 2–3 µm thick walls in M. foliacea versus 4–6 µm diam. with 0.5–1 µm thick walls in A. tristicula) and in the hyphae associated with the goniocysts (3–5 µm thick in M. foliacea and 2–3 µm in A. tristicula). However, such differences in cell size are known from other lichenized Ascomycota, for instance with regard to excipular cells in the genus Coenogonium (Rivas Plata et al. 2006), and generally there is great variation in hyphal thickness in both the Ascomycota and Basidiomycota (Maheshwari 2016), so this parameter cannot be used to diagnostically differentiate between the two phyla. The characteristic papillae on the cortical cells of A. tristicula (Coppins and Bennell 1979) were not observed in the material of Marchandiomphalina foliacea, but these are not present in all species of Agonimia (Orange and Purvis 2009).
While Santesson (1989) reported some thalli of M. foliacea to carry the lichenicolous fungi Lichenopeltella minuta and Stigmidium joergensenii (Fig. 5I–J), all thalli had perithecia presumably of the lichenicolous fungus Norrlinia peltigericola, although molecular data showed these perithecia to represent a species of Agonimia (Muggia et al. 2010), not conspecific with N. peltigericola, supporting the phenotypical differences already observed by Santesson (1989). In all collections seen by us except one, these perithecia were regularly present (Fig. 5C–I). Compared with specimens of A. tristicola and A. globulifera M. Brand & Diederich, the disposition of the perithecia on the thalli of M. foliacea agree well with that of the perithecia in the Agonimia species (Fig. 5K–L). In addition, sterile thallus lobes of M. foliacea collected in Colombia yielded mtSSU sequences identical to those found in the perithecia of material from Ecuador. We therefore conclude that the thallus of M. foliacea is not formed by a basidiomycete but belongs to the same fungus as the commonly encountered perithecia, representing a previously unrecognized Agonimia (not conspecific with the actual lichenicolous fungus Norrlinia peltigericola growing on Peltigera). Coincidentally, the specimen from Ecuador from which both the thallus and perithecial sequences were first generated (Palice 4369) was initially identified as a potentially new species of Agonimia, labeled “macrophylla”. This interpretation is also consistent with the fact that Corticiaceae contain numerous lichenicolous lineages but otherwise no lichen-formers (Diederich and Lawrey 2007; Lawrey et al. 2007, 2008).
Nomenclatural consequences
Our results pose an interesting case as to the correct use of the names previously applied to the species and genera involved. With our new biological interpretation of Marchandiomphalina foliacea representing a species of Agonimia, the original material of Omphalina foliacea P.M. Jørg. from Venezuela (Jørgensen 7545) presumably contains four different fungi: the fungus forming the thallus (Jørgensen 7545) and the regular perithecia previously identified as Norrlinia peltigericola (Jørgensen 7545b), both belonging to Agonimia, two lichenicolous fungi manifested by their fruiting bodies, namely Lichenopeltella minuta (Jørgensen 7545c) and Stigmidium joergensenii (Jørgensen 7545d; Santesson 1989), and presumably an unmanifested basidiomycete which was not specifically found in this material but is present in other sequenced specimens from Colombia and Ecuador. Initially, the thallus mycobiont was described under the name O. foliacea (Jørgensen 1989) and the perithecia formed in the same material were simultaneously identified with the lichencolous fungus Norrlinia peltigericola (Nyl.) Theiss. & Syd. (Santesson 1989). Since there is no reason to assume that any discernable hyphae belong to a basidiomycete, the structures described in the original material have three names available: Omphalina foliacea, Lichenopeltella minuta, and Stigmidium joergensenii; the fourth name associated with the material, Norrlinia peltigericola, is based on an entirely different specimen unrelated to the taxa in question and represents a misidentification.
ICN Art. 9.14 states: “When a type (herbarium sheet or equivalent preparation) contains parts belonging to more than one taxon (see Art. 9.11), the name must remain attached to the part (specimen as defined in Art. 8.2) that corresponds most nearly with the original description or diagnosis.” Therefore, since all described morphological and anatomical thallus features in the type material are linked to the name Omphalina foliacea, that name cannot be used for the phenotypically unmanifested basidiomycete. Instead, the name applies to the unnamed Agonimia, including both the thallus mycobiont and the perithecia incorrectly identified as Norrlinia peltigericola, and the new combination A. foliacea is required for this lichen fungus. As a consequence, the name Marchandiomphalina, which is based on the name O. foliacea and ultimately on its type, becomes a synonym of Agonimia in the Verrucariaceae.
Agonimia Zahlbr., Öst. Bot. Z. 59: 350 (1909).
Index Fungorum Number: IF 86.
Type: Agonimia tristicula (Nyl.) Zahlbr., Öst. Bot. Z. 5: 351 (1909; lectotype fide Clements and Shear 1931, p. 289).
Index Fungorum Number: IF 121594.
Synonym nov.: Marchandiomphalina Diederich, Manfr. Binder & Lawrey in Diederich & Lawrey, Mycol. Progr. 6: 73 (2007).
Index Fungorum Number: IF 29171.
Type: Marchandiomphalina foliacea (P.M. Jørg.) Diederich, Manfr. Binder & Lawrey in Diederich & Lawrey, Mycol. Progr. 6(2): 73 (2007; holotype).
Index Fungorum Number: IF 529037.
Agonimia foliacea (P.M. Jørg.) Lücking & Moncada, comb. nov.
Index Fungorum Number: IF 553227.
Basionym: Omphalina foliacea P.M. Jørg., Nordic J. Bot. 9: 89 (1989). Type: VENEZUELA. Mérida: Sierra Nevada de Santo Domingo, towards Laguna Negra; 3500 m; terrestial over bryophytes; 9 January 1979, P. Jørgensen 7545 (BG holotype; MERF, US! isotypes).
Index Fungorum Number: IF 136715.
Description. Thallus (Fig. 5A–J) terrestrial, usually between bryophytes, rarely epiphytic at the base of trees or shrubs, macrosquamulose to minutely foliose, orbicular to irregularly shaped, up to 3 cm diam., bright green when fresh, becoming green-grey to olive or brown-grey in the herbarium. Lobes orbicular to flabellate, becoming crenulate to irregularly lacerate, 1–5 mm wide, with the margins often downturned. Upper surface smooth to uneven; lower surface (Fig. 5B) appearing arachnoid to pruinose, bearing numerous goniocysts, light green to whitish. Thallus in Sect. 150–230 µm thick, with 30–50 µm thick, paraplectenchymatous cortex of 7–15 × 5–10 µm large, perpendicularly oriented cells with thick walls (2–3 µm), 30–60 µm thick photobiont layer composed of several layers of algal cells, and a lower layer of up to 100 µm wide, rounded to irregular goniocysts associated with 3–5 µm thick hyphae. Ascomata (Fig. 5C–I) perithecia, sessile, subglobose to ovoid, 0.08–0.13 mm diam., 0.01–0.15 mm high, brown-black to black; wall more or less paraplectenchymatous, 25–35 µm thick, outer parts dark brown, inner parts light brown to hyaline. Basal paraphyses absent but ostiolar area with numerous ostiolar paraphyses, up to 20 × 2.5 µm large; asci formed in bundles at the base of the cavity, broadly clavate, 80–120 × 35–45 µm, I–, KI + blue. Ascospores 2 per ascus, muriform, ellipsoid, 50–70 × 17–25 µm, hyaline. Secondary chemistry: no substances detected by TLC.
Notes. Agonimia foliacea differs from all other species of the genus by the size of the thallus and lobes. Two-spored asci are known, for instance, from A. opuntiella (Buschardt & Poelt) Vězda and A. tristicula, but apart from the much smaller lobes, A. opuntiella differs by the hairy proliferations and A. tristicula has much larger ascospores (Breuss 2002; Orange and Purvis 2009; Hafellner 2014). Another species with 2-spored asci, A. vouauxii (B. de Lesd.) M. Brand & Diederich, is microsquamulose to granulose (Sérusiaux et al. 1999).
Agonimia foliacea is a terrestrial species occasionally growing on the bases of shrubs and trees between bryophytes. It is known from Costa Rica and the northern Andes [Colombia, Venezuela, Ecuador; see below, additional specimens cited in Jørgensen 1989)], occurring between 3200 and 4250 m altitude in wet páramos.
Hafellner (2014) reports facultatively lichenicolous growth for Agonimia opuntiella and A. tristicula, which at first glance might trigger another look at the interpretation of the biology of the lichen fungi involved with A. foliacea. However, in these cases, the actual lichens grow on other lichens, often accidentally so, forming their own, autonomous thallus. Such lichenicolous lichens, which have been reported from many other genera (Lawrey and Diederich 2016), are biologically very different from lichenicolous fungi associating with the photobiont of the host lichen. Thus far, we have not seen a single collection in which thalli of A. foliacea would be associated with Normandina thalli or growing on each other, although Normandina is frequently found accidentally growing on other lichens.
Material examined: COSTA RICA. San José: Reserva Río Macho, Cerro Sábila; 09°35′N, 83°45′W, 3400 m; saxicolous over mosses; 24 November 1999, H. Sipman & L. Umaña 46483 (B). COLOMBIA. Cundinamarca: Páramo de Guasca; terrestrial; 18 August 2011, Lücking & Moncada 33368a, b, c (B, UDBC). Carretera Páramo de Palacio a Río Chuza; terrestrial; 1 September 1972, A. Cleef 5387 (B, COL). Páramo de Chingaza; 9 November 2011, Lücking & Moncada 34080 (B, UDBC). Páramo de Palacio, Lagunas de Buitrago; 3555 m; terrestrial; 27 April 1973, A. Cleef 9596 (B, COL). Páramo de Sumapaz, Chisacá; 3700 m; terrestrial; 11 May 1972, A. Cleef 3582 (B, COL). Guasca, Parque Nacional Natural Chingaza; 04°46′N, 74°41′W, 3200 m; terrestrial; 15 October 1988, H. Sipman & J. Aguirre 27339 (B). Meta: Páramo de Sumapaz, Cerro Nevado del Sumapaz; 4250 m; terrestrial; 11 January 1973, A. Cleef 7634b (B, COL). Nariño: Pasto, Corregimiento de La Laguna, Alto Zapallurco; 3250 m; epiphytic on trunk base; 25 July 1997, B. Ramírez & D. Salas 10974 (B). Risaralda: Santa Rosa de Cabal, 1 km NE of Finca La Sierra; 3750 m; terrestrial; 17 September 1984, J. Aguirre & H. Sipman 5500 (B, COL). ECUADOR. Carchi: Volcán Chiles, E-ESE of Laguna Verde; 00°48′N, 77°55′W, 3950 m; terrestrial (on soil over rocks); 29 September 2000, Z. Palice & Z. Soldán 4369 (B).
Material of Agonimia tristicula examined for comparison: GREECE. Dodecanese, Karpathos Island, summit area of Mt. Kali Limni; 35°36′N, 27°07′E, 1100 m; between mosses on branches of Crataegus; 17 September 2007, H. Sipman & T. Raus 55732 (B).
This leaves the basidiomycete fungi present in the thalli of Agonomia foliacea and in Normandina species without a name, a case quite similar to the undescribed, yet phylogenetically well-delimited species of Cyphobasidium present mostly in Parmeliaceae (Spribille et al. 2016). Presumably, the Code does not allow the formal description of species for which no tangible type specimen is available. Sequencing phenotypically unmanifested contaminants with Sanger technology is frequent; however, the bulk of new fungal taxa is discovered through environmental high throughput sequencing (HTS), with a staggering amount of information (Kodama et al. 2012). Currently, the Sequence Read Archive (SRA; Leinonen et al. 2011), using the query “(fungal OR Fungi) AND (internal transcribed spacer OR ITS1 OR ITS2)”, returns 179 studies, 1822 biosamples (= environmental samples), and 14,334 experiments (= HTS runs), containing 928 million fungal ITS reads, with an average length of 353 bases (SRA: https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?view=search_obj; accessed 12 March 2017). In contrast, using the same query, GenBank (Benson et al. 2013) returns a total 993,987 sequences obtained predominantly through Sanger sequencing (GenBank: https://www.ncbi.nlm.nih.gov/genbank; accessed 12 March 2017). Thus, at present there are almost 1000 times more HTS reads than Sanger sequences for the fungal barcoding marker. Only three years ago, this ratio amounted to 18:1 (Lücking 2014), and so it is expected to further grow exponentially within the next few years. Very clearly, traditional taxonomy and nomenclature is unfit to properly catalogue the enormous number of unrecognized taxa detected in these data and, regardless of any reservations one might have to formally describe fungi based on DNA sequence data only, a solution is urgently needed and cannot be postponed for another six years until the next IBC nomenclature section, a time when the ratio of HTS to Sanger sequences might have grown to 2.5 million(!) to one.
This problem has been amply discussed (Hawksworth et al. 2011; Hibbett et al. 2011; Lücking 2014; Redhead et al. 2014; Hibbett 2016; de Beer et al. 2016), and various suggestions have been put forward to name such taxa informally or even formally (Taylor 2011; Hawksworth et al. 2011; Hibbett et al. 2011). The recently established, yet invalid species Hawksworthiomyces sequentia Z.W. de Beer, T.A. Duong, M.J. Wingf. sp. nov. ENAS (de Beer et al. 2016), with the suffix ENAS (Environmental Nucleic Acid Sequence), denotes a species based on a sequence as type. A formal proposal was made to allow sequence data as types for such ecologically cryptic fungi (Hawksworth et al. 2016). Unfortunately, the Nomenclature Committee for Fungi does not (yet) support this proposal (Turland and Wiersema 2017), and the Rapporteurs expressed the concern about a presumed “… lack of control as to the type sequence being an informative sequence. Many taxa could have the same sequence.” (Turland and Wiersema 2017: 225). In our opinion this argument does not apply. First, an ample body of literature has shown that sequence data, while not without problems, as a whole by far outperform phenotype features in the correct delimitation of natural taxa, as shown by much improved fungal classifications including lichen fungi (e.g., Hibbett et al. 2007; Jaklitsch et al. 2016; Lücking et al. 2017; and works cited therein). Hence, reservations one could have about the usefulness of sequence data would apply even more to any other feature that could be diagnosed in a physical type specimen. Second, the statement that many taxa could have the same sequence is ill-defined, as it is precisely the sequence data that are used to delimit the taxa. There are only three possibilities that two entities could have the same sequence: they belong to the same taxon, which is analogous to a specimen belonging to the same taxon as its type (which would make a sequence a perfect type since the identity can be easily and objectively checked); they represent a conserved marker in which case that marker would not be used to delimit taxa in the first place; or they arose through homoplasy. Notably, in fungi, homoplasy has been shown to be a massive issue with non-molecular features of physical type specimens (and yet, physical specimens are allowed as types), whereas homoplasy in sequence data, to the point that sequences of unrelated taxa would be identical or highly similar, is statistically highly unlikely, unless one deals with horizontal gene transfer or with an evolving species complex with incomplete lineage sorting (e.g., Maddison and Knowles 2006; Stewart et al. 2014). In such cases, the potential impact of wrongly defined taxa would be neglectable compared to the impact phenotype homoplasy has had throughout history on wrong classifications of fungi at any given level from kingdom down to species. Likewise, other potential problems such as inadvertently describing artifactual sequences (low quality reads, chimaeras), would be few compared to the benefits of being able to formally describe possibly hundreds of thousands of good fungal species that would otherwise for sure be left undescribed, an untenable prospect.
Admittingly, there is one serious practical problem with a formal nomenclature based on single-marker sequence reads, as obtained through HTS technologies, and that is the fact that phylogenies based on separate markers cannot yet be consolidated. Therefore, such an approach must be restricted to a single marker, and the ITS barcoding locus (Schoch et al. 2012) is the obvious choice, despite some shortcomings. It is usually overlooked in this context that the aim of formal environmental sequence nomenclature cannot be to accurately resolve difficult species complexes, but to put names on hundreds of thousands of species of fungi that would otherwise be left undescribed. It does not matter whether a small part of these then represent improperly defined species complexes or infraspecific lineages. Allowing more than one marker, as suggested by de Beer et al. (2016), would not be feasible, due to the danger of establishing parallel classifications of the same species; however, additional markers could be introduced once HTS technologies are capable of sequencing multiple markers or the entire genome from a single template.
While there seems to be reluctance about formally accepting sequences as types, and there is little hope this issue will be resolved within a reasonable time frame, the Code already has a provision for validly describing ecologically cryptic fungi based on sequence data only. According to Art. 8.1., the “type (holotype, lectotype, or neotype) of a name of a species or infraspecific taxon is either a single specimen conserved in one herbarium or other collection or institution, or an illustration (but see Art. 8.5; see also Art. 40.4 and 40.5).” An illustration is thereby defined as “… a work of art or a photograph depicting a feature or features of an organism, e.g. a picture of a herbarium specimen or a scanning electron micrograph.” Art. 40.4 provides that an illustration can serve as type “prior to 1 January 2007” and “on or after that date, the type must be a specimen (except as provided in Art. 40.5).” Thus, in cases to which Art. 40.5 applies, the type can be an illustration, and that article states: “For the purpose of Art. 40, the type of a name of a new species or infraspecific taxon of microscopic algae or microfungi (fossils excepted: see Art. 8.5) may be an effectively published illustration if there are technical difficulties of preservation or if it is impossible to preserve a specimen that would show the features attributed to the taxon by the author of the name.”
The explicit citation of “microfungi” and the provisions of “technical difficulties of preservation” and “impossible to preserve a specimen that would show the features” apply precisely to the situation of environmental sequencing and also to the present case, where a specific fungus can be phylogenetically identified but the sequence data cannot be attributed to an exact physical specimen of that fungus. This “illustration clause” was mentioned by de Beer et al. (2016) and Turland and Wiersema (2017), and even Reynolds and Taylor (1992) already suggested that a depiction of a sequence could serve as type. De Beer et al. (2016) considered this option a circumvention of the Code, but here we argue that rather than circumventing the Code, it can be applied “as written”.
The content of Art. 40.5. goes back to the original definition of the type in Art. 18, Note 2 of the Stockholm Code (Lanjouw et al. 1952): “A holotype (‘type’) is the one specimen or other element used by the author or designated by him as the nomenclatural type.” At that time, the phrase “other element” allowed illustrations at types, without restriction. While this article was moved to Art. 7. in subsequent versions of the Code, its content remained unchanged until including Art. 7.3. of the Leningrad Code (Stafleu et al. 1978). With the Sydney Code (Voss et al. 1983), the phrase “other element” was changed explicitly to “illustration”: “A holotype is the one specimen or illustration used by the author or designated by him as the nomenclatural type”, which remained so in the Berlin Code (Greuter et al. 1988). In the Tokyo Code (Greuter et al. 1994), the definition of what is allowed as type was narrowed down considerably; while Art. 8.1. stated: “The type of a name of a species or infraspecific taxon is a single specimen or illustration…”, Art. 8.3. specified: “If it is impossible to preserve a specimen as the type of a name of a species or infraspecific taxon of non-fossil plants, or if such a name is without a type specimen, the type may be an illustration.” In the St. Louis Code (Greuter et al. 2000), the content of Art. 8.3. was reissued as Art. 37.4. and further modified, stating that “… the type of a name of a new species or infraspecific taxon (fossils excepted: see Art. 8.5) may be an illustration if, and only if, it is impossible to preserve a specimen.” The current wording, specifically allowing illustrations as types only for microalgae and microfungi (excluding fossils) in the case of technical difficulties to preserve a type, was first implemented in Art. 37.5. of the Vienna Code (McNeill et al. 2006), following a rejected proposal to permit illustrations as types of microfossils (Traverse et al. 2004) and discussion on the floor at the Nomenclature Section of IBC XVII about the removal of the provision to allow illustrations as types after 1958 (McNeill and Turland 2005; McNeill et al. 2005; Flann et al. 2015).
An illustration which can serve as type must be “depicting a feature or features of an organism”. According to ICN Art. 38.2., a diagnosis (of a new taxon) is a “statement of that which in the opinion of its author distinguishes the taxon from other taxa”. It is not further specified what features can be used for a valid diagnosis, but from precedent it is obvious that features are allowable even if they can only be diagnosed with technical equipment, such as secondary chemistry, anatomical details, spores. For instance, scanning electron micrographs are explicitly cited as a possible content of an illustration serving as type, although these only allow indirect observation of a feature through a specific technology. By extension it follows that also sequence data are allowed by the Code as diagnostic features, and such data have already been implemented for the valid description of new taxa (e.g. Fliegerová et al. in Kirk 2012; Lücking et al. 2016), although there is some controversy as to the format in which sequence data need to be presented to qualify as validating diagnostic feature (Tripp and Lendemer 2012; de Beer et al. 2016). Since an illustration can be an artwork, the illustration of sequence data in an effectively published image can serve as only type for the description of new taxa of cryptic microfungi. Turland and Wiersema (2017), in their rapport on the aforementioned proposal to allow sequence data as types, discuss this issue, arguing that, while a DNA sequence might be “… analogous to an illustration”, it would be difficult to consider a sequence a work of art, although they admit the ambiguity of the definition of art. While that definition is indeed broad [https://en.wikipedia.org/wiki/Work_of_art], there are three elements that one could envision: non-text, intellectual property, and copyright. Thus, in a publication, to serve as type, an illustration must clearly be identified as such, separate of the text body, as Figure, Plate, etc. The illustration must be the individual work of a person or group of persons made specifically for this purpose; in that case, the copyright is implicit: “A work of art is automatically protected by copyright law as soon as it is created.” [http://www.wikihow.com/Copyright-Your-Artwork]. Thus, the sequence per se cannot be a work of art in the above sense, as it is the result of an abstraction of a naturally biological feature obtained through a purely technical process. In order to serve as type via an illustration, the sequence must be depicted in a graphical form that reflects the intellectual property of the author of the graphics.
While it would certainly be desirable to have an actual, accessioned sequence as type (de Beer et al. 2016; Hawksworth et al. 2016), below we take advantage of these provisions for the valid description of the cryptic basidiomycetes occurring in Agonimia foliacea and in Normandina species, with one new genus and seven new species. We are aware that there will be controversy about our approach and that there will be arguments that this interpretation of ICN Art. 40.5 is not as it was intended. However, law is to be followed “as written” and not “as intended”, and if it was not intended that way, it would have to be rewritten. Also, it is hard to conceive a case where this article should apply that would at the same time exclude environmental sequencing; if anything, environmental sequencing is the perfect example. Using ICN Art. 40.5 might open the door to the formal description of thousands of fungal species from environmental sequencing techniques without further changes to the Code, and this will hopefully stimulate a serious discussion leading to a quick solution to the problem of formally describing fungi from environmental sequence data.
Following other workers (e.g. de Beer et al. 2016), we take the opportunity to set a precedent for good practice when using this provision:
-
1.
A morphologically unmanifested, environmentally cryptic new fungal species that is being formally described using the illustration of a sequence as type must be defined in a phylogenetic context using an accepted barcode marker (e.g., ITS); simple blast or clustering techniques are not acceptable to delimit species; if appropriate, e.g. in the case of more complex topological patterns, a quantitative species recognition method should be used.
-
2.
If more than one sequence defines a new species, the most appropriate sequence should be selected for typification, using the criteria of quality (few, if any, ambiguous base calls, clear chromatograms in case of Sanger-derived sequences), length (longest sequence if other criteria apply equally), and position (basal in clade if other criteria apply equally). Formally named sequences should be marked for immediate release after submission to GenBank, to ensure their quick availability as reference sequences, and a specific prefix should be used for type sequences (e.g., ENAS_000000000); also, deposition in a curated database such as UNITE (Kõljalg et al. 2013) is highly recommended.
-
3.
For typification, the complete sequence must be illustrated in a published figure graphics designed for this purpose; the image should also be supplemented as PDF to allow direct copying of the sequence data, in addition to citing the GenBank accession number.
-
4.
The diagnostic features of the sequence providing the type illustration must be detailed by comparison with the sister clade of the new species, preferably with a diagnostic illustration. Here we follow Tripp and Lendemer (2012); the argument put forward by de Beer et al. (2016) that this approach is not feasible, as topologies and relationships change with new data, does not apply, since a diagnosis is always specific between taxa and based on current knowledge in the moment of publication. For instance, the diagnosis that a new species differs from its (at the time known) closest, named relative by larger ascospores or red instead of white caps is not invalidated a posteriori by the subsequent discovery that an even more closely related species has yet another ascospore size or yellow caps. Independent of the features used, the Code requires either a diagnosis or a description, and a diagnosis simply stating that a new taxon differs from a known one in ascospore size or cap color is not sufficient, because such a statement is not directly verifiable from the protologue, unless otherwise detailed.
-
5.
In case of Sanger-derived sequences (as in the present study or in the work on Cyphobasidium; Spribille et al. 2016), the original chromatograms could be provided as supplementary files or submitted to GenBank with the sequence data, to provide for quality control.
Lawreymyces Lücking & Moncada, gen. nov.
Index Fungorum Number: IF 553219.
Type: Lawreymyces palicei Lücking & Moncada (holotype; see below).
Etymology: Dedicated to our esteemed colleague, James D. Lawrey, for his important contributions to lichen ecology and to our knowledge of lichenicolous fungi.
Diagnosis: A cryptic, lichenicolous basidiomycete in the family Corticiaceae, order Corticiales, class Agaricomycetes, occurring on lichens of the family Verrucariaceae, known specifically from the genera Agonimia and Normandina; consistently differing from the currently resolved sister clade formed by the related genera Erythricium and Marchandiobasidium in 50 parsimony-informative alignment positions of the internal transcribed spacer (ITS), including 41 substitutions (AC = 4, AG = 3, AT = 3, CG = 2, CT = 6, GA = 5, GC = 1, GT = 4, TA = 6, TC = 5, TG = 2) and nine indels (Fig. 6).
Lawreymyces bogotensis Lücking & Moncada, sp. nov.
Index Fungorum Number: IF 553220.
Type: Fig. 7A (holotype acc. to ICN Art. 8.5., 40.5.); illustrated diagnostic sequence graphics taken from sequence MF070062 obtained from the following host specimen of Normandina sp. growing on Sticta sp.: COLOMBIA. Bogotá, D.C.: Pasquilla; 3 December 2015, Lücking & Moncada 41028a (B).
Etymology: Growing in thalli of an unidentified Normandina collected within the capital district of Bogotá.
Diagnosis: Differing from the sister species, L. columbiensis, in seven parsimony-informative alignment positions of the internal transcribed spacer (ITS), all substitutions (AG = 1, CT = 2, TC = 4; Fig. 8F, G).
Illustration of the diagnostic ITS sequence features for the seven new species of Lawreymyces, showing the parsimony-informative columns only. The corresponding alignment file was obtained from the original alignment (Suppl. File S6) by deleting the outgroup taxa and subsequently deleting all constant and parsimony-uninformative columns
Lawreymyces columbiensis Lücking & Moncada, sp. nov.
Index Fungorum Number: IF 553221.
Type: Fig. 7B (holotype acc. to ICN Art. 8.5., 40.5.); illustrated diagnostic sequence graphics taken from sequence MF070060 obtained from the following host specimen of Normandina columbiensis: COLOMBIA. Cauca: 30 March 2016, Moncada 10521 (B, UDBC).
Etymology: Growing on specimens of Normandina columbiensis, a new species described elsewhere (Moncada and Lücking in prep.).
Diagnosis: Differing from the sister species, L. bogotensis, in seven parsimony-informative alignment positions of the internal transcribed spacer (ITS), all substitutions (CT = 4, GA = 1, TC = 2; Fig. 8F, G).
Lawreymyces confusus Lücking & Moncada, sp. nov.
Index Fungorum Number: IF 553222.
Type: Fig. 7C (holotype acc. to ICN Art. 8.5., 40.5.); illustrated diagnostic sequence graphics taken from sequence MF070057 obtained from the following host specimen of Normandina aff. columbiensis: COLOMBIA. Cauca: 30 March 2016, Moncada 10567 (B, UDBC).
Etymology: The epithet refers to the confusion caused by the first detected aberrant ITS sequence of this basidiomycete from a sample of Normandina.
Diagnosis: Differing from the sister clade formed by the species L. bogotensis and L. columbiensis in 11 parsimony-informative alignment positions of the internal transcribed spacer (ITS), all substitutions (AG = 2, CG = 1, CT = 2, GA = 1, TC = 3, TG = 2; Fig. 8E–G).
Lawreymyces foliaceae Lücking & Moncada, sp. nov.
Index Fungorum Number: IF 553223.
Type: Fig. 7D (holotype acc. to ICN Art. 8.5., 40.5.); illustrated diagnostic sequence graphics taken from sequence MF070054 obtained from the following host specimen of Agonimia foliacea: COLOMBIA. Cundinamarca: Páramo de Guasca; 18 August 2011, Lücking & Moncada 33368 (B, UDBC).
Etymology: Growing in thalli of Agonimia foliacea.
Diagnosis: Differing from the sister species, L. palicei, in nine parsimony-informative alignment positions of the internal transcribed spacer (ITS), six substitutions (AT = 1, CT = 2, CG = 2, TA = 1) and three indels (Fig. 8A, B).
Lawreymyces palicei Lücking & Moncada, sp. nov.
Index Fungorum Number: IF 553224.
Type: Fig. 7E (holotype acc. to ICN Art. 8.5., 40.5.); illustrated diagnostic sequence graphics taken from sequence AY542865 obtained from the following host specimen of Agonimia foliacea: ECUADOR: Volcán Chiles, 29 September 2000, Z. Palice & Z. Soldán 4369 (F).
Etymology: Named after Zdeněk Palice, the collector of the original material from which the sequences were obtained, and who first assessed the status of Omphalina foliacea as a species of Agonimia correctly.
Diagnosis: Differing from the sister species, L. foliaceae, in nine parsimony-informative alignment positions of the internal transcribed spacer (ITS), six substitutions (AT = 1, GC = 2, TA = 1, TC = 2) and three indels (Fig. 8A, B).
Lawreymyces pulchellae Lücking & Moncada, sp. nov.
Index Fungorum Number: IF 553225.
Type: Fig. 7F (holotype acc. to ICN Art. 8.5., 40.5.); illustrated diagnostic sequence graphics taken from sequence MF070063 obtained from the following host specimen of Normandina pulchella: COLOMBIA. Cundinamarca: Chipaque-Marilandia; November 2016, Moncada 10624 (B, UDBC).
Etymology: Growing in thalli of Normandina pulchella.
Diagnosis: Differing from the sister clade formed by the species L. bogotensis, L. columbiensis, and L. confusus, in ten parsimony-informative alignment positions of the internal transcribed spacer (ITS), nine substitutions (AG = 2, CG = 1, CT = 1, GA = 1, GC = 1, TC = 3) and one indel (Fig. 8D–G).
Lawreymyces spribillei Lücking & Moncada, sp. nov.
Index Fungorum Number: IF 553226.
Type: Fig. 7G (holotype acc. to ICN Art. 8.5., 40.5.); illustrated diagnostic sequence graphics taken from sequence MF070056 obtained from the following host specimen of Agonimia foliacea: COLOMBIA. Cundinamarca: Villapinzón, Páramo de Guacheneque; 12 May 2012, Moncada 5410 (B, UDBC).
Etymology: This new species honours our colleague Toby Spribille, for his important contributions to our understanding of the lichen symbiosis and the discovery of the potential role of cryptic Basidiomycota in ascolichen symbioses.
Diagnosis: Differing from the sister clade formed by the species L. foliaceae and L. palicei in 23 parsimony-informative alignment positions of the internal transcribed spacer (ITS), 18 substitutions (AC = 1, AG = 3, CT = 7, GA = 5, TC = 2) and five indels (Fig. 8C, A–B).
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Aptroot A (1991) A conspectus of Normandina (Verrucariaceae, lichenized Ascomycetes). Willdenowia 21:263–267
Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2013) GenBank. Nucleic Acids Res 41(D1):D36–D42
Binder M, Hibbett DS, Larsson KH, Larsson E, Langer E, Langer G (2005) The phylogenetic distribution of resupinate forms across the major clades of mushroom-forming fungi (Homobasidiomycetes). Syst Biodivers 3:113–157
Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, Madden TL, Matten WT, McGinnis SD, Merezhuk Y, Raytselis Y, Sayers EW, Tao T, Ye J, Zaretskaya I (2013) BLAST: a more efficient report with usability improvements. Nucleic Acids Res 41:W29–W33
Breuss O (2002) Agonimia. In: Nash TH III, Ryan BD, Gries C, Bungartz F (eds) Lichen Flora of the Greater Sonoran Desert Region. I. Lichens Unlimited. Arizona State University, Tempe, pp 90–91
Clements FE, Shear CL (1931) The genera of fungi. Wilson, New York
Coppins BJ, Bennell AP (1979) Thallus surface features in Agonimia tristicula. Lichenologist 11:107–108
Crespo A, Kauff F, Divakar PK, del Prado R, Pérez-Ortega S, de Paz GA, Ferencova Z, Blanco O, Roca-Valiente B, Núñez-Zapata J, Cubas P, Argüello A, Elix JA, Esslinger TL, Hawksworth DL, Millanes AM, Molina MC, Wedin M, Ahti T, Aptroot A, Barreno E, Bungartz F, Calvelo S, Candan M, Cole MJ, Ertz D, Goffinet B, Lindblom L, Lücking R, Lutzoni F, Mattsson JE, Messuti MI, Miądlikowska J, Piercey-Normore MD, Rico VJ, Sipman HJM, Schmitt I, Spribille T, Thell A, Thor G, Upreti DK, Lumbsch HT (2010) Phylogenetic generic classification of parmelioid lichens (Parmeliaceae, Ascomycota) based on molecular, morphological and chemical evidence. Taxon 59:1735–1753
David JC, Hawksworth DL (1989) Lauderlindsaya, a new genus in the Verrucariales for Sphaerulina chlorococca (Leighton) R. Sant. Sydowia 41:108–121
De Beer ZW, Marincowitz S, Duong TA, Kim JJ, Rodrigues A, Wingfield MJ (2016) Hawksworthiomyces gen. nov. (Ophiostomatales), illustrates the urgency for a decision on how to name novel taxa known only from environmental nucleic acid sequences (ENAS). Fungal Biol 120:1323–1340
Diederich P, Lawrey JD (2007) New lichenicolous, muscicolous, corticolous and lignicolous taxa of Burgoa s.l. and Marchandiomyces s.l. (anamorphic Basidiomycota), a new genus for Omphalina foliacea, and a catalogue and a key to the non-lichenized, bulbilliferous basidiomycetes. Mycol Prog 6:61–80
Diederich P, Sérusiaux E (1993) A nomenclatural note on Lauderlindsaya (Ascomycotina, Verrucariales). Lichenologist 25:97–100
Divakar PK, Crespo A, Wedin M, Leavitt SD, Hawksworth DL, Myllys L, McCune B, Randlane T, Bjerke JW, Ohmura Y, Schmitt I (2015) Evolution of complex symbiotic relationships in a morphologically derived family of lichen-forming fungi. New Phytol 208:1217–1226
Flann C, McNeill J, Barrie FR, Nicolson DH, Hawksworth DL, Turland NJ, Monro AM (2015) Report on botanical nomenclature—Vienna 2005. XVII international botanical congress, Vienna: nomenclature Section, 12–16 July 2005. PhytoKeys 45:1–341
Frisch A, Ohmura Y (2015) The phylogenetic position of Normandina simodensis (Verrucariaceae, Lichenized Ascomycota). Bull Natl Mus Nat Sci 41:1–7
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rust. Mol Ecol 2:113–118
Greuter W, Burdet HM, Chaloner WG, Demoulin V, Grolle R, Hawksworth DL, Nicolson DH, Silva PC, Stafleu FA, Voss EG, McNeill J (1988) International Code of Botanical Nomenclature adopted by the Fourteenth International Botanical Congress, Berlin, July–August 1987. Regnum Veg 118:i–xiv,1–328
Greuter W, Barrie FR, Burdet HM, Chaloner WG, Demoulin V, Hawksworth DL, Jørgensen PM, Nicolson DH, Silva PC, Trehane P, McNeill J (1994) International Code of Botanical Nomenclature (Tokyo Code), adopted by the Fifteenth International Botanical Congress, Yokohama, August-September 1993. Regnum Veg 131:i–xviii,1–389
Greuter W, McNeill J, Barrie FR, Burdet HM, Demoulin V, Filguerias TS, Nicolson DH, Silva PC, Skog E, Trehane P, Turland NJ, Hawksworth DL (2000) International Code of Botanical Nomenclature (Saint Louis Code), adopted by the Sixteenth International Botanical Congress, St Louis, Missouri, July-August 1999. Regnum Veg 138:i–xviii,1–474
Hafellner J (2014) Distributional and other data for some Agonimia species (Verrucariales, lichenized Ascomycota). Fritschiana 78:25–46
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Hall TA (2011) BioEdit: an important software for molecular biology. GERF Bull Biosci 2:60–61
Hawksworth DL, Crous PW, Redhead SA, Reynolds DR, Samson RA, Seifert KA, Taylor JW, Wingfield MJ (2011) The Amsterdam declaration on fungal nomenclature. IMA Fungus 2:105–112
Hawksworth DL, Hibbett DS, Kirk PM, Lücking R (2016) (308–310) Proposals to permit DNA sequence data to serve as types of names of fungi. Taxon 65:899–900
Hibbett DS (2016) The invisible dimension of fungal diversity. Science 351:1150–1151
Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lücking R, Thorsten Lumbsch H, Lutzoni F, Matheny PB, McLaughlin DJ, Powell MJ, Redhead S, Schoch CL, Spatafora JW, Stalpers JA, Vilgalys R, Aime MC, Aptroot A, Bauer R, Begerow D, Benny GL, Castlebury LA, Crous PW, Dai YC, Gams W, Geiser DM, Griffith GW, Gueidan C, Hawksworth DL, Hestmark G, Hosaka K, Humber RA, Hyde KD, Ironside JE, Kõljalg U, Kurtzman CP, Larsson KH, Lichtwardt R, Longcore J, Miadlikowska J, Miller A, Moncalvo JM, Mozley-Standridge S, Oberwinkler F, Parmasto E, Reeb V, Rogers JD, Roux C, Ryvarden L, Sampaio JP, Schüssler A, Sugiyama J, Thorn RG, Tibell L, Untereiner WA, Walker C, Wang Z, Weir A, Weiss M, White MM, Winka K, Yao YJ, Zhang N (2007) A higher-level phylogenetic classification of the fungi. Mycol Res 111:509–547
Hibbett DS, Ohman A, Glotzer D, Nuhn M, Kirk P, Nilsson RH (2011) Progress in molecular and morphological taxon discovery in Fungi and options for formal classification of environmental sequences. Fung Biol Rev 25:38–47
Hoffmann N, DePriest PT (2000) Fruiting Normandina pulchella versus Lauderlindsaya borreri: phylogenetic analysis of molecular data clarifies the species concept. In: The Fourth IAL Symposium, Progress and Problems in Lichenology at the Turn of the Millennium. Book of Abstracts: 95
Honegger R, Conconi S, Kutasi V (1996) Field studies on growth and regenerative capacity in the foliose macrolichen Xanthoria parietina (Teloschistales, Ascomycotina). Bot Acta 109:187–193
Jaklitsch WM, Baral HO, Lücking R, Lumbsch HT (2016) Ascomycota. In: Frey W (ed) Syllabus of Plant Families—Adolf Engler’s Syllabus der Pflanzenfamilien. Borntraeger, Stuttgart
Jørgensen PM (1989) Omphalina foliacea, a new basidiolichen from America. Nord J Bot 9:89–95
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Mentjies P, Drummond A (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Kirk PM (2012) Nomenclatural novelties. Index Fungorum 1:1
Kirk PM, Cannon PF, Minter DW, Stalpers JA (2008) Dictionary of the fungi, 10th edn. CAB International, Wallingford
Kodama Y, Shumway M, Leinonen R (2012) The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res 40(D1):D54–D56
Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AF, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson KH (2013) Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5277
Lanjouw J, Baehni C, Merrill ED, Rickett HW, Robyns W, Sprague TA, Stafleu FA (1952) International code of botanical nomenclature adopted by the Seventh international botanical congress, Stockholm, July 1950. Regnum Veg 3:1–228
Lawrey JD, Diederich P (2003) Lichenicolous fungi: interactions, evolution, and biodiversity. Bryologist 106:81–120
Lawrey JD, Diederich P (2016) Lichenicolous fungi—worldwide checklist, including isolated cultures and sequences available. http://www.lichenicolous.net. Accessed 31 Jan 2017
Lawrey JD, Binder M, Diederich P, Molina MC, Sikaroodi M, Ertz D (2007) Phylogenetic diversity of lichen-associated homobasidiomycetes. Mol Phyl Evol 44:778–789
Lawrey JD, Diederich P, Sikaroodi M, Gillevet PM (2008) Remarkable nutritional diversity of basidiomycetes in the Corticiales, including a new foliicolous species of Marchandiomyces (anamorphic Basidiomycota, Corticiaceae) from Australia. Am J Bot 95:816–823
Leinonen R, Sugawara H, Shumway M (2011) The sequence read archive. Nucleic Acids Res 39(D1):D19–D21
Lücking R (2014) A phylogenetic classification system for unvouchered environmental fungal sequences of unknown taxonomic affiliation. In: The 10th International Mycological Congress, Bangkok, Thailand. IMC10 Book of Abstracts: O 8.6.1, Abstract ID ABS0123. http://www.fabinet.up.ac.za/newsitem/112-IMC10eBookofAbstracts.pdf
Lücking R, Nelsen MP, Aptroot A, Klee RB, Bawingan PA, Benatti MN, Binh NQ, Bungartz F, Cáceres MES, Canêz LS, Chaves J-L, Ertz D, Esquivel RE, Ferraro LI, Grijalva A, Gueidan C, Hernández JE, Knight A, Lumbsch HT, Marcelli MP, Mercado-Diaz JA, Moncada B, Morales EA, Naksuwankul K, Orozco T, Parnmen S, Rivas Plata E, Salazar-Allen N, Spielmann AA, Ventura N (2016) A phylogenetic framework for reassessing generic concepts and species delimitation in the lichenized family Trypetheliaceae (Ascomycota: Dothideomycetes). Lichenologist 48:739–762
Lücking R, Hodkinson BP, Leavitt SD (2017) The 2016 classification of lichenized fungi in the Ascomycota and Basidiomycota—approaching one thousand genera. Bryologist 119:361–416
Lumbsch HT, Schmitt I, Lindemuth R, Miller A, Mangold A, Fernandez F, Huhndorf S (2005) Performance of four ribosomal DNA regions to infer higher-level phylogenetic relationships of inoperculate euascomycetes (Leotiomycota). Mol Phyl Evol 34:512–524
Maddison WP, Knowles LL (2006) Inferring phylogeny despite incomplete lineage sorting. Syst Biol 55:21–30
Maheshwari R (2016) Fungi: experimental methods in biology, 2nd edn (Mycology). CRC Press, Boca Raton
Mares D, Fasulo MP, Bruni A (1993) Contribution to the study of Normandina pulchella: a cytological approach. Orsis Org Sist 8:33–40
McCune B (1997) Lauderlindsaya, a parasitic fungus on Normandina, new to North America. Evansia 14:13
McNeill J, Turland NJ (2005) Synopsis of proposals on Botanical Nomenclature—Vienna 2005. A review of the proposals concerning the International Code of Botanical Nomenclature submitted to the XVII international botanical congress. Taxon 54:215–250
McNeill J, Stuessy TF, Turland NJ, Hörandl E (2005) XVII international botanical congress: preliminary mail vote and report of congress action on nomenclature proposals. Taxon 54:1057–1064
McNeill J, Barrie FR, Burdet HM, Demoulin V, Hawksworth DL, Marhold K, Nicolson DH, Prado JS, Silva PC, Skog JE, Wiersema, JH, Turland NJ (2006) International Code of Botanical Nomenclature (Vienna Code), adopted by the seventeenth international botanical congress, Vienna, Austria, July 2005. Regnum Veg 146:i–xviii, 1–568
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the gateway computing environments workshop (GCE), pp. 1–8, New Orleans
Muggia L, Gueidan C, Grube M (2010) Phylogenetic placement of some morphologically unusual members of Verrucariales. Mycologia 102:835–846
Orange A, Purvis OW (2009) Agonimia Zahlbr. (1909). In: Smith CW, Aptroot A, Coppins BJ, Fletcher A, Gilbert OL, James PW, Wolseley PA (eds) The lichens of Great Britain and Ireland. British Lichen Society, London, pp 136–138
Ott S (1987) Reproductive strategies in lichens. Biblioth Lichenol 25:81–93
Palice Z, Schmitt I, Lumbsch HT (2005) Molecular data confirm that Omphalina foliacea is a lichen-forming basidiomycete. Mycol Res 109:447–451
Penn O, Privman E, Ashkenazy H, Landan G, Graur D, Pupko T (2010a) GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Res 38:W23–W28
Penn O, Privman E, Landan G, Graur D, Pupko T (2010b) An alignment confidence score capturing robustness to guide-tree uncertainty. Mol Biol Evol 27:1759–1767
Rambold G, Triebel D (1992) The inter-lecanoralean associations. Biblioth Lichenol 48:1–201
Redhead SA, Lutzoni F, Moncalvo JM, Vilgalys R (2002) Phylogeny of agarics: partial systematics solutions for core omphalinoid genera in the Agaricales (Euagarics). Mycotaxon 83:19–57
Redhead SA, Demoulin V, Hawksworth DL, Seifert KA, Turland NJ (2014) Fungal nomenclature at IMC10: report of the nomenclature sessions. IMA Fungus 5:449–462
Reynolds DR, Taylor JW (1992) Article 59: reinterpretation or revision? Taxon 41:91–98
Rivas Plata E, Lücking R, Aptroot A, Sipman HJM, Chaves JL, Umaña L, Lizano D (2006) A first assessment of the Ticolichen biodiversity inventory in Costa Rica: the genus Coenogonium (Ostropales: Coenogoniaceae), with a world-wide key and checklist and a phenotype-based cladistic analysis. Fung Div 23:255–321
Santesson R (1989) Parasymbiotic fungi on the lichen-forming basidiomycete Omphalina foliacea. Nord J Bot 9:97–99
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA 109:6241–6246
Sérusiaux E, Diederich P, Brand AM, van den Boom P (1999) Lichens et champignons nouveaux ou intéressants pour la flore de la Belgique et du G.-D. de Luxembourg. VIII. Lejeunia 162:1–95
Spribille T, Tuovinen V, Resl P, Vanderpool D, Wolinski H, Aime MC, Schneider K, Stabentheiner E, Toome-Heller M, Thor G, Mayrhofer H, Johannesson H, McCutcheon JP (2016) Basidiomycete yeasts in the cortex of ascomycete macrolichens. Science. doi:10.1126/science.aaf8287
Stafleu FA, Demoulin V, Greuter W, Hiepko P, Linczevski IA, McVaugh R, Meikle RD, Rollins RC, Ross R, Schopf JM, Voss EG (1978) International Code of Botanical Nomenclature, adopted by the Twelfth International Botanical Congress, Leningrad, July 1975. Regnum Veg 97:i–xiv, 1–457
Stamatakis A (2015) Using RAxML to infer phylogenies. Unit 6.14. Current protocols in bioinformatics. Wiley, New York
Stewart JE, Timmer LW, Lawrence CB, Pryor BM, Peever TL (2014) Discord between morphological and phylogenetic species boundaries: incomplete lineage sorting and recombination results in fuzzy species boundaries in an asexual fungal pathogen. BMC Evol Biol 14:38
Swindell SR, Plasterer TN (1997) SEQMAN: Contig assembly. In: Swindell SR (ed) Sequence data analysis guidebook. Methods in molecular medicine, vol 70. Springer, New York, pp 75–89
Taylor JW (2011) One fungus = one name: DNA and fungal nomenclature twenty years after PCR. IMA Fungus 2:113–120
Thüs H, Muggia L, Pérez-Ortega S, Favero-Longo SE, Joneson S, O’Brien H, Nelsen MP, Duque-Thüs R, Grube M, Friedl T, Brodie J (2011) Revisiting photobiont diversity in the lichen family Verrucariaceae (Ascomycota). Eur J Phycol 46:399–415
Traverse A, Jansonius J, Nichols DJ (2004) (208–212) Proposals to amend Arts. 8, 9, 37, and 38 to provide that the type of the name of a plant microfossil may be an illustration. Taxon 53:849–850
Tripp EA, Lendemer JC (2012) (4–5) Request for binding decisions on the descriptive statements associated with Mortierella sigyensis (fungi: Mortierellaceae) and Piromyces cryptodigmaticus (fungi: Neocallimastigaceae. Taxon 61:886–888
Tscherrnak-Woess E (1981) Zur Kenntis der Phycobionten von Lobaria linita und Normandina pulchella. Nova Hedwigia 35:63–73
Turland NJ, Wiersema JH (2017) Synopsis of proposals on nomenclature—Shenzhen 2017: a review of the proposals concerning the International Code of Nomenclature for algae, fungi, and plants submitted to the XIX international botanical congress. Taxon 66:217–274
Voss EG, Burdet HM, Chaloner WG, Demoulin V, Hiepko P, McNeill J, Meikle RD, Nicolson DH, Rollins RC, Silva PC, Greuter W (1983) International Code of Botanical Nomenclature, adopted by the thirteenth international botanical congress, Sydney, August 1981. Regnum Veg 111:i–xv, 1–472
White TJ, Bruns TD, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols. Academic Press, San Diego, pp 315–322
Zhou S, Stanosz GR (2001) Primers for amplification of mt SSU rDNA, and a phylogenetic study of Botryosphaeria and associated anamorphic fungi. Mycol Res 105:1033–1044
Zoller S, Scheidegger C, Sperisen C (1999) PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming ascomycetes. Lichenologist 31:511–516
Acknowledgements
This study was partially supported by two grants from the National Science Foundation: Neotropical Epiphytic Microlichens—An Innovative Inventory of a Highly Diverse yet Little Known Group of Symbiotic Organisms (DEB 0715660 to The Field Museum; PI R. Lücking), and Phylogenetic Diversity of Mycobionts and Photobionts in the Cyanolichen Genus Dictyonema, with Emphasis on the Neotropics and the Galapagos Islands (DEB 0841405 to George Mason University; PI J. Lawrey; Co-PIs: R. Lücking, P. Gillevet). The Verein der Freunde des Botanischen Gartens und Botanischen Museums Berlin-Dahlem e.V. (https://www.bgbm.org/de/BGBM/freunde/index.html) supported molecular sequencing work for specimens collected as part of the Pilotprojekt Kooperation mit dem Botanischen Garten Bogotá (BMBF, see below). The Universidad Distrital Francisco José de Caldas, Bogotá, is thanked for the support to the lichen herbarium and the curatorial work of the UDBC collections, and the Field Museum and the BGBM kindly provided the logistics for the molecular work. The Jardín Botánico de Bogotá José Celestino Mutis organized the field trip to Pasquilla (Bogotá), by agreement with the Botanical Garden and Botanical Museum Berlin, partially financed through the German Federal Ministry of Education and Research (BMBF; Pilotprojekt Kooperation mit dem Botanischen Garten Bogotá; Förderkennzeichen: 01DN13030). Paul Diederich is warmely thanked for placing a photograph of Agonimia globulifera at our disposal.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lücking, R., Moncada, B. Dismantling Marchandiomphalina into Agonimia (Verrucariaceae) and Lawreymyces gen. nov. (Corticiaceae): setting a precedent to the formal recognition of thousands of voucherless fungi based on type sequences. Fungal Diversity 84, 119–138 (2017). https://doi.org/10.1007/s13225-017-0382-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13225-017-0382-4