Abstract
This study was carried out to understand the probiotic features, ability to utilize non-digestible carbohydrates and comparative genomics of anti-inflammatory Bifidobacterium strains isolated from human infant stool samples. Bacterial strains were isolated from the stool samples using serial dilution on MRS agar plates supplemented with 0.05% l-cysteine hydrochloride and mupirocin. Molecular characterization of the strains was carried out by 16S rRNA gene sequencing. Anti-inflammatory activity was determined using TNF-α and lipopolysaccharide (LPS) induced inflammation in Caco2 cells. Probiotic attributes were determined as per the established protocols. Isomaltooligosaccharides (IMOS) utilization was determined in the broth cultures. Whole genome sequencing and analysis was carried out for three strains. Four obligate anaerobic, Gram positive Bifidobacterium strains were isolated from the infant stool samples. Strains were identified as Bifidobacterium longum Bif10, B. breve Bif11, B. longum Bif12 and B. longum Bif16. The strains were able to prevent inflammation in the Caco2 cells through lowering of IL8 production that was caused by TNF-α and LPS treatment. The strains exhibited desirable probiotic attributes such as acid and bile tolerance, mucin binding, antimicrobial activity, bile salt hydrolase activity, cholesterol lowering ability and could ferment non-digestible carbohydrates such as isomaltooligosaccharides and raffinose. Furthermore, Isomaltooligosaccharides supported the optimum growth of the strains in vitro, which was comparable to that on glucose. Strains could metabolize IMOS through cell associated α-glucosidase activity. Genomic features revealed the presence of genes responsible for the utilization of IMOS and for the probiotic attributes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Shifting eating habits from traditional to the westernized diets and irrational use of antibiotics causes dysbiosis in the gut microbiota and associated diseases, such as obesity, insulin resistance, type 2 diabetes, cancer, mental illnesses, acute and chronic inflammatory conditions, such as ulcerative colitis, irritable bowel disease (IBD) (Woodmansey 2007; Valdes et al. 2018). Foods that are rich in dietary fibers, probiotics, prebiotics and their combination as synbiotics help in the restoration or replenishment of the microbiota and decreasing the disease burden (Kondepudi et al. 2012; Gagliardi et al. 2018). Probiotics are the live microorganisms and when administered in an adequate amount has been suggested to improve the host’s health. Among the widely studied probiotic bacteria, Bifidobacterium genus inhabit the gut of humans and animals with higher abundance of B. longum, B. breve, and B. bifidum during human infancy, while ageing population has higher abundance of B. catenulatum, B. adolescentis and B. longum spp due to high intake of plant-derived carbohydrates (Arboleya et al. 2016). Bifidobacterium is largely known to boost the innate and adaptive immunity (Ruiz et al. 2017), suppress pathogen invasion by competitive exclusion (Bermudez-Brito et al. 2012), production of antimicrobial peptides (Cazorla et al. 2018) and short chain fatty acids (Usta-Gorgun and Yilmaz-Ersan 2020), besides utilizing non-digestible carbohydrate polymers and oligomers (Fu et al. 2019). Studies on animals and humans suggested the protective efficacy of some of the probiotic strains against enteric infections, antibiotic associated diarrhea (AAD), intestinal bowel disease IBD, ulcerative colitis and colorectal cancer studies (Kajander et al. 2008). Recent studies also suggest that Bifidobacterium strains could promote metabolic and mental health by preventing the loss of intestinal barrier function and suppressing gut inflammation (Sarkar et al. 2016). Some of the studies on Indian origin Bifidobacterium strains includes that on acid resistant and immuno-modulatory Bifidobacterium breve NCIM 5671, Bifidobacterium longum NCIM 5672 and Bifidobacterium bifidum NCIM 5697 isolated from Indian infants (Achi et al. 2019; Rohith and Halami, 2021; Sundararaman et al. 2021) and a Bifidobaacterium enriched ice cream (Kataria et al. 2018).
Prebiotics are non-digestible substances, especially oligosaccharides, which are metabolized by the gut microbes and modulate the composition and/or activity of the gut microbiome, thus conferring a beneficial physiological effect on the host (Guarino et al. 2020). International Society for Probiotics and Prebiotics recommended a consensus definition for prebiotics as a substrate which is selectively utilized by the host microorganisms (Gibson et al. 2017). Gut microbial action on prebiotics generate acetate, butyrate and propionate, the key secondary metabolites, in the gut and those can help in maintaining gut-barrier homeostasis, mucus production, gut hormone secretion, systemic beneficial effects, improving calcium and magnesium availability and majorly inhibition of enteric pathogens (Turroni et al. 2016). Several studies suggested that the prebiotics promote the bifidobacterial growth (bifidogenic effect) in the gut (Meyer and Stasse-Wolthuis 2009; Kumar et al. 2020). As there are limited studies on the utility of Bifidobacterium strains isolated from Indian infants for their health promoting properties, the present study was aimed at the isolation of Bifidobacterium strains that could prevent TNF-α and LPS induced intestinal epithelial cell inflammation using in vitro studies, their probiotic characterization and prebiotic preferences.
Materials and methods
Materials
MRS broth, MRS agar, phenol, lactic acid (HPLC grade) and other chemicals were purchased from HiMedia Laboratories, Mumbai, India. The DNA isolation kit was purchased from ZR Zymo (Zymo Research Corporation, CA, USA). Mucin type III from porcine stomach, fructooligosaccharides (FOS), inulin from chicory, carboxyfluorescein diacetate (CFDA), 2-Nitrophenyl α‐d-galactopyranoside (PNP-α‐d-galactose), 2-Nitrophenyl β‐d-galactopyranoside (PNP-β‐d-galactose) and volatile Short chain fatty acid Mix (HPLC) were purchased from Sigma-Aldrich, St Louis, USA. Isomaltooligosaccharides (IMOS) was a gift from BioNeutra, Alberta, Canada. Xylene, chloroform, hexane, and hydrochloric acid were purchased from Merck, Kenilworth, USA. API CHL 50 kit purchased from BioMérieux, Craponne, France. Ethanol estimation kit was purchased from Megazyme International Ireland Limited (Pro Lab Marketing PVT. LTD., New Delhi, India). Agilent Hi-Plex columns (Zorbax Hi-Plex H, 300 × 7.7 mm HPLC column; 8 μm particle size and Hi-Plex H cartridge, 3 × 5 mm internal diameter) were purchased from Agilent technologies, Santa Clara, USA. Caco-2 cell line was purchased from the National Centre for Cell Science, Pune, India. Anoxomat and anoxomat jars used for creating anaerobic conditions were purchased from Mart Microbiology (Model no: AN2CTS), Drachten, The Netherlands.
Methods
Isolation and identification of Bifidobacterium strains
Fecal samples were collected from Indian infants of the Tricity region (2–4 months, born by natural delivery) as per the laws and guidelines of the Post Graduate Institute of Medical Education and Research (PGIMER), Chandigarh and after the institute’s ethical committee approval (No. P-641) and informed consent from the parents. Fecal samples were thoroughly homogenized and were serially diluted on MRS agar plates supplemented with 0.05% (w/v) each of l-Cysteine hydrochloride and mupirocin. Plates were incubated under anaerobic conditions (80% N2, 10% CO2 and 10% H2) in an anoxomat jar at 37 °C for 48 h. A single colony of the strains was streaked on the fresh MRSC agar plates to obtain the pure cultures and the isolated strains were subcultured and stored as glycerol stock cultures at − 80 °C. A loop full of glycerol stock of the strains was activated on MRSC plates and incubated at 37 °C for 48 h under anaerobic conditions. A single colony of the strain was inoculated into fresh pre-reduced MRSC broth and incubated at 37 °C for 24 h and transferred three times. The cultures were centrifuged at 9000×g for 10 min at 4 °C and the pellets obtained were washed and suspended in sterile phosphate buffer saline with 0.05% l-cysteine (PBSC) and used in all the experiments.
Molecular characterization: Strains were grown in the MRSC broth till logarithmic growth and the genomic DNA was extracted using ZR Zymo DNA isolation kit as per manufacturer’s instructions. The presence of fructose 6-phosphate phosphoketolase gene was determined using PCR amplification using XFPF 5′-CGGCCACGGCTGGGGCC-3′ and XFPR 5′-TCCTGACGCCAGACGTGGG-3′ primers. PCR amplification of the 16S rRNA gene was carried out using universal primers 27F 5′-AGAGTTTGATCCTGGCTGAG 3′, 1492R-5′ GGTTACCTTGTTACGACTT 3′ primers. The amplicons were sequenced and the sequences were subjected to nucleotide blast using the NCBI database server. Accession numbers for the 16S rRNA gene sequences for B. longum Bif10, B. breve Bif11, B. longum Bif12 and B. longum Bif16 were MK850096, MK850097, MK850098 and MK910584, respectively, and are available in the GenBank. B. longum Bif10, B. breve Bif11 and B. longum Bif16 were deposited under Budapest treaty at Microbial Type Culture Collection (MTCC), CSIR-IMTECH, Chandigarh with MTCC 25245, MTCC 25246 and MTCC 25247 as the accession numbers (Supplementary Table 1).
Biochemical tests
Catalase, Haemolytic pattern and antibiotic susceptibility were determined as per the standard biochemical assays for bacteria. Degradation of gastric mucin by the Bifidobacterium strains was evaluated using an agar plate method developed by Zhou et al. (2001). Carbohydrate fermentation pattern of the strains was determined using API CHL 50 kit as per the manufacturer’s instructions and the change in colour of the medium was recorded.
In vitro protective effect on human intestinal epithelial cell (Caco2) inflammation: Caco2 cells were cultured in DMEM containing 10% fetal bovine serum and 1% penicillin–streptomycin at 37 °C under 5% CO2 in a humidified incubator. Media was changed on every second day until confluency was attained (15 days). Caco2 cells were treated with the four Bifidobacterium strains for 24 h and the viability was determined using the MTT assay kit as per the manufacturer’s instructions. The ability of the strains to prevent the TNF-α and LPS-induced inflammation in the Caco2 cells was determined as described by Singh et al. (2018). Briefly, the confluent Caco2 cells were treated with TNF-α (100 ng/mL) for 24 h followed by LPS alone (to induce inflammation) or with a combination of LPS and test strains of OD600nm of 1.0 (OD600nm 1.0 corresponds to 1010 CFU as determined by plate count method) in DMEM for another 24 h. Later, the supernatants were collected from respective treatments and the level of IL-8 was determined using commercially available IL-8 measurement kit as per the manufacturer’s instructions.
In-vitro probiotic characterization:
Acid and bile tolerance, anti-microbial activity, cholesterol lowering activity, bile salt hydrolase (BSH) activity, adhesion to mucin, auto-aggregation and salt aggregation were determined as described elsewhere (Singh et al. 2018). Detailed description is given in Supplementary methods.
Prebiotic Profiling
This was determined for newly isolated strains by agar plate and batch fermentation assays. B. breve Bif40 isolated from VSL#3 was used as a reference strain for B. breve Bif11 and B. longum JCM 1217 procured from Riken Culture Collection, Japan was used as reference strain for B. longum Bif10, B. longum Bif12 and B. longum Bif16.
Agar plate assay
Single colony of the strains was spotted on 0.5% (v/v) prebiotic–MRSC plates (pH 6.8) supplemented either with IMOS, FOS, inulin, starch or resistant starch (RS). Color change from red to yellow of phenol red as pH indicator was recorded and a score was given from 1 to 3 depending upon width (mm) of the zone around the spot.
Batch Fermentation in IMOS and raffinose containing medium
Bacterial strains were inoculated into MRSC-basal broth (MRSC-BB) supplemented with either 1% of IMOS or raffinose as a prebiotic source. Growth of the strains in MRSC-BB supplemented with 1% glucose was used as a positive control, while growth in MRSC-BB without any carbohydrate source was used as a negative control.
Growth profiles in IMOS
The bacterial cells were adjusted to an initial OD600nm of 0.05 units in 5 mL of MRSC-BB supplemented with 1% each of glucose, IMOS and raffinose separately and were incubated under anaerobic conditions in anoxomat jars at 37 °C. Growth was determined by measuring absorbance at OD600nm for every 12 h interval up to 72 h. Doubling time in each carbohydrate substrate was determined in 96 well plates by measuring OD600nm for every 4 h up to 16 h.
Doubling time was calculated using following formula:
where r is the growth rate which is calculated using formula:
Change in pH was determined in the broth after 72 h of incubation for each strain. The relative bacterial growth (%) for each strain in IMOS and raffinose was determined by comparing the OD600nm values of glucose grown cells, which was considered as 100%.
Biomass (g/L) of the strains obtained in MRSC broth supplemented with 1% either of glucose or IMOS was determined by subtracting the initial dry weight of washed cells at time zero from the dry weight of the cells in the log phase of growth (36 h). Total carbohydrate utilization by the strains in glucose and IMOS was determined using phenol sulphuric acid method (Dubois et al. 1956).
Production of hydrolytic enzymes: Bacterial strains were grown in MRSC-BB supplemented with either 1% of glucose, IMOS or raffinose under anaerobic conditions at 37 °C for 24 h. Bacterial pellets were washed, resuspended in 1 mL of PBSC and the optical density was adjusted to 1.0. Live cells were sonicated (QSonica, LLC, 53 Church Hill RD. Newtown, CT, USA) and centrifuged to obtain the intracellular and cell debris fractions. α-Glucosidase, α-galactosidase and β-galactosidase activities were determined in the whole cells, intracellular extract and cell debris. Briefly, to a mixture of 125 µl of 2.0 mM PNP-α‐d-galactose, PNP-α‐d-glucose or PNP-β‐d-galactose, 75 µl autoclave MQ water, a 50 µl of bacterial suspension or intracellular extracts or cell debris was added and incubated at 37 °C in a circulating water bath for 30 min. The reaction was stopped by adding 1 mL of 1.0 M sodium carbonate solution and the absorbance was measured at 405 nm. Results obtained were expressed as 1 μmol of nitrophenol released per minute under the assay conditions.
Production of acetate, lactate and ethanol
Acetate and lactate production by the Bifidobacterium strains were determined using high performance liquid chromatography (HPLC) as described by Singh et al. (2016). Full methodology is given in Supplementary methods. Ethanol production was estimated using commercially available ethanol estimation kit as per the manufacturer’s instructions.
Effect of growth in IMOS on acid and bile tolerance, adhesion to mucin and cell surface hydrophobicity:
Bifidobacterium strains were grown in 1% either of glucose or IMOS. Acid and bile tolerance, mucin adhesion and cell surface hydrophobicity were determined as described by Singh et al. (2018). Full methodology is given in Supplementary methods.
Genome sequencing, assembly, and gene annotations and comparative genomic analysis:
As the strain B. longum Bif12 and B. longum Bif16 were isolated from the same fecal sample, strains B. longum Bif10, B. breve 11 and B. longum Bif16 were short listed for whole genome sequencing and for in-silico studies. The genomic DNA isolated from these strains was sequenced using the Illumina-HiSeq 1000 technology and the genome assembly was performed in CLC Genomics Workbench (Chander et al. 2020). The genome annotations for B. longum Bif10, B. breve Bif11 and B. longum Bif16 were performed using Rapid Annotation Server and Technology (RAST) (Aziz et al. 2008). Comparative genomics to evaluate presence or absence of a particular gene and in silico search of candidate genes responsible for probiotic traits was carried out in RAST annotated genomes and was further confirmed by BLASTn to analyze genes for probiotic properties. BLASTp was used to predict the presence of specific genes or their homologues in respective genome sequences available at the genome database of NCBI. CRISPRCasFinder was used to predict CRISPR sequences in the genome sequences (Couvin et al. 2018). Furthermore, antiSMASH tool version 2.0 (Blin et al. 2019), bacteriocin genome mining tool and BAGEL version 3 (Van Heel et al. 2018) web tools were used for identifying the gene clusters for secondary metabolites, bacteriocin, or lantibiotics. Locus IDs of IMOS utilization by Bifidobacterium genes were deduced from their whole genomes and prediction of genes were based on CAZymes pipeline and BLASTp of the genes available in public domain. The SWISS-MODEL was used for predicting the function of each gene present in the locus. The signalP-5.0 server was used for predicting the signal peptide in each gene in the locus. The assembly and draft genome sequence of B. longum Bif10, B. breve Bif11 and B. longum Bif16 are available as a whole-genome shotgun (WGS) project in GenBank under accession numbers QCZM00000000, QELD00000000 and QCZN00000000.
Statistical analysis
PRISM-8.0 software was used (GraphPad software Inc., CA, USA) for all the statistical analyses. All the values were expressed as Mean ± SEM.
For the experiments on Caco2 cell viability and IL-8 production, cholesterol reduction, antimicrobial activity and relative bacterial growth, the data were analyzed using one-way ANOVA followed by Tukey’s post-hoc test. For mucin binding assays, doubling time, biomass, acetate, lactate and ethanol production, the data were analyzed using t test. For acid and bile tolerance in presence of IMOS and Glucose grown cells, the data were analyzed using two-way ANOVA followed by Tukey’s Multiple comparisons test. In all the experiments P ≤ 0.05 was considered as significant.
Results
Isolation and molecular characterization of Bifidobacterium strains
Four bacterial strains designated as Bif10, Bif11, Bif12 and Bif16 were isolated from the feces of healthy human infants. The strains were pale white, circular, convex, glistening and opaque colonies with entire margin. Bif10 and Bif11 colonies were non-mucilaginous and powdery in texture, while Bif12 and Bif16 colonies were mucilaginous and sticky in texture. All strains were anaerobic, Gram positive and showed Y- to pleomorphic shapes under the microscope (40× and 100× oil emulsion). Biochemical analysis indicated that the strains were catalase negative, γ-haemolytic and do not ferment mucin, when grown on mucin agar or in mucin broth. The presence of fructose 6-phosphate phosphoketolase gene confirmed the strains were Bifidobacterium spp. The 16S rRNA gene sequencing and blast analysis of Bif10, Bif11, Bif12 and Bif16 showed 99% homology with B. longum, B. breve, B. longum and B. longum, respectively, and they were named as B. longum Bif10, B. breve Bif11, B. longum Bif12 and B. longum Bif16, respectively (Supplementary Table 1).
Effect on intestinal epithelial cell inflammation: Human intestinal epithelial cells (Caco2 cells) showed 100% viability when treated with B. longum Bif10, B. breve Bif11, B. longum Bif12 and B. longum Bif16 (Fig. 1A). Furthermore, the four strains curtailed the TNF-α and LPS induced inflammation in the human intestinal epithelial cells as there was a reduction in IL-8 production which otherwise stimulated by TNF-α and LPS in the colonic cells (Fig. 1B).
Biochemical characterization
Antibiotics susceptibility
The effect of antibiotic treatment on the four strains is given in the Supplementary Table 2. B. longum Bif10 was sensitive to all the antibiotics except sulphatriad; B. breve Bif11 was sensitive towards all the antibiotics except lincomycin, ciprofloxacin, co-trimoxazol and sulphatriad; and B. longum Bif12 was sensitive towards all antibiotics except cloxacillin, ciprofloxacin, co-trimoxazol and levofloxacin. B. longum Bif 16 was sensitive towards all antibiotics except ciprofloxacin, co-trimoxazole and sulphatriad.
Fermentation towards different carbohydrates
Strain variations in the utilization of various carbohydrates was evident among B. longum Bif10, B. breve Bif11, B. longum Bif12 and B. longum Bif16 in API-CHL 50 assay (Supplementary Table 3). B. longum Bif10 fermented 15 carbohydrates, B. breve Bif11 fermented 22 carbohydrates, B. longum Bif12 and B. longum Bif16 fermented 16 carbohydrates (Supplementary Table 3).
Probiotic attributes
Acid and bile tolerance
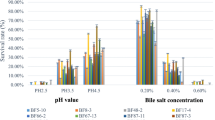
In acidic growth conditions, B. longum Bif10 showed 50, 82 and 84% survivability at pH 2.5, 3.0, and 3.5. B. breve Bif11 showed 57, 90 and 92% survivability at pH 2.5, 3.0, and 3.5. B. longum Bif12 showed 55, 74 and 76% survivability at pH 2.5, 3.0, and 3.5. B. longum Bif16 showed 62, 81 and 91% survivability at pH 2.5, 3.0, and 3.5, respectively (Table 1).
In presence of 0.2 and 0.4% bile, the viability of B. longum Bif10 was 81 and 87%. Viability of B. breve Bif11 was 92 and 93%, B. longum Bif12 was 82 and 84% and. B. longum Bif16 was 80 and 89% viability in 0.2% bile and 0.4%, respectively (Table 1).
Adhesion to gastric mucin: There was a strain dependent binding to porcine gastric mucin. B. longum Bif10, B. longum Bif12 and B. longum Bif16 showed 10, 25 and 13% each of mucin binding, whereas B. breve Bif11 showed maximum of 56% mucin binding (Fig. 2A). None of the strains showed mucin degradation on mucin agar plates or liquid broth.
Probiotic attributes of Bifidobacterium strains A mucin binding; B antimicrobial activity and C in vitro cholesterol reduction. Note: Data A were analyzed using unpaired t test, B and C were analyzed using one-way ANOVA followed by Tukey’s Multiple comparisons test. For mucin binding assay, *P < 0.05 was considered as significant between each strain grown in MRSC–glucose and MRSC–IMOS, respectively. For antimicrobial activity, *P < 0.05 was considered as significant between the isolated strains and the reference strain B. breve Bif40 grown in presence of S. typhi, E. coli and S. aureus, respectively
Cell surface properties
Auto-aggregation
Ability of the strains to auto-aggregate is given in Supplementary Table 4. B. longum Bif10, B. breve Bif11, B. longum Bif12 and B. longum Bif16 showed 41, 7, 22, and 5% auto-aggregation, respectively.
Cell surface hydrophobicity: B. longum Bif10 showed 61, 55 and 58%, B. breve Bif11 showed 16, 17 and 14%, B. longum Bif12 showed 40, 28 and 56% and B. longum Bif16 showed 10, 8 and 12% hydrophobicity in xylene, hexane, and chloroform, respectively (Supplementary Table 4).
Congo red binding
Bifidobacterium longum Bif10 showed 22 and 25%, B. breve Bif11 showed 29 and 23%, Bif12 showed 20 and 31% and B. longum Bif16 exhibited 17 and 52% of Congo red binding in agar grown and broth grown cells, respectively (Supplementary Table 4).
Salt aggregation test (SAT)
Bifidobacterium longum Bif10 showed SAT at all molarities (0.2 to 4 M) of ammonium sulphate solution, while B. breve Bif11, B. longum Bif12 and B. longum Bif16 showed aggregation at 3.2 and 4 M of ammonium sulphate solution for the agar and broth grown cells (Supplementary Table 4). Interestingly, there was no significant difference in cell surface properties of glucose and IMOS grown cells (Supplementary Table 6).
BSH activity
Bifidobacterium longum Bif10, B. breve Bif11, B. longum Bif12 and B. longum Bif16 showed positive BSH activity towards TDCA as there was a white precipitate formation surrounding the bacterial colonies as observed in the agar plates. B. longum Bif10 showed strong BSH activity as the zone of precipitate formation was 3 mm MRSC–TDCA agar plates, while B. breve Bif11 and B. longum Bif12 showed moderate activity of 2 mm each. Furthermore, B. longum Bif10 also showed a weak BSH activity (1 mm) on MRSC–TCA agar plates, while rest of the strains showed no BSH activity on MRSC–TCA agar plates.
Cholesterol lowering activity
B. longum Bif16 showed a maximum cholesterol reduction of 45% followed by B. longum Bif12 that showed 38% reduction. This was followed by B. longum Bif10 which showed 34% and B. breve Bif11 showed 28% of cholesterol reduction (Fig. 2C).
Anti-microbial activity
Antimicrobial potential of bacterial culture supernatants of B. longum Bif10, B. breve Bif11, B. longum Bif12 and B. longum Bif16 against E. coli (MTCC-3222), S. aureus (ATCC-9144) or S. typhimurium (NCTC-74) is given in the Fig. 2B. Maximum anti-bacterial activity was exhibited by B. longum Bif16 as it showed 20, 18 and 19 mm zone of inhibition against S. typhimurium, E. coli and S. aureus, respectively.
Prebiotic profiling
Agar plate assay suggested that B. longum Bif10, B. breve Bif11 and B. longum Bif16 were able to ferment IMOS. B. longum Bif10 alone fermented FOS, while B. breve Bif11 alone produced a halo around the colonies on SS and RS agar plates upon staining with iodine solution due to the hydrolysis of SS and RS (Supplementary Table 5). Inulin was not metabolized by any of the strains (Supplementary Table 5).
Growth profiles in IMOS
Bifidobacterium longum Bif10, B. breve Bif11, B. longum Bif12 and B. longum Bif16 showed comparable growth in 1% IMOS in MRSC-BB relative to that in glucose (Fig. 3). The doubling times of B. longum Bif10, B. breve Bif11, B. longum Bif12 and B. longum Bif16 grown in glucose supplemented broth were 8.9, 8.5, 11.1 and 9.6 h, while in IMOS, the doubling time were 11.0, 10.3, 14.1 and 13.4 h, respectively (Fig. 3I). In the MRSC-BB supplemented with either IMOS or glucose, the pH was reduced to 4.5 from an initial pH of 7.0. Relative bacterial growth of B. longum Bif10, B. breve Bif11, B. longum Bif12 and B. longum Bif16 in IMOS were 89, 84, 86 and 85%, respectively (Fig. 3E, F). Cell mass of B. longum Bif10, B. breve Bif11, B. longum Bif12 and B. longum Bif16 obtained was 2.8, 3.3, 3.0, 3.6, 2.55 and 2.5 g/L, respectively, in MRSC-BB supplemented with IMOS, while on glucose the cell mass obtained were 2.4, 2.6, 3.1, 2.8, 2.36 and 2.11 g/L, respectively (Fig. 3J). In presence of IMOS, the total carbohydrate utilization was 71, 66, 59 and 63% for B. longum Bif10, B. breve Bif11, B. longum Bif12 and B. longum Bif16, respectively, while in glucose, the carbohydrate utilization was 85, 79, 84 and 81% at the end of 72 h of fermentation. B. longum JCM 1217 and B. breve Bif40 showed 65% utilization of IMOS, while glucose utilization was 78 and 77%.
Effect of IMOS and Raffinose on the growth of Bifidobacterium strains. Growth curves (A–F); relative bacterial growth; (G–L), doubling time (M) and dry weight (N) of B. longum Bif10, B. breve Bif11, B. longum Bif12, B. longum Bif16, B. longum JCM 1217 and B. breve Bif40, respectively. Note: Data were analyzed using one-way ANOVA followed by Tukey’s post-hoc test. *P < 0.05 was considered as significance between each strain grown in MRSC–glucose and MRSC–IMOS, respectively. For calculating the relative growth, the growth of strains in MRSC-BB supplemented with glucose was considered as 100%
Hydrolytic enzyme production
α-Glucosidase activity
Whole cells of B. longum Bif10, B. breve Bif11, B. longum Bif12, B. longum Bif16, B. longum JCM 1217 and B. breve Bif40 showed 0.19, 0.15, 0.13, 0.13, 0.04 and 0.18 µM of PNP released per min from respective PNP-glycoside, respectively, when grown on IMOS. Glucose grown culture of B. longum Bif10, B. breve Bif11, B. longum Bif12, B. longum bif16 and B. longum JCM 1217 and B. breve Bif40 showed 0.10, 0.11, 0.08, 0.12, 0.05 and 0.15 µM of PNP formation, respectively (Supplementary Table 7). Intracellular α-glucosidase activity of IMOS grown B. longum Bif10, B. breve Bif11, B. longum Bif12, B. longum Bif16, B. longum JCM 1217 and B. breve Bif40 was 0.10, 0.12, 0.14, 0.16, 0.10 and 0.18 µM of PNP release, respectively, while glucose grown cultures showed 0.05, 0.12, 0.10, 0.16, 0.03 and 0.13 µM of PNP release, respectively (Supplementary Table 7).
α-Galactosidase activity
Strains exhibited very less or no extracellular α-galactosidase activity in the culture supernatants. However, whole cells of B. longum Bif10, B. breve Bif11, B. longum Bif12, B. longum Bif16, B. longum JCM 1217 and B. breve Bif40 grown in IMOS showed 0.10, 0.031, 0.035, 0.033, 0.027, and 0.04 µM of PNP formation, respectively, while the glucose grown cells showed 0.08, 0.03, 0.09, 0.08, 0.02, and 0.03 µM of PNP formation, respectively (Supplementary Table 7).
β-Galactosidase activity
Whole cell β-galactosidase activity in B. longum Bif10, B. breve Bif11, B. longum Bif12, B. longum Bif16, B. longum JCM 1217 and B. breve Bif40 in IMOS was 0.04, 0.05, 0.04, 0.02, 0.06, and 0.05 µM of PNP released, respectively, while the glucose grown cultures showed 0.03, 0.02, 0.04, 0.03, 0.06 and 0.04 µM of PNP production, respectively (Supplementary Table 7).
Production of secondary metabolites
In IMOS broth, acetate production was 24.5, 26.8, 23.4, 24.6, 25.8 and 26.3 mM/mL; lactate production was 14.9, 14.8, 14.32, 14.9, 14.8 and 17.6 mM/mL and ethanol production were 181.3, 180.2, 168.7, 178.4, 206.9 and 177.6 mg/L, respectively, by B. longum Bif10, B. breve Bif11, B. longum Bif12, B. longum Bif16, B. longum JCM 1217 and B. breve Bif40, respectively. In glucose broth, acetate production was 25.6, 26.6, 26.9, 27.4, 26.8, 24.6 mM/mL; lactate production was 15.7, 15.0, 15.3, 15.2, 16.4 and 17.2 mM/mL and ethanol production was 161.7, 175.5, 190.9, 221.2, 172.3 and 154.8 mg/L, respectively, for B. longum Bif10, B. breve Bif11, B. longum Bif12, B. longum Bif16, B. longum JCM 1217 and B. breve Bif40, respectively (Fig. 4A–C). In case of B. longum Bif12 and B. longum Bif16 acetate production was higher in IMOS grown cells as compared to that in glucose.
Mucin binding assay
Bifidobacterium longum Bif10 showed 11 and 30.8%, B. breve Bif11 showed 56 and 58%, B. longum Bif12 showed 25 and 32.8 and B. longum Bif16 showed 13 and 31.4% mucin binding in media supplemented with glucose and IMOS, respectively. B. breve Bif40 showed 54.7 and 62% and B. longum JCM 1217 showed 48.6 and 54.2% of mucin binding in glucose and IMOS supplemented media, respectively (Fig. 2A).
Genomic features of the Bifidobacterium strains
Genome based taxonomy for the three strains using ANI with their type strain reference genome was performed which confirmed their species status. The general genomic features of all the strains are represented in Table 2. Among the three sequenced genomes, B. longum Bif10 had largest genome size (2,631,972 bp), followed by B. breve Bif11 (2,494,489) and B. longum Bif16 (2,469,902) with typical GC content to their species. Functional classification of genes using RAST subsystem showed the most abundant functional subsystems are amino acids and derivatives, carbohydrates and protein metabolism in all the 3 strains. CRISPRCasFinder had identified 4 CRISPR sequences in B. longum Bif10, 6 in B. breve Bif11 and 5 in B. longum Bif16.
Acid/Gastric response
The genomes of the three strains harbored the genes for multi antimicrobial extrusion protein Na( +)/drug antiporter, MATE family of MDR efflux pumps, glyceraldehyde-3-phosphate dehydrogenase, heat shock protein DnaK and chaperones, such as dnaJ, groEL and groES responsible for the acid tolerance (Supplementary Table 8).
Bile/intestinal response
All the three strains showed the presence of tlyC1 (encoding a hemolysin-like protein), Choloylglycine hydrolase (responsible for bile hydrolysis), gene for the Bl0920 protein, and gene coding for cholate efflux transporter having 97–100% homology with that of B. longum BBMN68. The gene for Multidrug resistance protein belongs to the permeases of the major facilitator superfamily and its homologue was present in the three strains, with 62% similarity (296/475). Furthermore, MDR ABC transporters were present in the genomes of the three strains. Thus, 11 such transporters were present in B. breve Bif11 and B. longum Bif16. Although, strain B. longum Bif10 lacks these transporters but it had other unique genes for Multiple sugar ABC transporter, such as membrane-spanning permease protein MsmG, membrane-spanning permease protein MsmF and substrate-binding protein that may perform such functions (Supplementary Table 8).
Genome based metabolic capabilities: carbohydrate utilization
The three strains were able to utilize l-arabinose and d-Ribose that corroborates with genomic features. Ten genes responsible for d-ribose utilization were present in the three strains. Glucose was utilized by the three strains and 9 genes were reported in three strains in the subsystem “Glycolysis and Gluconeogenesis”. Similarly, the three strains harbored fructose-bisphosphate aldolase class II and fructose-6-phosphate phosphoketolase encoding proteins responsible for D-Fructose metabolism (Supplementary Table 9). Batch fermentation study showed that the three strains could utilize raffinose. Gene annotation predicted that all three strains had genes encoding for sucrose phosphorylase and α-galactosidase which cleaves sucrose and melibiose, respectively. Besides this, the genomes of the strains showed various amylases that may help in the utilization of starch and related carbohydrates (Supplementary Table 10).
Genomic determinants for metabolism of maltose and isomaltooligosaccharides
All three strains have maltose and maltodextrin utilization locus containing 20 genes. Some of them encode α-glucosidase and associated transporters required for maltose and maltodextrin utilization. Besides these, B. breve Bif11 and B. longum Bif16 had maltodextrin glucosidase, for maltose and maltodextrin utilization (Supplementary Table 11).
Genes for galactose, lactose and oligosaccharides utilization
There were six genes that help in the galactose uptake and utilization and 11 genes were involved in d-Lactose utilization in the subsystem lactose and galactose uptake and utilization (Supplementary Table 12). There were five genes responsible for galactose utilization in strain B. longum Bif16. These genes code for α-galactosidase, β-galactosidase, α-galactosidase precursor, evolved β-d-galactosidase (beta subunit) and evolved β-d-galactosidase (alpha subunit). However, B. longum Bif10 lacks a gene that encodes for α-galactosidase precursor B. breve Bif11 contains only α-galactosidase and β-galactosidase.
Since all three strains have found to be grown on IMOS, we have nailed down the locus that can encode a set of genes, associating with breakdown of the sugars at extracellular environment and then transport them into cytoplasm, response regulators and a gene for oligo-1,6-glucosidase (GH13_31). Two loci for B. longum Bif10 and one each locus for B. breve Bif11 and B. longum Bif16 were identified. Besides this, multiple oligo-1,6-glucosidase genes were present observed. In case of B. breve Bif11 and B. longum Bif16, a GH13 hydrolase (α-amylase) degrades large molecules of starch was also observed.
Genomic characterization of Adhesion, Mucin binding and Mucin degradation
Subsystem Sortase
Three genes responsible for pilus assemblage and mucin binding were present in the three Bifidobacterium strains that encompassed Sortase A (LPXTG specific), cell wall surface anchor family protein and NPQTN specific sortase B.
Within the sialic acid metabolism subsystem, B. breve Bif11 contains 15 genes, whereas B. longum Bif10 and B. longum Bif16 had nine genes each (Supplementary Table 12) responsible for colonization of bacteria in the host gut. Importantly, B. breve Bif11 had two important genes, sialic acid utilization regulator of RpiR family and sialidase that were absent in B. longum Bif16 and B. longum Bif10. Total four genes responsible for capsular polysaccharides biosynthesis and assembly were present in B. longum Bif16, while B. longum Bif10 and B. breve Bif11 genomes possess only two genes and lack O-antigen flippase Wzx and oligosaccharide repeat unit polymerase Wzy. In subsystem, rhamnose containing glycans and dTDP-rhamnose synthesis, total nine genes were present in B. breve Bif11 and B. longum Bif16, while B. longum Bif10 had 7 genes (Supplementary Table 12).
Moonlighting proteins
DnaK, transaldolase, and enolase were present in all the three genomes. However, EF-Tu (elongation factor thermo unstable) was present in B. breve Bif11 and B. longum Bif16 and absent in B. longum Bif10.
Secondary metabolites prediction
RAST subsystem predicted the genes for secondary metabolism in B. longum Bif16 strain. Furthermore, both antiSMASH and Bagel results predicted the presence of lanthipeptide of putative class II type in B. longum Bif16 strain in contig 22 of the assembled genome with complete cluster of size nearly 23 Kb. B. longum Bif10 also harbored lanthipeptide cluster of size approximately 20 Kb as predicted by Bagel, which was located in contig 8. The clusters in all the three strains comprised of genes important for immunity, transport & leader cleavage of the predicted lanthipeptide.
Discussion
Majority of the chronic diseases stems due to loss of intestinal barrier function as a result of altered gut microbiota composition and translocation of microbial associated molecular patterns and resultant inflammation (Belkaid and Hand 2014; Singh et al. 2020). In the present study, a decrease in IL-8 production by the intestinal epithelial cells, an inflammatory marker, suggested that the Bifidobacterium strains were able to reduce the inflammation caused due to LPS and TNF-α treatment. Our study is in alignment with an earlier report on reduction in inflammation in LPS stimulated HT-29 cells by Bifidobacterium strains through reduction in IL-8 production (Li et al. 2019).
Acid tolerance is the major challenge in case of Bifidobacteria and, therefore, is vital for its survival, adaptation to the host and for exerting probiotic functions. In this study, the four Bifidobacterium strains showed high resilience towards gastric and bile conditions which are considered as the primary selection criteria for a probiotic strain (Ganguly et al. 2011). Strains in the present study showed 50–90% survivability at a pH of 2.5, 3.0 and 3.5. Genome annotations revealed the existence of F0/F1-ATPase, production of ammonia via cysteine–cystathionine cycle, GAPDH, a moonlighting protein, membrane transporters, such as major facilitator super family (MFS) and multiple antimicrobial extrusion protein family (MATE) transporter families that helps the bacteria to withstand the acid stress (Corcoran et al. 2005; Antikainen et al. 2007; Lee and O’Sullivan 2010; Andriantsoanirina et al. 2013; Alnaseri et al. 2015; Schindler and Kaatz 2016). Furthermore, the three strains harboured genes for efflux pumps, multidrug transporters, Cholate efflux transporters and bile salt hydrolases that helps the bacteria from bile toxicity and rendering them more tolerant to bile toxicity and making them robust candidates for GIT delivery (Lye et al. 2010). Interestingly, the presence of BSH activity was confirmed using in vitro assays, where the three strains showed positive BSH activity towards TDCA, while B. longum Bif10 could degrade TCA.
Besides stress tolerance, adhesion to intestinal epithelial cells and mucus are also the preferred probiotic attributes as suggested by FAO/WHO, 2002. The four strains showed adhesion to epithelial cells and mucin but do not degrade mucin as shown by earlier researchers (Abe et al. 2010). Earlier studies suggested the presence of two and in majority of the cases more types of sortase genes in Gram positive bacteria (Mandlik et al. 2008). Occurrence of Sortase A (LPXTG specific), cell wall surface anchor family protein and NPQTN specific sortase B genes in the three genomes would help in cross-linking individual pilin monomers and thus help in anchoring the bacteria to the intestinal cells (Mandlik et al. 2008). The three strains contained Wzx and Wzy genes encoding flippase that helps in the translocation of O-antigen across the membrane to the cell surface (Islam et al. 2010). Rhamnose-rich O-antigen is responsible for surface attachment, cell–cell aggregation, and biofilm maturation (Islam et al. 2010). B. breve Bif11 consists of sialic acid utilization regulator of RpiR family, sialidase and LPXTG that are covalently linked to sortase A. This helps in the colonization and adhesion of bacteria to cells, such as A549, HeLa and Caco-2 cells.
Antimicrobial activity is one of the desired probiotic features which have been vastly explored by many researchers. In this study, the acid supernatants and not the neutralized ones of the strains inhibit the tested pathogens, which is due to the combined effect of acetic acid, ethanol, lactic acid and H2O2 as reported by previous researchers (Marianelli et al. 2010). Interestingly, the genomes of B. longum Bif10 and B. longum Bif16 contained locus for lanthipeptide and its functional presence and time dependent secretion warrants further studies.
The ability of Bifidobacterium strains to utilize/metabolize the dietary and host-derived carbohydrates is highly strain specific and depends on the presence of specific carbohydrases, sugar transporters, such as ABC transporters, which were present in the three strains (Pokusaeva et al. 2011). Growth studies of the strains suggested IMOS as the most suitable non-digestible carbohydrate, as IMOS yielded comparable growth of the strains to that on glucose. Some of the probiotic attributes are improved when the strains were grown with IMOS, although the difference was not significant.
Genomic studies of the three strains suggested the presence of α-glucosidases, and oligo-1,6 glucosidases required for the breakdown of IMOS, maltose and maltodextrin into their monomeric units. In vitro results augmented the functional presence of cell associated α-glucosidase in the strains when grown on IMOS or glucose in the liquid cultures. Some studies also suggested that saccharides such as lactose, raffinose or IMOS in the fermentation media act as an inducer for α- and β-glucosidases production (Carevic et al. 2016). Since the strains have grown on IMOS, we have identified the locus that can encode for carbohydrases at extracellular environment, and then transport them into cytoplasm, response regulators and a gene for oligo-1,6-glucosidase (GH13_31). Two loci in B. longum Bif10 and one locus each in B. breve Bif11 and B. longum Bif16 was observed, while multiple genes encoding for oligo-1,6-glucosidase were also present. In B. breve Bif11 and B. longum Bif16, presence of a GH13 (α-amylase) helps in the hydrolysis of polymers such as starch into dextrin forms which are further hydrolysed by GH77 enzymes into oligosaccharides. These oligosaccharides can then be internalized and hydrolysed to monosaccharide units by the action of oligo-1,6-glucosidases and α-1,4-glucosidases in the cytoplasm for further processing in the energy fermentative pathways. The four strains produce lactate and ethanol besides acetate when grown on IMOS and glucose and hence are heterofermentative. Acetate, through co-feeding can be converted into butyrate by the butyrate producing microbes, a crucial step for achieving the healthy gut homeostasis (Rowland et al. 2018).
Conclusion
Bifidobacterium strains isolated in the present study are metabolically active as they ferment IMOS and galactose-based oligosaccharides and possess desirable probiotic attributes and are anti-inflammatory as they reduce LPS-induced inflammation in the human intestinal epithelial cells (Fig. 5). Hence, these strains could be blended with IMOS or galactose-based oligosaccharides to use as synergistic synbiotics for alleviating inflammatory gut conditions. Detailed studies on the molecular mechanisms for the beneficial effects observed through this study are in progress.
Summary of the in vitro protective effect of Bifidobacterium strains against TNF-α and LPS-induced inflammation in Caco2 cells and the organization of representative loci for utilization of isomaltooligosaccharides (IMOS) in Bifidobacteria and mode of IMOS utilization system in the Bifidobacteria. Note: Based on sequence homology from SWISS-MODEL and SignalP-5.0 Server, it has been predicted that IMOS such as isomaltose and isomaltotriose can be taken in the cells by ABC transporter and digested by oligo-1,6-glucosidase in to glucose. Prediction of ulilization of other sugars was also made from sequence homology
References
Abe F, Muto M, Yaeshima T, Iwatsuki K, Aihara H, Ohashi Y, Fujisawa T (2010) Safety evaluation of probiotic bifidobacteria by analysis of mucin degradation activity and translocation ability. Anaerobe 16:131–136. https://doi.org/10.1016/j.anaerobe.2009.07.006
Achi SC, Talahalli RR, Halami PM (2019) Prophylactic effects of probiotic Bifidobacterium spp. in the resolution of inflammation in arthritic rats. Appl Microbiol Biotechnol 103:6287–6296. https://doi.org/10.1007/s00253-019-09864-2
Alnaseri H, Arsic B, Schneider JET, Kaiser JC, Scinocca ZC, Heinrichs DE, McGavin MJ (2015) Inducible expression of a resistance-nodulation-division-type efflux pump in Staphylococcus aureus provides resistance to linoleic and arachidonic acids. J Bacteriol 197:1893–1905. https://doi.org/10.1128/JB.02607-14
Andriantsoanirina V, Allano S, Butel MJ, Aires J (2013) Tolerance of Bifidobacterium human isolates to bile, acid and oxygen. Anaerobe 21:39–42. https://doi.org/10.1016/j.anaerobe.2013.04.005
Antikainen J, Kupannen V, Lahteenmaki K, Korhonen TK (2007) pH-dependent association of enolase and glyceraldehyde-3-phosphate dehydrogenase of Lactobacillus crispatus with the cell wall and lipoteichoic acids. J Bacteriol 189:4539–4543. https://doi.org/10.1128/JB.00378-07
Arboleya S, Watkins C, Stanton C, Ross RP (2016) Gut bifidobacteria populations in human health and aging. Front Microbiol 7:1204. https://doi.org/10.3389/fmicb.2016.01204
Aziz RK, Bartels D, Best A, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. https://doi.org/10.1186/1471-2164-9-75
Belkaid Y, Hand TW (2014) Role of the microbiota in immunity and inflammation. Cell 157:121–141. https://doi.org/10.1016/j.cell.2014.03.011
Bermudez-Brito M, Plaza-Díaz J, Muñoz-Quezada S, Gómez-Llorente C, Gil A (2012) Probiotic mechanisms of action. Ann Nutr Metab 61:160–174. https://doi.org/10.1159/000342079
Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T (2019) AntiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87. https://doi.org/10.1093/nar/gkz310
Carevic M, Banjanac K, Corovic M, Jakovetic S, Milivojevic A, Vukasinovic-Sekulic M, Bezbradica D (2016) Selection of lactic acid bacteria strain for simultaneous production of α- and β-galactosidases. Zast Mater 57:265–273. https://doi.org/10.5937/zasmat1602265c
Cazorla SI, Maldonado-Galdeano C, Weill R, De Paula J, Perdigón GDV (2018) Oral administration of probiotics increases paneth cells and intestinal antimicrobial activity. Front Microbiol 9:736. https://doi.org/10.3389/fmicb.2018.00736
Chander AM, Singh S, Sharma S, Chaudhry V, Rajarammohan S, Mantri SS, Bishnoi M, Bhadada SK, Kondepudi KK (2020) Draft Genome Sequence of Bifidobacterium pseudocatenulatum Bif 4, Isolated from Healthy Infant Feces. Microbiol Resour Announc 9(26):e00561-e620. https://doi.org/10.1128/mra.00561-20
Corcoran BM, Stanton C, Fitzgerald GF, Ross RP (2005) Survival of probiotic lactobacilli in acidic environments is enhanced in the presence of metabolizable sugars. Appl Environ Microbiol 71:3060–3067. https://doi.org/10.1128/AEM.71.6.3060-3067.2005
Couvin D, Bernheim A, Toffano-Nioche C, Touchon M, Michalik J, Néron B, Rocha EPC, Vergnaud G, Gautheret D, Pourcel C (2018) CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res 46:W246–W251. https://doi.org/10.1093/nar/gky425
Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F (1956) Colorimetric method for determination of sugars and related substances. Anal Chem 28:350–356. https://doi.org/10.1021/ac60111a017
Fu X, Liu Z, Zhu C, Mou H, Kong Q (2019) Nondigestible carbohydrates, butyrate, and butyrate-producing bacteria. Crit Rev Food Sci Nutr 59:S130–S152. https://doi.org/10.1080/10408398.2018.1542587
Gagliardi A, Totino V, Cacciotti F, Iebba V, Neroni B, Bonfiglio G, Trancassini M, Passariello C, Pantanella F, Schippa S (2018) Rebuilding the gut microbiota ecosystem. Int J Environ Res Public Health 15:1679. https://doi.org/10.3390/ijerph15081679
Ganguly NK, Bhattacharya SK, Sesikeran B, Nair GB, Ramakrishna BS, Sachdev HPS, Batish VK, Kanagasabapathy AS, Muthuswamy V, Kathuria SC, Katoch VM, Satyanarayana K, Toteja GS, Rahi M, Rao S, Bhan MK, Kapur R, Hemalatha R (2011) ICMR-DBT Guidelines for evaluation of probiotics in food. Indian J Med Res 134:22–25. https://www.ijmr.org.in/text.asp?2011/134/1/22/83322
Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, Scott K, Stanton C, Swanson KS, Cani PD, Verbeke K, Reid G (2017) Expert consensus document: the international scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol 14:491–502. https://doi.org/10.1038/nrgastro.2017.75
Guarino MPL, Altomare A, Emerenziani S, Di Rosa C, Ribolsi M, Balestrieri P, Iovino P, Rocchi G, Cicala M (2020) Mechanisms of action of prebiotics and their effects on gastro-intestinal disorders in adults. Nutrients 12:1037. https://doi.org/10.3390/nu12041037
Islam ST, Taylor VL, Qi M, Lam JS (2010) Membrane topology mapping of the O-antigen flippase (Wzx), polymerase (Wzy), and ligase (WaaL) from Pseudomonas aeruginosa PAO1 reveals novel domain architectures. Mbio 1(3):e00189-10. https://doi.org/10.1128/mBio.00189-10
Kajander K, Myllyluoma E, Rajilić-Stojanović M, Kyronpalo S, Rasmussen M, Jarvenpaa S, Zoetendal EG, De Vos WM, Vapaatalo H, Korpela R (2008) Clinical trial: multispecies probiotic supplementation alleviates the symptoms of irritable bowel syndrome and stabilizes intestinal microbiota. Aliment Pharmacol Ther 27:48–57. https://doi.org/10.1111/j.1365-2036.2007.03542.x
Kataria A, Achi SC, Halami PM (2018) Effect of Encapsulation on viability of Bifidobacterium longum CFR815j and physiochemical properties of ice cream. Indian J Microbiol 58:248–251. https://doi.org/10.1007/s12088-018-0720-6
Kondepudi KK, Ambalam P, Nilsson I, Wadström T, Ljungh Å (2012) Prebiotic-non-digestible oligosaccharides preference of probiotic bifidobacteria and antimicrobial activity against Clostridium difficile. Anaerobe 18:489–497. https://doi.org/10.1016/j.anaerobe.2012.08.005
Kumar H, Collado MC, Wopereis H, Salminen S, Knol J, Roeselers G (2020) The bifidogenic effect revisited—ecology and health perspectives of bifidobacterial colonization in early life. Microorganisms 8:1–20. https://doi.org/10.3390/microorganisms8121855
Lee J-H, O’Sullivan DJ (2010) Genomic Insights into Bifidobacteria. Microbiol Mol Biol Rev 74:378–416. https://doi.org/10.1128/mmbr.00004-10
Li SC, Hsu WF, Chang JS, Shih CK (2019) Combination of Lactobacillus acidophilus and Bifidobacterium animalis subsp. Lactis shows a stronger anti-inflammatory effect than individual strains in HT-29 cells. Nutrients 11(5):969. https://doi.org/10.3390/nu11050969
Lye HS, Rusul G, Liong MT (2010) Removal of cholesterol by lactobacilli via incorporation and conversion to coprostanol. J Dairy Sci 93:1383–1392. https://doi.org/10.3168/jds.2009-2574
Mandlik A, Swierczynski A, Das A, Ton-That H (2008) Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol 16:33–40. https://doi.org/10.1016/j.tim.2007.10.010
Marianelli C, Cifani N, Pasquali P (2010) Evaluation of antimicrobial activity of probiotic bacteria against Salmonella enterica subsp. enterica serovar typhimurium 1344 in a common medium under different environmental conditions. Res Microbiol 161:673–680. https://doi.org/10.1016/j.resmic.2010.06.007
Meyer D, Stasse-Wolthuis M (2009) The bifidogenic effect of inulin and oligofructose and its consequences for gut health. Eur J Clin Nutr 63:1277–1289. https://doi.org/10.1038/ejcn.2009
Pokusaeva K, Fitzgerald GF, Van Sinderen D (2011) Carbohydrate metabolism in Bifidobacteria. Genes Nutr 6:285–306. https://doi.org/10.1007/s12263-010-0206-6
Rohith HS, Halami PM (2021) The combined effect of potential probiotic Bacillus licheniformis MCC 2514 and Bifidobacterium breve NCIM 5671 towards anti-inflammatory activity on HT-29 cell lines. Probiotics Antimicro Prot. https://doi.org/10.1007/s12602-021-09851-y
Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I, Tuohy K (2018) Gut microbiota functions: metabolism of nutrients and other food components. Eur J Nutr 57:1–24. https://doi.org/10.1007/s00394-017-1445-8
Ruiz L, Delgado S, Ruas-Madiedo P, Sánchez B, Margolles A (2017) Bifidobacteria and their molecular communication with the immune system. Front Microbiol 8:2345. https://doi.org/10.3389/fmicb.2017.02345
Sarkar A, Lehto SM, Harty S, Dinan TG, Cryan JF, Burnet PWJ (2016) Psychobiotics and the manipulation of bacteria–gut–brain signals. Trends Neurosci 39:763–781. https://doi.org/10.1016/j.tins.2016.09.002
Schindler BD, Kaatz GW (2016) Multidrug efflux pumps of Gram-positive bacteria. Drug Resist Updat 27:1–13. https://doi.org/10.1016/j.drup.2016.04.003
Singh D, Khare P, Zhu J, Kondepudi KK, Singh J, Baboota RK, Boparai RK, Khardori R, Chopra K, Bishnoi M (2016) A novel cobiotic-based preventive approach against high-fat diet-induced adiposity, nonalcoholic fatty liver and gut derangement in mice. Int J Obes 40:487–496. https://doi.org/10.1038/ijo.2015.197
Singh S, Bhatia R, Singh A, Singh P, Kaur R, Khare P, Purama RK, Boparai R, Rishi P, Ambalam P, Bhadada S, Bishnoi M, Kaur J, Kondepudi KK (2018) Probiotic attributes and prevention of LPS-induced pro-inflammatory stress in RAW264.7 macrophages and human intestinal epithelial cell line (Caco2) by newly isolated Weissella cibaria strains. Food Funct 9:1254–1264. https://doi.org/10.1039/C7FO00469A
Singh S, Bhatia R, Khare P, Sharma S, Rajarammohan S, Bishnoi M, Bhadada SK, Sharma SS, Kaur J, Kondepudi KK (2020) Anti-inflammatory Bifidobacterium strains prevent dextran sodium sulfate induced colitis and associated gut microbial dysbiosis in mice. Sci Rep 10:18597. https://doi.org/10.1038/s41598-020-75702-5
Sundararaman A, Bansal K, Sidhic J, Patil P, Prakash PM (2021) Genome of Bifidobacterium longum NCIM 5672 provides insights into its acid-tolerance mechanism and probiotic properties. Arch Microbiol 203:6109–6118. https://doi.org/10.1007/s00203-021-02573-3
Turroni F, Milani C, Duranti S, Mancabelli L, Mangifesta M, Viappiani A, Lugli GA, Ferrario C, Gioiosa L, Ferrarini A, Li J, Palanza P, Delledonne M, Van Sinderen D, Ventura M (2016) Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. ISME J 10:1656–1668. https://doi.org/10.1038/ismej.2015.236
Usta-Gorgun B, Yilmaz-Ersan L (2020) Short-chain fatty acids production by Bifidobacterium species in the presence of salep. Electron J Biotechnol 47:29–35. https://doi.org/10.1016/j.ejbt.2020.06.004
Valdes AM, Walter J, Segal E, Spector TD (2018) Role of the gut microbiota in nutrition and health. BMJ 361:36–44. https://doi.org/10.1136/bmj.k2179
Van Heel AJ, De Jong A, Song C, Viel JH, Kok J, Kuipers OP (2018) BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res 46:W278–W281. https://doi.org/10.1093/nar/gky383
Woodmansey EJ (2007) Intestinal bacteria and ageing. J Appl Microbiol 102:1178–1186. https://doi.org/10.1111/j.1365-2672.2007.03400.x
Zhou JS, Gopal PK, Gill HS (2001) Potential probiotic lactic acid bacteria Lactobacillus rhamnosus (HN001), Lactobacillus acidophilus (HN017) and Bifidobacterium lactis (HN019) do not degrade gastric mucin in vitro. Int J Food Microbiol 63:81–90. https://doi.org/10.1016/S0168-1605(00)00398-6
Acknowledgements
Authors would like to acknowledge National Agri-Food Biotechnology Institute (NABI); Department of Biotechnology, Government of India and Indian Council of Medical Research for providing financial support for this research. DBT-e-Library Consortium (DeLCON) is gratefully acknowledged for providing access to electronic journals.
Funding
This study was funded by Core Grant from NABI, DBT, Government of India and Indian council of Medical Research: File No: 5/9/1312/2020-Nut.
Author information
Authors and Affiliations
Contributions
SS1, SS2, KK and MB conceived the research and designed the experiments. SS2, KK and SKB isolated the Bifidobacterium strains; SS1, SS2, RM and PR conducted the experiments. VC, SM, AC, SR and RS performed the bacterial genomic characterization and in-silico data analysis. SS1, SS2, MB, SKB and KK analyzed the data. SS1, SS2, VC, SM, AC, MB and KK wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflict of interest.
Data availability
The data sets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request. Whole genome sequencing data presented in this study are openly available in The National Centre for Biotechnology Information (NCBI).
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Consent for publication
Authors undersigned and give the consent for the publication of the given research material in the above journal.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sharma, S., Singh, S., Chaudhary, V. et al. Isomaltooligosaccharides utilization and genomic characterization of human infant anti-inflammatory Bifidobacterium longum and Bifidobacterium breve strains. 3 Biotech 12, 89 (2022). https://doi.org/10.1007/s13205-022-03141-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-022-03141-2