Abstract
This study was conducted to investigate the γ-aminobutyric acid (GABA) production ability of 20 Lactobacillus and 25 Bifidobacterium strains which were previously isolated in our laboratory. Effect of initial pH, incubation time, monosodium glutamate (MSG), and pyridoxal-5′-phosphate (PLP) concentration for highest GABA production by two potent bacterial strains, Levilactobacillus brevis LAB6 and Limosilactobacillus fermentum LAB19 were optimized in the MRS media. A threefold increase in GABA production at an initial pH 4.0, incubation time of 120 h in medium supplemented with 3% MSG and 400 μM of PLP for LAB6 and 300 μM for LAB19 lead to the production of 19.67 ± 0.28 and 20.77 ± 0.14 g/L of GABA, respectively. Coculturing both strains under optimized conditions led to a GABA yield of 20.02 ± 0.17 g/L. Owing to potent anti-inflammatory activity in-vitro, as reported previously, and highest GABA production ability of LAB6 (MTCC 25662), its whole-genome sequencing and bioinformatics analysis was carried out for mining genes related to GABA metabolism. LAB6 harbored a complete glutamate decarboxylase (GAD) gene system comprising gadA, gadB, and gadC as well as genes responsible for the beneficial probiotic traits, such as for acid and bile tolerance and host adhesion. Comparative genomic analysis of LAB6 with 28 completely sequenced Levilactobacillus brevis strains revealed the presence of 95 strain-specific genes-families that was significantly higher than most other L. brevis strains.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As one of the few neurotransmitters that has been preserved throughout the coevolution of the host and gut microbiome, GABA facilitates communication between phylogenetically very different life forms by acting as a shared language, assisting in the homeostatic regulation of the enteric bacteria, and perhaps even influencing brain and behavior function. Gamma-aminobutyric acid (GABA) is a four-carbon ubiquitous and abundant non-protein amino acid which is extensively distributed in microbes, plants, and animals serving different functions in different organisms (Rashmi et al. 2018). Owing to its myriad physiological roles in humans like neuro-inhibition (Mann and Paulsen 2007), regulation of stress and cognition (Prévot and Sibille 2021), and regulation of the cardiovascular system (Zhu et al. 2019) GABA-producing probiotics are in good demand. As the direct supplementation of synthetic GABA in food is considered unsafe and is not permissible (Li et al. 2010; Chua et al. 2019); therefore, much effort in nutraceutical formulations is implied on in-situ production of GABA through microbial fermentation. Furthermore, microbial-derived GABA is associated with ‘gut–microbiota–brain axis’ signaling and homeostasis (Bauer et al. 2016). Majorly, lactic acid bacteria (LAB) and bifidobacteria are utilized as probiotics because of being fundamentally recognized as safe (GRAS); however, not all these have been given GRAS status (Hoover 2014).
Microbes produce GABA in response to acid stress via α-decarboxylation of L-glutamic acid or monosodium glutamate (MSG) catalyzed by the glutamate amino decarboxylase (GAD) [EC: 4.1.1.15] and functionally implicated in spore germination (Dhakal et al. 2012). Although the GAD gene is widely distributed in most LAB and bifidobacteria species, only a few of these produce GABA because of high variability in the genetic organization of this gene (Cui et al. 2020). Production of GABA is affected by culture conditions like concentration of MSG, incubation time and temperature, initial pH, and pyridoxal-5′-phosphate (PLP) supplementation (Dhakal et al. 2012). Recently, research to optimize and increase GABA yield via culturing LAB with monosodium glutamate (MSG) supplemented media has progressed spectacularly, since similar culture conditions cannot be applied to all bacterial strains (Diana et al. 2014). Lactic acid bacteria possess enormous interspecific diversity, and among those reported strains, typically Levilactobacillus brevis strains showed the highest GABA productivity and are mostly isolated from traditional fermented foods (Li et al. 2010; Dhakal et al. 2012).
Various mechanisms for GABA synthesis can be elucidated by genomic data analysis depending upon the cellular physiology of different microbial species. Metabolically active bacterial cells are indispensably reliant on homeostasis of cytoplasmic pH as majority of enzymes have distinctive pH ranges within which they can function normally (Krulwich et al. 2011). Remarkably, the GAD system is considered one of the most effective mechanisms to mitigate acid stress amid numerous other types of acid responses and tolerance mechanisms in bacteria (De Biase et al. 1999). The GAD operon contains three key elements that are responsible for GABA production and glutamate utilization in bacteria: gadR encoding positive transcriptional regulator, gadC encoding glutamate/GABA antiporter, and gadA or gadB that encodes for glutamate decarboxylase (Wu et al. 2017). Interestingly, while in most LAB strains, there is a presence of either gadA or gadB in the GAD system, the only known species that carries both genes in its operon is Levilactobacillus brevis. gadA is located considerably downstream from the operon; however, both genes encode biochemically similar isoforms of the GAD protein (Lyu et al. 2018). Additionally, GAD operon also consists of a transcriptional regulator, gadR, which positively monitors the production of GABA in a glutamate-dependent manner and is requisite for glutamate to GABA conversion (Gong et al. 2019).
In this study, previously isolated Lactobacillus and Bifidobacterium strains from different food sources and infant fecal samples in our laboratory were screened for GABA-producing ability. Major fermentative parameters affecting the GABA production by LAB6 and LAB19, such as initial pH, incubation time, MSG, and PLP concentration, in the culture medium were evaluated. Further, the whole-genome sequence of potential GABA-producing putative probiotic LAB6 was reported and scrutinized for GAD gene assembly and comparative genomic analysis of related L. brevis strains.
Materials and methods
Bacterial strains and culture conditions
A total of 45 bacterial strains (20 Lactobacillus and 25 Bifidobacterium strains) were used in the study, which were previously isolated in our lab from various sources (Singh et al 2020; Sharma et al. 2022; Bhatia et al. 2023) (Table S1). Frozen stock cultures ( – 80 °C) were streaked on MRS (deMan Rogosa and Sharpe) agar for Lactobacillus and MRS with L-cysteine hydrochloride (MRSc) agar for Bifidobacterium strains and incubated at 37 °C for 48 h under aerobic conditions for Lactobacillus strains and under anaerobic conditions for Bifidobacterium strains. A single colony of each strain was sub-cultured two times in MRS/MRSc broth for 24 h and incubated at 37 °C. Following the third transfer, the cultures were centrifuged at 7000 × g for 10 min at 4 °C, and obtained pellets were washed and resuspended in sterile PBS. Cells of each bacterial strain was set to an optical density of 0.5 and used for further experiments.
Screening of GABA-producing LAB
Activated bacterial cultures were inoculated in MRS/MRSc broth supplemented with 2.0% (w/v) of monosodium glutamate (MSG) and incubated under static conditions at 37 °C for 48 h, and the culture supernatants were collected by centrifugation at 6,000 × g for 12 min. Consequently, the samples were assessed for the presence of GABA using thin-layer chromatography (TLC) as described by Cho et al. (2011) with Silicagel 60 F254 TLC plates (Merck, Darmstadt, Germany). A 0.5 µl of cell-free supernatant was spotted on TLC plates compared with 0.5% (w/v) of standard GABA and MSG. Chromatographic separation was done using a solvent mix of butanol:acetic acid:water, in the ratio of 4:1:1 v/ v as a mobile phase. The plates were then sprayed with a 1.0% (w/v) ninhydrin solution and heated at 100 °C for 5–10 min, or until the development of spots.
Glutamate decarboxylase (GAD) colorimetric assay
Rapid GAD colorimetric assay was adopted to evaluate the change in pH as an indicator for the GAD activity of bacterial strains (Lacroix et al. 2013). A 5 mL overnight grown culture in MRS media at 37 °C was centrifuged at 5,000 × g for 20 min at 25 °C and washed with 5 mL saline under similar centrifugal conditions. The cell pellet obtained was homogenized in 500 µL GAD solution consisting of 1 g of L-glutamic acid, 90 g of NaCl, 300 µL of Triton X-100, and 0.05 g of bromocresol green in 1 L of distilled water adjusted to pH 4.0. Subsequently, the strains were incubated for 4 h under anaerobic conditions at 37 °C. No change in color (yellow) indicated negative result, while conversion to blue color indicated positive GAD active strains.
Morphological analysis of selected strains by FE-SEM
Morphological analysis of GABA-producing strains was done using field emission scanning electron microscopy (FE-SEM ThermoFisher Apreo HiVac). After incubation for 24 h at 37 °C, the cells were rinsed twice, and the pellet obtained was fixed with 2% glutaraldehyde solution for 4 h. The fixed cells were rinsed two times with 1X PBS followed by staining with 1% tannic acid for 1 h. After washing the cells two times with 1X PBS, the samples were dehydrated in increasing concentrations of ethanol from 10%, 30%, 50%, 70%, 90%, and 100% v/v with intermediate incubation of 5 min. The obtained sample was casted on silicon wafers, dried at room temperature overnight, and analyzed under FE-SEM.
Molecular detection of GAD gene
GABA-producing bacterial strains were cultivated in MRS broth until logarithmic growth was reached, and then, genomic DNA was isolated using the ZR Zymo DNA isolation kit according to the product manual. The identification of the GAD gene in the selected strains was evaluated using primers CoreF (5′-CCTCGAGAAGCCGATCGCTTAGTTCG-3′) and CoreR (5′-TCATATTGACCGGTATAAGTGATGCCC-3′), constructed from highly conserved regions of GAD gene (Siragusa et al. 2007) and the PCR protocol was carried out as described by Demirbaş et al. (2017). The amplicons obtained were visualized under GelDoc.
Determination of molecular weight of presumptive GABA by Q-TOF
The presence of presumptive GABA in the supernatant was validated by direct infusion of supernatant into a quadrupole time-of-flight (AB SCIEX TripleTOF 5600) fitted with an electrospray ionizer (ESI). With a curtain gas flow (CUR) of 30 psi, an ion spray voltage (ISVF) of 5500 V, an ion source (TEM) temperature of 40 °C, and an ion source gas (GSI) of 30 psi, the Q-TOF was operated in positive ion mode. The flow rate of the syringe was adjusted to 5 μL min−1. Both MS and MS/MS spectra of standard GABA and test samples were recorded.
Quantification of GABA by high-performance liquid chromatography
Bacterial culture supernatants obtained by centrifugation (6000 × g for 12 min at 4 °C) were used for high-performance liquid chromatography (HPLC). HPLC system consisted of an Agilent Technologies 1260 Infinity with a DAD detector (G1315D- 1260 DAD VL). The system was equipped with a 5 µm particle size (150 mm × 4.6 mm, ID) Zorbax Eclipse Plus C18 analytical column. The mobile phase comprised of 0.05 M sodium acetate (3.40 g in 500 ml MQ), tetrahydrofuran, and methanol (50:1:49, v/v) adjusted to pH 4.0 (Zhuang et al. 2018). Before use, the mobile phase was vacuum degassed and passed through Millipore 0.45 µm Durapore membrane filters. A 20 µl of the supernatant or standard GABA was mixed with 180 µl of methanol/water (85:15, v/v), 40 µL of freshly prepared methanolic o-phthalaldehyde OPA (10 mg/mL), 150 µL borate buffer (pH 9.9), and 10 µL 2-mercapto ethanol. The resultant reaction mixture was vortexed and analyzed after 2 min at room temperature. Chromatographic evaluations were carried out at 30 ± 2 °C. At a flow rate of 0.8 mL/min for 12 min, compounds were isocratically eluted. The UV detector was set at 340 nm. For the chromatographic data analysis, an integrator (EZ Chrome Elite) was employed. Obtained values were reported in g/L.
Optimization of culture conditions for GABA production
One-factor-at-a-time approach was followed to study the effect of alterations in culture media on GABA production (Thuy et al. 2020). Effect of initial medium pH (4, 5, 6, 7, and 8), MSG concentration (0, 1, 2, 3, 4, and 5%), time of fermentation (24, 48, 72, 96, and 120 h), and PLP supplementation (0, 100, 200, 300, 400, and 500 µM) on GABA production was evaluated. The amount of GABA produced in the culture medium was quantified using HPLC as mentioned above.
Genome sequencing, assembly, and gene annotations
Even though LAB6 and LAB19 showed GABA-producing ability, LAB6 showed better probiotic attributes and protective effects against LPS induced NO production in-vitro (Bhatia et al. 2023); hence, it was shortlisted for whole-genome sequencing and in-silico analysis. Utilizing the Illumina (NOVASEQ 6000) technology, the genomic DNA extracted from LAB6 was sequenced. CLC Genomics Workbench 21 was used for genome assembly, and Rapid Annotation Server and Technology (RAST) was used for genome annotation (Aziz et al. 2008). Existence of rRNA and tRNA, protein-coding genes were determined, and functions to detected genes were assigned. CRISPRCasFinder 4.2.20 tool was used to identify clustered regularly interspaced short palindromic repeats (CRISPRs) in the genome (Couvin et al. 2018). The presence of any antibiotic resistance genes was determined using Comprehensive Antibiotic Resistance Database (CARD) (Alcock et al. 2020). A circular genome map of the draft genome was generated using CGView server's Proksee software (Grant and Stothard 2008). The draft genome and assembly of the strain was deposited to GenBank (accession no. JARWBX000000000).
Amino acid comparison of GAD system genes
GAD operon homologous genes in the LAB6 genome were identified using the gadR, gadC, gadB, gts, and gadA gene sequences of L. brevis ATCC367 (GenBank: CP000416). Gene loci of putative GAD system genes in LAB6 were analyzed with the BLASTx program against NCBI databases. The GAD system protein sequences from various L. brevis strains were obtained from NCBI, and the protein sequences were aligned using MUSCLE alignment. Furthermore, a maximum-likelihood tree was built with 1000 replications in the bootstrap test using MEGA 11 (Tamura et al. 2021). Gene pathways associated with GABA, glutamate, and glutamine metabolism in LAB6 were identified using the KEGG database (Kanehisa and Goto 2000). The genomic features of LAB6 and 28 reference lactobacilli strains used in this study are listed in Table S3.
Pan/core-genome analysis
The BPGA tool was used to perform the pan-genome analysis (Chaudhari et al. 2016). To cluster the sequences, the USEARCH algorithm was used, with a sequence identity cut-off of 50% (Edgar 2010). For the generation of the Pan and Core-genome phylogeny, multiple sequence alignment was performed using MUSCLE, and the UPGMA tree was constructed (Edgar 2004). To identify the functional categories, clusters were annotated against KEGG (Kanehisa and Goto 2000) and COG (Tatusov et al. 2000) databases. The core cluster was defined as present in all genomes, the accessory cluster in more than one but not all, and the unique cluster as present in only a single genome.
Statistical analysis
GraphPad Prism 8.0 software (GraphPad Software Inc., CA, USA) was used for all the statistical analysis. Values were expressed as Mean ± SEM. All experiments to determine GABA production from the selected LAB strains were done in triplicates, and the data were analyzed using two-way ANOVA followed by Tukey's post hoc test.
Results and discussion
Identification of GABA-producing LAB
Among the 45 bacterial strains, isolates LAB6 (Levilactobacillus brevis) and LAB19 (Limosilactobacillus fermentum) clearly depicted GABA production with brightest spot based on the TLC, while LAB7 (Lactiplantibacillus plantarum) and LAB31 (Lactiplantibacillus plantarum) showed a hazy spot of GABA (Fig. 1); however, none of the Bifidobacterium strains could produce GABA. Further, to confirm the TLC results, GAD colorimetric assay was performed to confirm the GAD activity that catalyses the conversion of glutamate to GABA. This colorimetric assay employs green bromocresol dye, which is particularly sensitive to the utilization of proton and shift of pH, which is indicated by a transition of color from green to blue, qualitatively reveals the strains producing GABA (Lacroix et al. 2013; Santos-Espinosa et al. 2020). Upon conversion of glutamate to GABA, GAD expends one proton ion from the cytoplasm altering cytoplasmic pH, thereby sensitizing the indicator dye. The degree of color change indicates the activity of GAD and indirectly ability of bacterial strains to transform glutamate to GABA. Consequently, four GABA-producing strains were unambiguously allied to the presence and activation of the GAD enzyme as indicated by rapid GAD colorimetric assay. The selected strains appeared as regular smooth rods of about 1.2–2.2 µm long, with some shorter forms and occasional lengthened cells as observed in FE-SEM (Fig. 2).
TLC profiles of γ-aminobutyric acid (GABA) and monosodium glutamate (MSG) standards, MRS media supplemented with MSG, and different LAB strains supplemented with 2% MSG. Spots encircled with red represent strains with the highest GABA-producing ability and blue color encircled spots represent moderate GABA producers
GABA quantitation and detection of glutamate decarboxylase (GAD) gene from selected bacterial strains
Quantitative estimation of GABA yield from selected strains revealed conversion efficiencies of MSG to GABA were 63.4% (6.34 ± 0.05 g/L), 66% (6.60 ± 0.05 g/L), 6.2% (0.62 ± 0.08 g/L), and 4.4% (0.44 ± 0.05 g/L) in MRS supplemented with 10 g/L MSG by LAB6, LAB19, LAB7, and LAB31, respectively. Studies suggest that GABA yield is highest for L. brevis strains compared to other LAB species. GABA production range by L. brevis ranges from 15.0 mg/L to 205 g/L (Cui et al. 2020). Further, L. fermentum is less documented for the production of GABA, with a production range of 2.11 to 5.15 g/L (Woraharn et al. 2015; Lin et al. 2017). Figure S1 depicts the distinctive HPLC chromatographic peak of GABA in the standard and in the bacterial broth.
Further, the above-mentioned GABA-producing strains were investigated for gad gene. Primers built from a highly conserved region of the gad gene produced a PCR amplicon of approximately 540 bp (Fig. S2) for all strains that have positive GABA profiles (Siragusa et al. 2007). This is in congruence with the other studies reported earlier.
Despite the fact that the product was identified as GABA using TLC and HPLC, to authenticate the compound, its molecular weight must be determined. MS analysis was used to characterize the molecular weight of GABA. GABA emerged as [M + H]+ (hydrogen adduct of molecule 'M') under specific conditions. The mass spectra of the GABA standard revealed a prominent peak at 104.2853 m/z. The presence of a similar characteristic peak at 104.2852 m/z in the underivatized L. brevis LAB6 supernatant confirmed the production of GABA (Fig. S3A). The outcomes of the daughter scan from the precursor ions of [M + H]+ are shown in Fig. S3B which indicated a similar fragmentation pattern as that of standard GABA (Inoue et al. 2016).
Influence of culture conditions on GABA productivity in the MRS broth
Although bacterial glutamate decarboxylases are usually present in the cytoplasm, GABA and glutamate can interchange across membranes, causing GABA to accumulate extracellularly, although the molecular mechanisms remain unclear (Higuchi et al. 1997). The amount of extracellular GABA that bacteria produce varies depending on many fermentation variables. The most frequent and essential ones were optimized to produce the best yield of GABA, including pH, precursor concentration (MSG concentration), cultivation duration, and PLP supplementation.
Effect of initial pH
The bioconversion of glutamate to GABA in microbes is primarily pH driven phenomenon; therefore, it has the most significant effect during fermentation. By consuming H+ ions, bacterial amino acids decarboxylase is thought to play a fundamental role in maintaining an acidic pH (De Biase et al. 1999). The end product GABA is released stoichiometrically, and a proton is consumed due to the decarboxylation of glutamate in LAB. The overall result of this process is to make the cytoplasm and surroundings more alkaline to cope with acid stress. Evident from GAD physiology under acidic conditions suggesting the consumption of H + ion for GABA production, maintaining pH levels in the medium to the acidic range will significantly increase GABA production. To study the effect of pH on the production of GABA by L. brevis LAB6, L. fermentum LAB19 and its coculture, the initial pH of the media was adjusted to 4, 5, 6, 7, and 8 containing 2% MSG and incubated for 48 h at 37 °C. As shown in Fig. 3, pH plays a vital role in the production of GABA. The highest GABA production by L. brevis LAB6, L. fermentum LAB19, and its coculture reached 11 ± 0.22 g/L, 10.63 ± 0.33 g/L, and 10.45 ± 0.18 g/L, respectively when the strains were grown at pH 4.
Effect of initial pH of the growth medium on GABA production by L. brevis LAB6, L. fermentum LAB19 and its coculture. The data were analyzed by two-way ANOVA followed by Tukey’s post hoc test. The data were conducted in triplicate and expressed as mean ± SEM. a significant vs. pH 4, b significant vs. pH 5, c significant vs. pH 6, whereas d significant vs. pH 7 of LAB6, LAB19, and LAB6 + 19. The P value was < 0.0001 for all the groups. An increase in pH associated with a decline in GABA production could be observed
Further increase in the pH decreased GABA production drastically. These findings were consistent with the previous observations that the pH range between 4.0 and 5.0 was ideal for maintaining the GAD system by the LAB strains (Huang et al. 2007). A partial decrease in GAD activity may result from higher or lower pH levels. Generally, at higher pH, there was maximum loss of GAD activity, and GABA production was significantly less (Yao et al. 2021). Conclusively, initial pH affects the final yield of GABA, which is to be maintained adequately (Dhakal et al. 2012).
Effect of MSG concentration
Glutamate is the precursor molecule and an indispensable media additive for the formation of GABA by GAD in several LAB (Li and Cao 2010). The purpose of increasing MSG is to stimulate GAD to produce GABA through the GABA shunt route. Generally, the addition of MSG is more beneficial when added at the beginning of fermentation (0 h) rather than in between incubation (Lu et al. 2009). However, excessive supplementation with MSG could constrain cell growth and reduce GABA production (Villegas et al. 2016; Cui et al. 2020). The highest GABA was produced at 3% MSG supplementation with concentrations of 7.43±0.08 g/L, 8.76±0.093 g/L, and 7.68±0.08 g/L by L. brevis LAB6, L. fermentum LAB19, and its coculture when incubated for 48h at 37 °C. Coculturing of strains may not always increase the GABA yield (Jitpakdee et al. 2022), as seen in this study. This could be due to the concomitant usage of limited media additives by both the strains. Further increase in the MSG concentration was negatively correlated with the GABA yield, as illustrated in Fig. 4. Higher concentration of glutamate in the media poses toxic effect on the cells and suppresses the expression of gadB (Yogeswara et al. 2020).
Effect of MSG on GABA yield by L. brevis LAB6, L. fermentum LAB19 and its coculture. The data were analyzed by two-way ANOVA followed by Tukey’s post hoc test. The data were conducted in triplicate and expressed as mean ± SEM. a significant vs. 0% MSG, b significant vs. 1% MSG, c significant vs. 2% MSG, d significant vs. 3% MSG, whereas e significant vs. 4% MSG of LAB6, LAB19, and LAB6 + 19. The P value is < 0.0001 for all the groups
Effect of fermentation time
Fermentation time is another crucial factor of postbiotic production. The ability of the strains to produce GABA was assessed using a time-dependent measurement of extracellular GABA content in a culture medium-containing 2% MSG and incubated at 37 °C. Time-dependent yield of GABA is depicted in Fig. 5. The highest yield was obtained after 120 h of inoculation with concentrations of 9.91 ± 0.15 g/L, 9.41 ± 0.05 g/L, and 9.55 ± 0.17 g/L by L. brevis LAB6, L. fermentum LAB19, and its coculture. Similar to our observation, time-dependent production of GABA was observed in L. brevis CRL 1942 with maximum production at 120 h (Villegas et al. 2016).
Effect of incubation time on GABA yield by L. brevis LAB6, L. fermentum LAB19 and its coculture. The data were analyzed by two-way ANOVA followed by Tukey’s post hoc test. The data were conducted in triplicate and expressed as mean ± SEM. a significant vs. 24 h, b significant vs. 48 h, whereas c significant vs. 72 h of LAB6, LAB19, and LAB6 + 19. The P value is < 0.0001 for all the groups. Time-dependent increase in GABA production could be observed by individual strains and their coculture
Effect of pyridoxal 5′-phosphate (PLP) supplementation
The irreversible α-decarboxylation of glutamate by glutamic acid decarboxylase (GAD, EC 4.1.1.15) results in the production of GABA and PLP serves as a cofactor for the GAD (Huang et al. 2018). Hence, it is conceivable that adding PLP to the culture medium would enhance GABA synthesis. Analysis of the extracellular GABA content in L. brevis and L. fermentum culture broth with PLP at various concentrations containing 2% MSG and incubated for 48 h at 37 °C was evaluated to determine the effect of PLP supplementation. The highest concentration of GABA reached 9.53 ± 0.11 g/L and 9.20 ± 0.18 g/L by L. brevis and L. fermentum when grown with 400 and 300 μM PLP, respectively. Coculturing of strains resulted in a slight enhancement of GABA production of 9.71 ± 0.05 at 400 μM PLP (Fig. 6). Comparable to our findings, successive increase of GABA production with increasing PLP concentration was observed in L. brevis TCCC 13007 with a maximum yield at 300 μM PLP supplementation (Shi et al. 2017). Augmentation of GAD activity upon PLP supplementation was observed in L. fermentum YS2 (Lin et al. 2017). The outcomes demonstrated that PLP augmented the GABA yield and that its yield peaked at a given concentration of PLP, because an excess of PLP hindered the enzyme process and prevented it from binding with additional MSG (Fan et al. 2012).
Effect of PLP on GABA yield by L. brevis LAB6, L. fermentum LAB19 and its coculture. The data were analyzed by two-way ANOVA followed by Tukey’s post hoc test. The data were conducted in triplicate and expressed as mean ± SEM. a significant vs. 0 μM PLP, b significant vs. 100 μM PLP, c significant vs. 200 μM PLP of LAB6, LAB19, and LAB6 + 19. The P value is < 0.0001 for all the groups
Upon optimization of these factors, a threefold increase in GABA production was observed at pH 4, incubation time of 120 h in a medium supplemented with 3% of MSG and 400 μM of PLP for LAB6, and 300 μM of PLP for LAB19, LAB6, and LAB19 accumulated 19.67 ± 0.28 and 20.77 ± 0.14 g/L of GABA, respectively. Coculturing of both strains under optimized conditions did not further enhance GABA production beyond 20.02 ± 0.17 g/L.
Genomic analysis of LAB6
The principal features of the LAB6 genome are represented in Fig. 7. The genomic size of LAB6 was found to be 2,606,217 bp with a typical GC content of 45.6%, similar to its species based on RAST annotation. Genome annotation of LAB6 revealed the existence of 2552 predicted coding sequences (CDS), 6 rRNAs and 64 tRNAs out of total 70 RNAs, and 8 CRISPR sequences were detected by CRISPRCasFinder. The most prevalent functional subsystems, as determined by the functional classification of genes using the RAST subsystem, are carbohydrates, protein metabolism and cofactors, vitamins, prosthetic groups, and pigments Fig. 7B. It is imperative to monitor if a probiotic strain might horizontally transfer antibiotic resistance genes to the opportunistic or pathogenic microbes. A comprehensive scan of LAB6 genome against the CARD database of antibiotic resistance genes and mutations in antibiotic targets known to confer resistance was performed using the Resistance Gene Identifier programme. There were no such identified resistance genes found in the LAB 6 genome. The assembled genome of strain LAB6 was deposited in GenBank with accession number JARWBX000000000. LAB6 was deposited in the Microbial Type Culture Collection (MTCC), Chandigarh, India, under Budapest Treaty and was assigned MTCC 25662 as the culture accession number.
General genome feature of LAB6 having genome size of 2,606,217 bp with a typical GC content of 45.6%; A subsystem category distribution having carbohydrates, protein metabolism and cofactors, vitamins, prosthetic groups, and pigments as most prevalent functional systems (B). The green bar represents the percentage of proteins that could be annotated by the RAST Server, and blue bar represents the proteins that were not annotated. The pie chart illustrates the percentage of proteins annotated to each subsystem category, and adjacent to it are subsystem categories listed in the legend from top to bottom and A graphical circular map of the genome performed with the Proksee tool (C). The outermost ring 1 (Forward strand) and ring 3 (Reverse strand) represent the protein-coding regions; rRNA, tRNA, and tmRNA are indicated. Ring 2 (gray) represents the contigs. Ring 4 represents the G + C content, and ring 5 represents GC skew
Genes responsible for probiotic attributes
Genes for survival under gastrointestinal conditions
A probiotic strain must endure several challenges to thrive and exhibit intended biological actions in the gastrointestinal tract of the host (Garcia-Gonzalez et al. 2022). The expression of bile salt hydrolase genes or the capacity to withstand stressful conditions imposed by the host are essential considerations when looking for suitable probiotic bacterial candidates. In silico analysis suggested that LAB6 genome encodes genes for several stress-related proteins, such as the F0F1-ATPases, GAD, HrcA-DnaK-DnaJ-GrpE operon, and GroES-GroEL chaperonin as the principal regulators of intracellular pH. Besides this, LAB6 genome possesses two choloylglycine hydrolases, two Na + /H + antiporters, and tlyC1 genes to thrive under bile stress (Table S2).
Host adhesion proteins
Surface adhesion is an essential paradigm in the survival of the commensal organism to implicate its beneficial effects and for the competitive exclusion of opportunistic pathogens from adhesion. Genes encoding adhesion proteins to adhere to collagen, fibronectin, mucus, and epithelial cells were also present in the LAB6 genome (Table S2).
GAD system in L. brevis LAB6
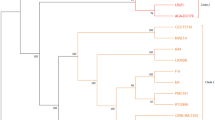
The genetic elements of the GAD arrangement in various reference strains of L. brevis were characterized. A maximum-likelihood tree was built to study the GAD genes of lactobacilli using the bootstrap test with 1000 replications (Fig. 8A). The architecture of GAD operon in LAB6 is distinct from the GAD gene clusters that have been previously identified in L. lactis (Sanders et al. 1998), L. acidophilus (Azcarate-Peril et al. 2004) and L. reuteri (Teixeira et al. 2014). Sequence analysis of LAB6 revealed the presence of two distinct GAD-encoding genes, gadA and gadB, that are characteristic of L. brevis strains (Wu et al. 2017). Monosodium glutamate is converted to GABA by the two isoforms, gadA or gadB that share comparable activities. Additionally, the two isoforms of the GAD were biochemically similar (De Biase et al. 1999). gadA is positioned apart from the other GAD genes, while gadB is connected to the glutamate/GABA antiporter gene (gadC) in the GAD operon of L. brevis (Lyu et al. 2018); however, the genome of L. brevis M10 strain had no gadA/gadB gene.
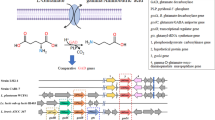
Analysis of GAD operon in LAB6 and reference strains of L. brevis. The maximum-likelihood tree is based on amino acid sequences of GAD genes (gadA, gadB, gadC) (A). Bootstrap values higher than 70% are shown at branch points. The length of GAD genes and its GenBank accession numbers from each strain are shown in brackets. Arrangements of GAD operon genes from LAB6 and Levilactobacillus brevis ATCC 367 strain. gadR, transcriptional regulator; antiporter gene gadC, glutamate/GABA antiporter gene; gadA/gadB, GAD genes; gts, glutamyl-tRNA synthetase gene (B) and gene pathways in LAB6 as identified via KEGG (https://www.genome.jp/kegg/kegg2.html) associated with GABA, glutamate, and glutamine metabolism, and essential genes for the enzymes needed to produce GABA from glutamate and glutamine were found in LAB6 (C)
The genes responsible for bioconversion of glutamate to GABA in L. brevis, including the canonical GAD operon, gadA (468 aa) and gadB (480 aa), and a glutamate/GABA antiporter gene, gadC (501 aa) are shown in Fig. 8B. Moreover, glutamyl-tRNA synthetase (gts, 504 aa) and transcriptional regulator (gadR, 197 aa) also presented in the GAD operon of LAB6 (Fig. 8B). RNA polymerase sigma factor (rpoD) that directly or indirectly regulates transcription of GAD (Tramonti et al. 2002) was also present in LAB6 genome. Glutamate is taken up by bacteria through the surface protein transporter GadC, and is converted to GABA by glutamate decarboxylase. Additionally, glutamate and glutamine are interconvertible (EC 6.3.1.2 and 1.4.1.13, respectively). The genes for the enzymes needed to produce GABA from glutamate and glutamine were found in LAB6 (Fig. 8C).
Comparative genomic analysis
A comparative genomic analysis of LAB6 with 28 completely sequenced L. brevis strains was performed. Pan-genome analysis revealed a core-genome set of 924 genes-families, predominantly encoding essential proteins for carbohydrate metabolism (Fig. S4), along with a dispensable genome set of 4264 gene families, resulting in a pan-genome of 5188 gene families. Figure 9A shows a reduction in core-genome size while pan-genome size continued to increase. The power fit equation's y value of 0.29 suggests an open pan-genome for the analyzed L. brevis strains (it can also be observed in the new gene plot Fig. S5) (Sun et al. 2015; Gao et al. 2019). LAB6 possesses 95 strain-specific genes-families, which is significantly higher than the majority of other L. brevis strains (Fig. 9B).
Conclusions
This study led to the identification of two high GABA-producing strains, Levilactobacillus brevis LAB6 (MTCC 25662) and L. fermentum LAB19 that were earlier isolated from soy-based fermented food and infant feces, respectively. Under optimized production parameters of initial pH of 4.0 and 3% MSG, LAB6 was able to produce 19.7 ± 0.3 g/L of GABA with 400 μM of PLP while LAB19 produced 20.8 ± 0.1 g/L of GABA with 300 μM of PLP in MRS broth incubated for 120 h at 37 °C under aerobic conditions. The whole-genome sequencing of LAB6 revealed the presence of canonical GAD operon genes gadA, gadB, and gadC for GABA production, genes for acid, bile tolerance, and genes for host adhesion, making it a potential probiotic candidate intended for modulating the host’s physiological processes.
Data availability
The data sets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request. Whole genome sequencing data presented in this study are openly available in The National Centre for Biotechnology Information (NCBI).
References
Alcock BP, Raphenya AR, Lau TTY et al (2020) CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48:D517–D525. https://doi.org/10.1093/nar/gkz935
Azcarate-Peril MA, Altermann E, Hoover-Fitzula RL et al (2004) Identification and inactivation of genetic loci involved with Lactobacillus acidophilus acid tolerance. Appl Environ Microbiol 70:5315–5322. https://doi.org/10.1128/AEM.70.9.5315-5322.2004
Aziz RK, Bartels D, Best AA et al (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genom 9:75. https://doi.org/10.1186/1471-2164-9-75
Bauer KC, Huus KE, Finlay BB (2016) Microbes and the mind: emerging hallmarks of the gut microbiota-brain axis: emerging hallmarks of the gut microbiota-brain axis. Cell Microbiol 18:632–644. https://doi.org/10.1111/cmi.12585
Bhatia R, Singh S, Maurya R et al (2023) In vitro characterization of lactic acid bacterial strains isolated from fermented foods with anti-inflammatory and dipeptidyl peptidase-IV inhibition potential. Braz J Microbiol 54:293–309. https://doi.org/10.1007/s42770-022-00872-5
Chaudhari NM, Gupta VK, Dutta C (2016) BPGA- an ultra-fast pan-genome analysis pipeline. Sci Rep 6:24373. https://doi.org/10.1038/srep24373
Cho SY, Park MJ, Kim KM et al (2011) Production of high γ-aminobutyric acid (GABA) sour kimchi using lactic acid bacteria isolated from mukeunjee kimchi. Food Sci Biotechnol 20:403–408. https://doi.org/10.1007/s10068-011-0057-y
Chua J-Y, Koh MKP, Liu S-Q (2019) Gamma-aminobutyric acid. In: Sprouted Grains. Elsevier, pp 25–54
Couvin D, Bernheim A, Toffano-Nioche C et al (2018) CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res 46:W246–W251. https://doi.org/10.1093/nar/gky425
Cui Y, Miao K, Niyaphorn S, Qu X (2020) Production of gamma-aminobutyric acid from lactic acid bacteria: a systematic review. Int J Mol Sci 21:995. https://doi.org/10.3390/ijms21030995
De Biase D, Tramonti A, Bossa F, Visca P (1999) The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol Microbiol 32:1198–1211. https://doi.org/10.1046/j.1365-2958.1999.01430.x
Demirbaş F, İspirli H, Kurnaz AA et al (2017) Antimicrobial and functional properties of lactic acid bacteria isolated from sourdoughs. Lebenson Wiss Technol 79:361–366. https://doi.org/10.1016/j.lwt.2017.01.067
Dhakal R, Bajpai VK, Baek K-H (2012) Production of gaba (γ - Aminobutyric acid) by microorganisms: a review. Braz J Microbiol 43:1230–1241. https://doi.org/10.1590/S1517-83822012000400001
Diana M, Quílez J, Rafecas M (2014) Gamma-aminobutyric acid as a bioactive compound in foods: a review. J Funct Foods 10:407–420. https://doi.org/10.1016/j.jff.2014.07.004
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Fan E, Huang J, Hu S et al (2012) Cloning, sequencing and expression of a glutamate decarboxylase gene from the GABA-producing strain Lactobacillus brevis CGMCC 1306. Ann Microbiol 62:689–698. https://doi.org/10.1007/s13213-011-0307-5
Gao D, Chang K, Ding G et al (2019) Genomic insights into a robust gamma-aminobutyric acid-producer Lactobacillus brevis CD0817. AMB Express 9:72. https://doi.org/10.1186/s13568-019-0799-0
Garcia-Gonzalez N, Bottacini F, van Sinderen D et al (2022) Comparative genomics of Lactiplantibacillus plantarum: Insights into probiotic markers in strains isolated from the human gastrointestinal tract and fermented foods. Front Microbiol 13:854266. https://doi.org/10.3389/fmicb.2022.854266
Gong L, Ren C, Xu Y (2019) Deciphering the crucial roles of transcriptional regulator GadR on gamma-aminobutyric acid production and acid resistance in Lactobacillus brevis. Microb Cell Fact 18:108. https://doi.org/10.1186/s12934-019-1157-2
Grant JR, Stothard P (2008) The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36:W181–W184. https://doi.org/10.1093/nar/gkn179
Higuchi T, Hayashi H, Abe K (1997) Exchange of glutamate and gamma-aminobutyrate in a Lactobacillus strain. J Bacteriol 179:3362–3364. https://doi.org/10.1128/jb.179.10.3362-3364.1997
Hoover DG (2014) Bifidobacterium. In: Encyclopedia of Food Microbiology. Elsevier, pp 216–222
Huang J, Mei L, Sheng Q et al (2007) Purification and characterization of glutamate decarboxylase of Lactobacillus brevis CGMCC 1306 isolated from fresh milk. Chin J Chem Eng 15:157–161. https://doi.org/10.1016/s1004-9541(07)60051-2
Huang J, Fang H, Gai Z-C et al (2018) Lactobacillus brevis CGMCC 1306 glutamate decarboxylase: crystal structure and functional analysis. Biochem Biophys Res Commun 503:1703–1709. https://doi.org/10.1016/j.bbrc.2018.07.102
Inoue K, Miyazaki Y, Unno K et al (2016) Stable isotope dilution HILIC-MS/MS method for accurate quantification of glutamic acid, glutamine, pyroglutamic acid, GABA and theanine in mouse brain tissues: HILIC-MS/MS assay of Glu analogs in mouse brain. Biomed Chromatogr 30:55–61. https://doi.org/10.1002/bmc.3502
Jitpakdee J, Kantachote D, Kanzaki H, Nitoda T (2022) Potential of lactic acid bacteria to produce functional fermented whey beverage with putative health promoting attributes. Lebenson Wiss Technol 160:113269. https://doi.org/10.1016/j.lwt.2022.113269
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. https://doi.org/10.1093/nar/28.1.27
Krulwich TA, Sachs G, Padan E (2011) Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol 9:330–343. https://doi.org/10.1038/nrmicro2549
Lacroix N, St-Gelais D, Champagne CP, Vuillemard JC (2013) Gamma-aminobutyric acid-producing abilities of lactococcal strains isolated from old-style cheese starters. Dairy Sci Technol 93:315–327. https://doi.org/10.1007/s13594-013-0127-4
Li H, Cao Y (2010) Lactic acid bacterial cell factories for gamma-aminobutyric acid. Amino Acids 39:1107–1116. https://doi.org/10.1007/s00726-010-0582-7
Li H, Qiu T, Huang G, Cao Y (2010) Production of gamma-aminobutyric acid by Lactobacillus brevis NCL912 using fed-batch fermentation. Microb Cell Fact 9:85. https://doi.org/10.1186/1475-2859-9-85
Lin Q, Li D, Qin H (2017) Molecular cloning, expression, and immobilization of glutamate decarboxylase from Lactobacillus fermentum YS2. Electron J Biotechnol 27:8–13. https://doi.org/10.1016/j.ejbt.2017.03.002
Lu X, Xie C, Gu Z (2009) Optimisation of fermentative parameters for GABA enrichment by Lactococcus lactis. Czech J Food Sci 27:433–442. https://doi.org/10.17221/45/2009-cjfs
Lyu C, Zhao W, Peng C et al (2018) Exploring the contributions of two glutamate decarboxylase isozymes in Lactobacillus brevis to acid resistance and γ-aminobutyric acid production. Microb Cell Fact 17:180. https://doi.org/10.1186/s12934-018-1029-1
Mann EO, Paulsen O (2007) Role of GABAergic inhibition in hippocampal network oscillations. Trends Neurosci 30:343–349. https://doi.org/10.1016/j.tins.2007.05.003
Prévot T, Sibille E (2021) Altered GABA-mediated information processing and cognitive dysfunctions in depression and other brain disorders. Mol Psychiatry 26:151–167. https://doi.org/10.1038/s41380-020-0727-3
Rashmi D, Zanan R, John S, et al (2018) γ-Aminobutyric Acid (GABA): Biosynthesis, Role, Commercial Production, and Applications. In: Studies in Natural Products Chemistry. Elsevier, pp 413–452
Sanders JW, Leenhouts K, Burghoorn J et al (1998) A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol Microbiol 27:299–310. https://doi.org/10.1046/j.1365-2958.1998.00676.x
Santos-Espinosa A, Beltrán-Barrientos LM, Reyes-Díaz R et al (2020) Gamma-aminobutyric acid (GABA) production in milk fermented by specific wild lactic acid bacteria strains isolated from artisanal Mexican cheeses. Ann Microbiol. https://doi.org/10.1186/s13213-020-01542-3
Sharma S, Singh S, Chaudhary V et al (2022) Isomaltooligosaccharides utilization and genomic characterization of human infant anti-inflammatory Bifidobacterium longum and Bifidobacterium breve strains. 3 Biotech 12:89. https://doi.org/10.1007/s13205-022-03141-2
Shi X, Chang C, Ma S et al (2017) Efficient bioconversion of L-glutamate to γ-aminobutyric acid by Lactobacillus brevis resting cells. J Ind Microbiol Biotechnol 44:697–704. https://doi.org/10.1007/s10295-016-1777-z
Singh S, Bhatia R, Khare P et al (2020) Anti-inflammatory Bifidobacterium strains prevent dextran sodium sulfate induced colitis and associated gut microbial dysbiosis in mice. Sci Rep 10:18597. https://doi.org/10.1038/s41598-020-75702-5
Siragusa S, De Angelis M, Di Cagno R et al (2007) Synthesis of gamma-aminobutyric acid by lactic acid bacteria isolated from a variety of Italian cheeses. Appl Environ Microbiol 73:7283–7290. https://doi.org/10.1128/AEM.01064-07
Sun Z, Harris HMB, McCann A et al (2015) Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun 6:8322. https://doi.org/10.1038/ncomms9322
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027. https://doi.org/10.1093/molbev/msab120
Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. https://doi.org/10.1093/nar/28.1.33
Teixeira JS, Seeras A, Sanchez-Maldonado AF et al (2014) Glutamine, glutamate, and arginine-based acid resistance in Lactobacillus reuteri. Food Microbiol 42:172–180. https://doi.org/10.1016/j.fm.2014.03.015
Thuy DTB, Nguyen A, Khoo KS et al (2020) Optimization of culture conditions for gamma-aminobutyric acid production by newly identified Pediococcus pentosaceus MN12 isolated from “mam nem”, a fermented fish sauce. Bioengineered 12:54–62. https://doi.org/10.1080/21655979.2020.1857626
Tramonti A, Visca P, De Canio M et al (2002) Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J Bacteriol 184:2603–2613. https://doi.org/10.1128/JB.184.10.2603-2613.2002
Villegas JM, Brown L, Savoy de Giori G, Hebert EM (2016) Optimization of batch culture conditions for GABA production by Lactobacillus brevis CRL 1942, isolated from quinoa sourdough. Lebenson Wiss Technol 67:22–26. https://doi.org/10.1016/j.lwt.2015.11.027
Woraharn S, Lailerd N, Sivamaruthi BS et al (2015) Evaluation of factors that influence the L-glutamic and γ-aminobutyric acid production during Hericium erinaceus fermentation by lactic acid bacteria. CyTA - J Food 14:47–54. https://doi.org/10.1080/19476337.2015.1042525
Wu Q, Tun HM, Law Y-S et al (2017) Common distribution of gad operon in Lactobacillus brevis and its GadA contributes to efficient GABA Synthesis toward Cytosolic near-neutral pH. Front Microbiol 8:206. https://doi.org/10.3389/fmicb.2017.00206
Yao L-L, Cao J-R, Lyu C-J et al (2021) Food-grade γ-aminobutyric acid production by immobilized glutamate decarboxylase from Lactobacillus plantarum in rice vinegar and monosodium glutamate system. Biotechnol Lett 43:2027–2034. https://doi.org/10.1007/s10529-021-03164-4
Yogeswara IBA, Kittibunchakul S, Rahayu ES et al (2020) Microbial production and enzymatic biosynthesis of γ-aminobutyric acid (GABA) using Lactobacillus plantarum FNCC 260 isolated from Indonesian fermented foods. Processes (basel) 9:22. https://doi.org/10.3390/pr9010022
Zhu Z, Shi Z, Xie C et al (2019) A novel mechanism of Gamma-aminobutyric acid (GABA) protecting human umbilical vein endothelial cells (HUVECs) against H2O2-induced oxidative injury. Comp Biochem Physiol C Toxicol Pharmacol 217:68–75. https://doi.org/10.1016/j.cbpc.2018.11.018
Zhuang K, Jiang Y, Feng X et al (2018) Transcriptomic response to GABA-producing Lactobacillus plantarum CGMCC 1.2437T induced by L-MSG. PLoS One 13:e0199021. https://doi.org/10.1371/journal.pone.0199021
Acknowledgements
The authors would like to thank the Department of Biotechnology (DBT), Government of India and National Agri-Food Biotechnology Institute (NABI) for providing the research grant and the facilities. KK, KC, SRJ, and MB would like to acknowledge DBT project NERBPMC (BT/PR16088/NE/95/69/2015 NER-DBT) for providing the research grant. DeLCON (DBT e-library consortium) is highly acknowledged for providing access to the journals to NABI.
Funding
This research was funded by the Department of Biotechnology (DBT), Government of India, for the research grant given to the National Agri-Food Biotechnology Institute (NABI), Kanthi Kiran Kondepudi & Mahendra Bishnoi, Kanwaljit Chopra and Santa Ram Joshi.
Author information
Authors and Affiliations
Contributions
TM conducted the experiments and performed the whole-genome analysis and in-silico data analysis; RB isolation of Lactobacillus strains; KK, MB, KC, and SRJ provided the financial support for carrying out the research; KK, MB, and KC conceptualized, designed the experiments, edited the manuscript, and got funding to carry out the study. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Not applicable.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Matta, T., Bhatia, R., Joshi, S.R. et al. GABA synthesizing lactic acid bacteria and genomic analysis of Levilactobacillus brevis LAB6. 3 Biotech 14, 62 (2024). https://doi.org/10.1007/s13205-024-03918-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-024-03918-7