Abstract
Insects can establish a variety of symbiotic associations with bacteria that can have a significant impact on their evolutionary ecology. Some bacterial lineages are particularly pervasive as symbiotic associates. This is the case of the Sodalis genus, whose members have established independent, maternally transmitted symbioses in diverse insect taxa. The first members of the genus were isolated and studied some thirty years ago in tsetse flies, where they evolved as heritable facultative symbionts. Since then, numerous symbiotic associations involving members of the genus have been documented, some of which have evolved into strictly host-dependent mutualistic associations. The genus also includes members circulating freely in the environment, which can be pathogenic, have extensive metabolic capabilities and constitute a potential reservoir of new insect symbionts. In this review, we cover more than thirty years of literature to highlight how the diversity of the Sodalis genus described so far embodies the different degrees of host dependence and anatomical integration that bacteria can experience over the course of their evolution with insects. We discuss the propensity of Sodalis bacteria to embrace an endosymbiotic lifestyle, how this feature can be used to understand the nascent stages of bacterial endosymbiosis, and how Sodalis bacteria can be used to address fundamental and applied research issues. Throughout the review, emphasis is placed on research gaps that need to be filled to better address these aspects. We also draw attention to previously overlook facets of the genus that deserve further investigation, such as the potential role of Sodalis bacteria in wood digestion in certain insects, or the nature of their interaction with plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Insects are engaged in a wide range of interactions with symbiotic bacteria. These prokaryotic partners can play an important role in the evolutionary ecology of these invertebrates, as they can affect host phenotypes in a variety of ways (Zientz et al. 2004; Feldhaar 2011; Engel and Moran 2013; Sudakaran et al. 2017). The nature of these interactions can be diverse and evolve rapidly along the parasitism-mutualism continuum, particularly as a function of ecological context (Zytynska et al. 2021; Kaur et al. 2021; Drew et al. 2021). While some insect symbionts are transient partners, others have become host-dependent associates that pass faithfully from one host generation to the next by vertical transmission (Frago et al. 2020). Depending on the host insect’s degree of dependence on heritable symbionts, these bacterial associates are generally classified as obligate or facultative. Obligate symbionts are essential for the survival and reproduction of their host, providing them with vital nutrients that are deficient in the host’s diet (Zientz et al. 2004) or ensuring the feasibility of specific developmental mechanisms (Dedeine et al. 2001). Facultative symbionts, on the other hand, are not essential to the survival and reproduction of their host but can affect their adaptative phenotypes by producing beneficial or detrimental effects whose expression is conditioned by the ecological context, or by selfishly manipulating their reproduction (Oliver et al. 2010; Zytynska et al. 2021; Kaur et al. 2021). The diversity of these symbiotic associations is also reflected in the multiplicity of host tissues that symbiotic bacteria can colonize: while certain symbionts reside on the insect surface (ectosymbionts) (Bruner-Montero et al. 2021; Ganesan et al. 2023), others evolve within the host body (endosymbionts), sometimes living intracellularly in specialized host cells called bacteriocytes (Baumann 2005; Engel and Moran 2013). Moreover, other symbionts can colonize the host gut and other tissues (Salem et al. 2017; Pons et al. 2022a).
Over the past two decades, the sequencing of host and symbiont genomes from a myriad of symbiotic systems has revealed the extraordinary functional diversity of insect-bacterial interactions (Chellappan and Ranjith 2021). Studies of insect symbiont diversity have highlighted the propensity of some bacterial genera and species to embrace endosymbiosis, often members of the class Gammaproteobacteria and in particular Enterobacterales, an order that includes many pathogenic bacteria (Moran et al. 2008). This suggests that heritable symbionts derive from pathogenic bacteria whose virulence has faded during their evolution towards a host-dependent lifestyle. Some genera and species of symbionts are associated exclusively with specific insect groups. This is typically the case for many obligate nutritional symbionts such as Buchnera aphidicola (Gammaprotobacteria) found only in aphids, even though it is related to the Erwiniaceae family, which includes many plant pathogens (Nováková et al. 2013). These nutritional partners acquired tens of millions of years ago, have undergone significant divergence from their original progenitors and tend to form well-defined monophyletic clades. Conversely, some symbiont genera and species evolve with a wider range of host species, with which they have established a wide variety of associations along the parasitism-mutualism continuum. Wolbachia pipientis (Alphaproteobacteria) is arguably the most iconic species that illustrates the propensity of some symbionts to exploit a wide range of phylogenetically distant insect hosts. Indeed, Wolbachia is formally the only species in the genus, but is present in over 60% of insect species and is featured by a diversity of strains with distinct associated phenotypes, ranging from parasitism to mutualism (Weeks et al. 2007; Zug and Hammerstein 2015; Landmann 2019).
However, beyond Wolbachia, other bacterial genera and species have proved remarkably pervasive in insect populations (Jousselin et al. 2013; Tláskal et al. 2021; Pons et al. 2022b). This is the case of the Sodalis symbionts (family Pectobacteriaceae, order Enterobacterales, class Gammaproteobacteria), which were first isolated from tsetse flies more than thirty years ago (Welburn et al. 1987), and whose ubiquity and diversity in insects have been repeatedly demonstrated in recent years. For instance, the genus includes members that are free-living (i.e., live independently of the insect host), some of which may even be opportunistic pathogens for humans (Dale and Maudlin 1999; Clayton et al. 2012; Chari et al. 2015; Tláskal et al. 2021), but also members that have established symbiotic associations with insects, either facultative (Toh et al. 2006; Arp et al. 2014; Matsuura et al. 2014), or obligate (Heddi et al. 1999; Husnik and McCutcheon 2016; Santos-Garcia et al. 2017). This diversity reflects the different degrees of host dependence and anatomical integration that insect symbionts can experience over the course of their evolution.
The present article focuses on the diversity of the Sodalis genus. Covering more than thirty years of literature, it aims to provide an overview of the genus’ tremendous diversity across the broad spectrum of interactions in which Sodalis bacteria can be involved, ranging from pathogenicity to mutualism and addressing the evolutionary transition from a free-living to a host-dependent lifestyle. This review focuses on what the multifaceted nature of Sodalis bacteria can tell us about the genesis of mutualistic symbiotic associations and their evolution, how symbiotic bacteria bypass host immunity to establish long-lasting relationships, and the role symbionts can play in the transmission of animal and human pathogens. We begin by briefly introducing the publicly available genomic resources for Sodalis members, which provides valuable information for exploring endosymbiosis evolution and will serve here as a roadmap for discussing the diversity of the genus and its evolution. We start with strains facultatively associated with insects, historically the first to have been discovered, and end with the free-living strains that have come to light more recently. Although Sodalis symbionts are pioneering models for the study of endosymbiosis in insects, there are still many gaps in knowledge regarding the exact nature of the associations these symbionts maintain with their insect hosts. We discuss these gaps and propose new avenues that leverage Sodalis members to grasp the many facets of bacterial symbiosis in insects.
2 Diversity and evolution of Sodalis bacteria: overview
The genus Sodalis has recently been classified in the family Pectobacteriaceae (Proteobacteria; Gammaproteobacteria, Enterobacterales), which consists mainly of plant pathogens, many members of which are characterized by their ability to hydrolyze pectin of the plants they infect (Adeolu et al. 2016; Motyka et al. 2017). Sodalis has repeatedly succeeded in entering new hosts to form maternally transmitted symbioses, and occurs in a wide variety of insect taxa, some of which are very distant from each other. Members of the genus were first described as symbiotic partners of tsetse flies (Diptera: Glossinidae) (Welburn et al. 1987), and have subsequently been identified in various insect orders, involved in either facultative associations or obligate mutualistic associations (Nováková and Hypša 2007; Fukatsu et al. 2007; Toju et al. 2010; Grünwald et al. 2010; Kaiwa et al. 2010; Toju and Fukatsu 2011; Boyd et al. 2016; Santos-Garcia et al. 2017; Rubin et al. 2018). Phylogenetic analyses indicate that these infections are the result of independent events, most likely through horizontal insect-to-insect transfer and/or acquisition from free-living strains in the insect’s immediate environment (Snyder et al. 2011; Clayton et al. 2012). In particular, the scenario of direct environmental acquisition is supported by the recent isolation of several free-living strains from non-insect material (Clayton et al. 2012; Tláskal et al. 2021). While certain Sodalis-allied insect symbionts are engaged in a long coevolution with their host, others have been acquired relatively recently, probably from free-living isolates, making the diversity of the genus a valuable resource for illuminating the different scenarios of bacterial symbiosis evolution in insects, in particular the transition from a free-living to a host-dependent lifestyle and from facultative to obligate symbiosis.
Sequencing and annotating the genome of bacterial symbionts is a pivotal step in deciphering the nature of the interactions in which they are involved and in sketching out the history they have with their host (Perreau and Moran 2022). To date, twenty-one genome assemblies referenced as belonging to the genus Sodalis are publicly available on the NCBI database. Table 1 documents the basic information of these assemblies, with the genomes ranked by size from largest to smallest. It should be noted, however, that the genus Sodalis may include other members whose genomes have been sequenced, but which have not been officially classified in the genus (Sloan and Moran 2012; Husnik and McCutcheon 2016; Santos-Garcia et al. 2017). This indicates that the taxonomy of the genus is not fully resolved. The diversity in genome size among Sodalis members reflects different degrees of genomic reduction, a typical process undergone by heritable symbionts, starting with a formerly free-living bacterium towards a host-dependent lifestyle involving a progressive genome remodeling (Fig. 1) (Moran and Bennett 2014). Free-living members are featured by a large genome (about 6 Mb) while ancient obligate symbionts have extremely small genomes, which can be less than 1 Mb. The genomes of facultative and recent obligate symbionts show an intermediate value.
Overview of the genome reduction process in endosymbionts. Free-living bacteria have few pseudogenes and mobile elements and regularly acquire new genetic material from the environment or from exchanges with other bacteria. Integration into the host of bacteria evolving as heritable symbionts leads to the accumulation of pseudogenes and mobile elements in their genome, which undergoes rearrangements. Over time, genes subject to relaxed selection are lost, while genes that play an essential role in the association are conserved. The result is an increase in host dependence for these bacteria. Gradually, the genomes become enriched in adenine and thymine (AT) nucleotides. Endosymbionts that evolve a high level of interdependence with the host end up with an extremely eroded genome, purged of mobile genetic elements and pseudogenes, retaining only the genetic information essential for symbiosis. As the level of interdependence between host and symbiont increases, so does the anatomical integration of the symbiont: it is condemned to an almost exclusively intracellular lifestyle in host-specific cells called bacteriocytes. By hindering genetic exchanges with other bacterial conspecifics, this intracellular lifestyle leads to asexuality, which also contributes to genomic degeneration
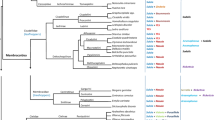
The diversity of available Sodalis genomes provides a valuable data resource to examine the evolution of bacterial symbioses in insects (Santos-Garcia et al. 2017). We performed a maximum likelihood phylogenetic analysis (1000 bootstraps) with the 21 assembled genomes of Sodalis bacteria publicly available in NCBI (Fig. 2). We included in the analysis other bacterial species that are not officially part of the genus but are considered “Sodalis-allied symbionts” (Sloan and Moran 2012; Husnik and McCutcheon 2016; Santos-Garcia et al. 2017). Phylogenomic analysis shows no evidence of co-speciation events: Sodalis symbionts associated with taxonomically close insect lineages do not form distinct clades and are scattered throughout the tree, suggesting that symbioses with Sodalis bacteria are the result of multiple independent acquisitions in different insect lineages, as previously reported (Husnik and McCutcheon 2016; Santos-Garcia et al. 2017). This raises the question of how Sodalis bacteria are acquired, their ability to pass from one insect lineage to another and to undergo horizontal transfers. Members of the genus Sodalis are also remarkable for exhibiting highly variable degrees of anatomical integration with their host (Table 1): while some members live independently of an insect host, others are host-dependent. Free-living strains of Sodalis can be cultured, providing access to experimental approaches to examine the processes governing the establishment of new symbiotic associations and their functioning, but also to develop paratransgenic approaches for disease vector control (Maltz et al. 2012; Enomoto et al. 2017; Munoz et al. 2020; Elston et al. 2022).
Rooted maximum likelihood phylogeny of the 21 Sodalis strains whose assembled genomes are publicly available in NCBI. Single-copy genes conserved between these Sodalis bacteria, bacteria considered as “Sodalis-allied symbionts” and the outgroups Dickeya dadanti and Pectobacterium carotovorum were obtained using OrthoFinder (Emms and Kelly 2019), and the concatenated sequences were aligned using MAFFT (Katoh and Standley 2013). The phylogenetic tree was constructed from the 50 conserved single-copy genes using maximum likelihood with 1,000 bootstrap replicates via IQ-Tree (http://iqtree.cibiv.univie.ac.at/). Bacterial species highlighted in blue do not formally belong to the Sodalis genus but are considered as “Sodalis-allied symbionts”. Bacteria used for the outgroup are highlighted in orange
3 Sodalis bacteria as facultative symbionts of insects
3.1 Sodalis glossinidius: a facultative associate of tsetse flies
3.1.1 An invasive heritable symbiont
The genus Sodalis was first established in the context of the study of the microbiota of tsetse flies (Welburn et al. 1987; Beard et al. 1993; Aksoy et al. 1997; Dale and Maudlin 1999). These insects are the vectors of trypanosomes, which cause Human African Trypanosomiasis (HAT, sleeping sickness) and Animal African Trypanosomiasis (AAT, Nagana). Tsetse flies (Glossina sp.) harbor an obligate nutritional symbiont, Wigglesworthia glossinidia (Gammaproteobacteria; Enterobacterales; Erwiniaceae), which supplements their host blood meal with folate (vitamin B9) and other B vitamins that are deficient in vertebrate blood (Aksoy 1995; Akman et al. 2002; Michalkova et al. 2014; Snyder and Rio 2015). W. glossinidia has a highly eroded genome (0.72 Mb) containing only those genes strictly necessary for the functioning of the symbiotic association, and resides intracellularly in bacteriocytes, which form the bacteriome organ in the anterior midgut, although it can also reside extracellularly in the milk gland lumen (Rio et al. 2012; Balmand et al. 2013) (Fig. 3). W. glossinidia is universally present in tsetse flies and is essential for multiple aspects of their biology, including digestion, reproduction, and immunity (Pais et al. 2008; Attardo et al. 2020). However, tsetse flies can harbor additional endosymbionts, including the reproductive manipulator W. pipientis that infects germ cells (Alam et al. 2011), and Sodalis glossinidius, a relatively recently facultative symbiont that can infect various host tissues (Dale and Maudlin 1999; Toh et al. 2006).
Schematics showing Sodalis bacteria’s localization and the diversity of its lifestyle. Free-living strains of the Sodalis bacterium have been found in humans and decomposing deadwood. In tsetse flies, S. glossinidius is a facultative symbiont localized intra- and extracellularly throughout the whole fly (red). W. glossinidia is the obligate symbiont found intracellularly in a bacteriome around the midgut, as well as extracellularly in the lumen of the milk glands (green). Both symbionts are maternally transmitted through milk feeding. In Sitophilus weevils, Sodalis pierantonius is an obligate symbiont localized in bacteriomes found at the apex of midgut mesenteric caeca, as well as at the apex of female ovaries, from which maternal transmission occurs (red) (inspired by (Zaidman-Rémy et al. 2018). In long-tailed mealybugs, Sodalis endolongispinus is a co-obligate symbiont (red) localized within the cytoplasm of the ancestral symbiont Tremblaya princeps (green). The nested endosymbiotic system is contained in bacteriocytes, and the Sodalis genome complements those of Tremblaya and its host for amino acid and vitamin biosynthesis pathways
Historically, S. glossinidius was the first reported member of the genus and the first insect endosymbionts to be isolated and cultured, first in the C6/36 cell line of Aedes albopictus (Welburn et al. 1987), then in pure culture (Dale and Maudlin 1999). The S. glossinidius genome is 4.3 Mb in size (Toh et al. 2006; Goodhead et al. 2020), which is quite large for a heritable insect symbiont. However, it contains many pseudogenes and is featured by a reduced coding capacity of around 50%, with a substantial number of pseudogenes in pathways that are no longer of critical importance in the context of the association with tsetse flies (Belda et al. 2010). With moderate genomic reduction and the ability to survive outside the host, S. glossinidius symbionts are in an early/intermediate stage between a free-living and a host-dependent lifestyle (Toh et al. 2006; Goodhead et al. 2020).
S. glossinidius is also featured by an invasive phenotype: it can exhibit both an extracellular and intracellular lifestyle and can colonize a wide range of host tissues, including the midgut, fat body, milk gland, uterus, and oviduct (Fig. 1) (Attardo et al. 2008; Balmand et al. 2013). Like W. glossinidia, S. glossinidius is transmitted vertically through the milk secretions produced by the female in the uterus, which are rich in the nutrients required for larval development (Attardo et al. 2008; Balmand et al. 2013). However, De Vooght et al. (2015) found that S. glossinidius can also be sexually transmitted to females through the male’s seminal secretions during mating in viviparous tsetse flies, and then passed on to the offspring (De Vooght et al. 2015). This paternal transmission pathway may partly explain the lack of phylogenetic congruence in tsetse-Sodalis associations and may drive genetic diversity in the symbiont via horizontal gene transfers (HGTs) between distinct strains co-infecting the same host that then spread within host populations via mixed modes of transmission (De Vooght et al. 2015). These genetic exchanges may be facilitated by the extracellular lifestyle of S. glossinidius.
3.1.2 Associated effects that are still unclear
The facultative nature of S. glossinidius has been evidenced both by field and experimental studies. Being an obligate symbiont (such as W. glossinidia) implies being fixed in the host species, i.e. having a 100% prevalence in host populations. On the other hand, facultative symbionts are not fixed in host species and have fluctuating prevalence in insect populations, as is the case with S. glossinidius. The symbiont exhibits a heterogeneous prevalence in tsetse fly field populations, which also varies among tsetse fly species (Maudlin et al. 1990; Farikou et al. 2010; Lindh and Lehane 2011; Alam et al. 2012; Wamwiri et al. 2013; Aksoy et al. 2014; Dennis et al. 2014; Tsagmo Ngoune et al. 2019; Mfopit et al. 2023; Malulu et al. 2023; El Yamlahi et al. 2023). While the effects associated with facultative symbionts have been clarified in some insects, for example aphids (Oliver et al. 2010; Zytynska et al. 2021), the exact nature of the relationship between S. glossinidius and tsetse flies remains unclear. Dale and Welburn (2001) reported that the selective elimination of the symbiont by antibiotics has a negative impact on the host lifespan (Dale and Welburn 2001). However, Weiss et al. (2006) found that S. glossinidius has neither negative nor positive effects on host fitness (Weiss et al. 2006). Studies about the impact of S. glossinidius on host fitness are quite limited, perhaps because antibiotic treatment of fertile flies often results in the elimination of the obligate symbiont W. glossinidia, leading to host sterility and making it challenging to generate fly lines lacking a specific symbiont. Furthermore, the nature of the relationship between S. glossinidius and tsetse fly species likely depends on a variety of factors, including strain-specific genotype, host species, coevolutionary processes that shaped the interaction, and environmental conditions, as has been shown in other well-studied model insects (Oliver et al. 2010; Zytynska et al. 2021). These aspects have been little studied in tsetse flies, although some S. glossinidius genomes are now available and some strains have been isolated and cultured, making it possible to experimentally test the symbiotic status of the facultative symbiont. This lack of knowledge is further compounded by the fact that most studies have focused on Glossina morsitans, whereas it is now clear that the nature of the relationship between a symbiont species and a host species can differ radically depending on the host-symbiont combination (Niepoth et al. 2018; Stoy et al. 2020). The Glossina genus is diverse (Moloo 1993; Dyer et al. 2008; Shaida et al. 2018) and it is likely that Sodalis symbionts hosted by tsetse flies have evolved differently according to host species and exhibit distinct associated phenotypic effects (Geiger et al. 2005).
3.1.3 A rich array of virulence factors
S. glossinidius was the first member of the genus to be documented (Dale and Maudlin 1999). Since it can be isolated and cultured, it is possible to used genetic engineering approaches to decipher the mechanisms used by symbiotic bacteria to infect their host and overcome immune barriers (Dale et al. 2002, 2005). Indeed, despite ongoing genome streamlining, the S. glossinidius genome retains genes encoding a wide range of virulence factors, which may explain the propensity of the symbiont to invade a wide range of host tissues (Akman et al. 2002; Toh et al. 2006). These virulence factors include several type III secretion systems similar to those found in free-living and parasitic bacterial species, but which have been altered during degenerative genome evolution (Dale et al. 2005; Toh et al. 2006). These modifications are assumed to be adaptive for a stabilized symbiotic relationship (Dale et al. 2002, 2005). Iron is an essential nutrient for bacterial survival and proliferation (Ratledge and Dover 2000). Genome analysis of S. glossinidius has also highlighted the role of iron chelation systems in host colonization by symbiotic bacteria. The S. glossinidius genome retains a series of genes involved in the biosynthesis of these systems, including siderophores, TonB-dependent transporters (TBDTs) and the hemin receptor HemR (Akman et al. 2002; Toh et al. 2006; Runyen-Janecky et al. 2010; Smith et al. 2013a; Hrusa et al. 2015). Using an RNA-seq approach complemented by a mutagenesis approach, Runyen-Janecky et al. (2022) recently demonstrated that S. glossinidius uses iron-uptake systems to colonize the host digestive tract, highlighting, in particular, the role of a phosphotransferase sugar (PTS) system component, a fucose transporter and bacterioferritin in the symbiont’s ability to reside in the heme-rich environment of the tsetse host’s midgut (Runyen-Janecky et al. 2022). Analysis of the S. glossinidius genome also suggests the importance of motility during host colonization. However, although the symbiont is virtually capable of biosynthesizing a complete flagellum structure, no apparent swimming motility has been observed (Akman et al. 2002; Toh et al. 2006; Goodhead et al. 2020). S. glossinidius is also endowed with an acylated homoserine lactone (AHL)-based quorum sensing system to modulate gene expression in a density-dependent manner and ensure coordination between symbiotic bacteria during host tissue invasion (Pontes et al. 2008). The key genes regulated by the quorum sensing system, although truncated, appear to have acquired a functionality that also reflects the adaptation of S. glossinidius to a stabilized endosymbiotic lifestyle. In addition, the lipopolysaccharide (LPS) structure of S. glossinidius is truncated and lacks the O-antigen, a major effector protein in Gram-negative bacteria recognized by the insect immune system (Toh et al. 2006). This may partly explain why, despite residing in the hemolymph and digestive tract, S. glossinidius elicits no apparent systemic or epithelial immune response (Hao et al. 2001; Trappeniers et al. 2019). Finally, the symbiont harbor a modified outer membrane protein (OmpA) that allows it not to be sensed as a foe by the host and is essential for the establishment of gut infections in tsetse flies through biofilm formation (Weiss et al. 2008; Maltz et al. 2012). In conclusion, examination of the role of S. glossinidius virulence factors in host colonization revealed that the bacterium’s propensity to invade a wide range of host tissues is explained in part by the conservation of genes encoding a wide range of colonization factors, but which appear to have been shaped for moderate virulence during genome streamlining to enable the bacterium to enter a long-lasting symbiotic lifestyle.
While S. glossinidius has been used as a model for understanding the arsenal of virulence factors that a facultative symbiont can use for host colonization, less attention has been paid to how the host regulates populations of this symbiont and how bacterial population dynamics evolve over the host’s life cycle (at least compared to other model insects such as aphids, stink bugs and weevils). Populations of W. glossinidia and S. glossinidius symbionts appear to fluctuate dramatically during Glossina morsitans morsitans development, with a rapid increase in the adult just after pupal hatching over a two-week period, before gradually declining (Rio et al. 2006). This dynamic is quite similar to that observed in other model insects (Vigneron et al. 2014; Simonet et al. 2018; Maire et al. 2020). However, the tissue distribution of S. glossinidius symbionts during tsetse fly development remains unknown and there is little data concerning factors that may influence symbiont population dynamics such as, for example, host genotype, host sex and environmental conditions (Rio et al. 2006; Hamidou Soumana et al. 2013). Rio et al. (2006) reported that fluctuations in relative humidity can have dramatic effects on the transmission and maintenance of tsetse-associated symbionts in subsequent generations (Rio et al. 2006). Temperature can play a major role in the dynamics of symbionts and symbiotic host organ development, and some facultative symbionts have protective effects for their host against heat stress (Burke et al. 2010; Kikuchi et al. 2016). However, nothing is known about the influence this environmental factor may have on S. glossinidius. Roma et al. (2019) examined the thermal stress responses of S. glossinidius, but only under in vitro conditions (Roma et al. 2019). Yet examining the impact of temperature on bacterial symbioses in tsetse flies is particularly crucial for understanding how climate change may affect the health of these insects, and for determining whether S. glossinidius may be associated with protective effects against heat stress, as is the case for some facultative symbionts in other insect species (Burke et al. 2010; Heyworth et al. 2020). Finally, as S. glossinidius may depend on W. glossinidia for access to certain nutrients such as thiamine (vitamin B1), the dynamics of the facultative symbiont during host development are assumed to mirror those of the obligate symbiont (Belda et al. 2010; Snyder et al. 2010; Wang et al. 2013; Hall et al. 2019). However, the study of the interaction between W. glossinidia and S. glossinidius during host development remains limited, and it is unclear how the two symbionts influence each other and are regulated. These are key aspects that future work should address, in particular to optimize S. glossinidius-based paratransgenic strategies for controlling trypanosome infection in tsetse flies.
3.1.4 S. glossinidius and trypanosome infections
S. glossinidius is a remarkable model for addressing fundamental questions concerning bacterial symbiosis, but also for applied purposes, as there appears to be a correlation between the presence of the symbiont and trypanosome infection in tsetse flies. Indeed, S. glossinidius influences the ability of tsetse flies to carry and transmit pathogenic trypanosomes (Trypanosoma brucei gambiense and Trypanosoma brucei rhodesiense): (1) the facultative symbiont increases tsetse fly susceptibility to trypanosomes (Welburn et al. 1993; Welburn and Maudlin 1999), (2) its selective elimination by antibiotics makes tsetse flies more resistant to trypanosome infection (Dale and Welburn 2001) and (3) the prevalence of parasitic infection is positively correlated with the presence of S. glossinidius in tsetse populations (Farikou et al. 2010; Hamidou Soumana et al. 2013; Kallu et al. 2023). However, the mechanisms behind this possible correlation remain uncertain, and the actual influence of S. glossinidus in the ability of tsetse flies to acquire and transmit the parasite is questioned by some studies (Geiger et al. 2005; Kanté Tagueu et al. 2018; Channumsin et al. 2018).
Despite these controversies, paratrangenic approaches (i.e. the genetic transformation of an organism’s symbionts to confer one or more specific functions that reduce the vector’s competence against pathogens) using S. glossinidus to control trypanosome transmission by tsetse flies have been set up (Aksoy et al. 2008; De Vooght et al. 2012; Vooght et al. 2014; Medlock et al. 2013; Ratcliffe et al. 2022). As the symbiont can be cultivated, genetic engineering manipulations are easily achievable. Paratransgenesis consists in genetically transforming cultivated S. glossinidius with genes encoding antitrypanosomal factors and reintroducing the modified symbiont into tsetse flies. The presence of S. glossinidius in host tissues is expected to establish an unsuitable environment for trypanosome multiplication, thus reducing their transmission capacity. Genetically modified S. glossinidius symbionts can deliver functional anti-trypanosome nanobodies (Nbs) in Glossina morsitans morsitans (De Vooght et al. 2012; Vooght et al. 2022). Their presence in the tsetse gut, in immediate proximity to pathogenic trypanosomes, promotes parasite exposure to antitrypanosomal factors (Geiger et al. 2015). However, this manipulation remains a proof of concept, not least because of the challenge of generating genetically modified strains that transmit vertically in a stable manner (De Vooght et al. 2012; Vooght et al. 2014, 2018). Bacteria expressing transgenes often suffer a fitness cost limiting the establishment of a stable relationship between the host insect and the engineered symbiont. This is an obstacle to be overcome for implementing effective biological control based on paratransgenesis (Elston et al. 2022). In this context, it is therefore crucial to clearly establish the nature of the interaction between S. glossinidius and tsetse flies because the impact of the symbiont on host fitness could influence the effectiveness of this type of approach.
3.2 Other cases of facultative Sodalis symbionts
S. glossinidius is undoubtedly the best-studied facultative symbiont of the genus and it is the only facultative symbiont of the genus whose genome has been sequenced and annotated. However, several studies report members of the genus as putative facultative partners, sometimes resembling S. glossinidius in terms of tissue tropism and cultivability. This is the case of Sodalis melophagi, a presumable facultative symbiont of the sheep ked Melophagus ovinus (Diptera: Hippoboscidae), a blood-feeding parasite of sheep related to tsetse flies (Chrudimský et al. 2012; Nováková et al. 2015). S. melophagi resembles S. glossinidius in many ways: (1) it cohabits with a bacteriocyte-associated endosymbiont of the genus Arsenophonus (A. melophagi), (2) it infects various host tissues, including the milk gland, the gut and the contiguous bacteriome and (3) it can live in insect cell culture as well as in cell-free medium. Unfortunately, there is a lack of experimental data to clarify the nature of the interaction between S. melophagi and M. ovinus and no complete annotated genome is available. Interestingly, given that S. melophagi encodes the same complete set of vitamin B pathways as the obligate endosymbiont A. melophagi, it has been suggested that it might be able to switch from a facultative to an obligate lifestyle and replace A. melophagi (Šochová et al. 2017). In the bloodsucking fly Craterina melbae (Diptera, Hippoboscoidea), the Sodalis symbiont is also a putative facultative associate (Nováková and Hypša 2007), but unfortunately no genomic data are yet publicly available. Finally, the 30% prevalence of Sodalis in Guimaraesiella lice feeding on songbirds also suggests a facultative relationship with these insects (Grossi et al. 2024).
Sodalis symbionts have been detected in a whole range of insects, sometimes belonging to distant taxa, and in which the biological significance of the symbiont’s presence is unclear. This is the case in stink bugs (Hemiptera), where members of Sodalis have been detected in different families (Acanthosomatidae, Pentatomidae, Scutelleridae, Urostylididae, Rhopalidae and Pyrrhocoridae), but sporadically, indicating that the bacteria are not fixed in these insect hosts and are facultative or even commensal partners (Kaiwa et al. 2010, 2011, 2014; Matsuura et al. 2014; Hosokawa et al. 2015; Fourie et al. 2023). Similarly, Sodalis bacteria have been sporadically detected in the potato psyllid Bactericera cockerelli Sulc (Hemiptera: Psylloidea: Triozidae), suggesting the facultative nature of the symbiont in this insect species (Arp et al. 2014; Cooper et al. 2022). Grünwald et al. (2010) investigated the gut flora of several species of bark- and wood-inhabiting cerambycid beetle larvae (Coleoptera: Chrysomeloidea: Cerambycidae) (Grünwald et al. 2010). Interestingly, they discovered that Sodalis massively colonizes the gut of the longicorn beetle Tetropium castaneum, a wood-boring insect that burrows into conifers. Although the authors describe the endosymbiont as a facultative partner, its associated effects are unknown. However, the massive presence of the symbiont in the insect’s digestive tract raises several questions. Is it a potentially pathogenic invasive partner, or, alternatively, does it fulfill a specific function in the digestive tract (e.g. a contribution to wood digestion)? The microbiota of wood-destroying beetles remains little studied, even though they could be teeming with bacteria associated with wood-decomposing properties. Sodalis bacteria have also been sporadically reported in archaeococcoid scale insects (Hemiptera: Coccoidea) (Dhami et al. 2013) and cicadas (Hemiptera: Cicadoidea: Cicadidae) (Matsuura et al. 2018; Zhang et al. 2023), again without the biological meaning of the symbiont’s presence in these insects being established. Interestingly, Sodalis bacteria have been detected in bacteriomes, fat bodies and spermatic tissue of the cicada Angamiana fuscula, but not in oocytes and sperms, suggesting that these are opportunistic strains acquired through horizontal transmission (Zhang et al. 2023). Sodalis bacteria are often found associated with social bees (Hymenoptera), but the nature of these interactions also remains unknown (Rubin et al. 2018). Finally, Sodalis symbionts have been frequently found associated with acorn weevils (Coleoptera: Curculionidae) (Toju et al. 2010, 2013; Toju and Fukatsu 2011). However, unlike Sodalis symbionts obligatorily associated with cereal weevils of the genus Sitophilus, in this case, they appear to be facultative associates.
4 Sodalis symbionts as obligate partners of insects
Most of the Sodalis symbionts whose genomes have been sequenced to date have been identified as obligate nutritional associates (Table 1). This means that they are permanent residents necessary for the survival and reproduction of the host insect, as they provide essential nutrients that the insect cannot find in sufficient quantities in its diet. These obligate Sodalis symbionts typically reside in bacteriocytes (Nováková and Hypša 2007; Fukatsu et al. 2007; Gruwell et al. 2010; Chrudimský et al. 2012; Koga et al. 2013; Smith et al. 2013b; Koga and Moran 2014). These symbiotic host cells, which often assemble to form an organ called a bacteriome, have specifically evolved to house nutritional symbionts and mediate metabolic exchanges between them and the insect host (Douglas 2014). This is achieved by a fine-tuning of the symbiont populations in these eukaryotic cells according to the insect’s nutritional needs (Login et al. 2011; Simonet et al. 2016; Whittle et al. 2021).
4.1 Sodalis pierantonius: the nutritional obligate symbiont of Sitophilus weevils
4.1.1 A recent obligate symbiont
The first cases of obligate symbioses involving associates of the genus Sodalis were identified in cereal weevils of the genus Sitophilus (Coleoptera: Curculionidae: Dryophthoridae) (Heddi et al. 1999; Plarre 2013). These cosmopolitan insects harbor an obligate nutritional endosymbiont, Sodalis pierantonius (formerly SOPE for Sitophilus oryzae primary endosymbiont), which is mainly studied in the context of its association with the rice weevil S. oryzae. S. pierantonius provides its host with vitamins (especially B vitamins) and amino acids (especially tyrosine and phenylalanine) that are lacking in cereals (Wicker and Nardon 1982; Wicker 1983; Oakeson et al. 2014; Vigneron et al. 2014). S. pierantonius boosts cuticle development, increasing its thickness and melanization. This cuticle-enhancing symbiont has been shown to provide desiccation resistance to its host (Kanyile et al. 2023). S. pierantonius is hosted in bacteriocytes that form a bacteriome surrounding the midgut of the insect (Fig. 3) (Zaidman-Rémy et al. 2018; Maire et al. 2020). In females, the symbiont also infects the apex of female ovaries, from which maternal transmission takes place.
The S. pierantonius genome has not undergone a drastic reduction in size: at 4.51 Mb (Table 1) it remains quite large compared to the genomes of most ancient obligate symbionts, which are typically between 2.0 and 0.4 Mb in size (Oakeson et al. 2014). Ancient obligate symbionts share specific genomic features, including small genomes, extreme biases in nucleotide composition, high coding density, and scarcity of mobile DNA (McCutcheon and Moran 2012) (Fig. 1). In S. pierantonius, these genomic features are less extreme. Almost half of the protein-coding genes of its genome are pseudogenes, indicating relaxed selection on many genes and ongoing genomic erosion (Oakeson et al. 2014). In addition, the symbiont has lost the ability to synthesize several essential amino acids and vitamins. However, similarly to S. glossinidius, retains many genes encoding virulence factors that have been tuned to suit the establishment of a mutualistic relationship. Moderate genome erosion, continuous gene inactivation with accumulation of pseudogenes, and strong modulation of genome dynamics with chromosomal rearrangements and proliferation of mobile genetic elements are genomic traits typically associated with facultative symbionts (Gil et al. 2008; Oliver et al. 2010; Lo et al. 2016). Despite differences in metabolic capabilities, S. pierantonius and S. glossinidius are very similar in terms of genomic evolution (Rio et al. 2003; Oakeson et al. 2014), and although obligate, S. pierantonius resembles a facultative symbiont. However, unlike the facultative symbiont of tsetse flies (and other facultative insect symbionts), S. pierantonius is compartmentalized into bacteriocytes, an anatomical integration that promotes metabolic exchange between the symbiont and the host and allows the insect to fine-tune symbiont populations (Masson et al. 2016). It is estimated that the free-living ancestor of S. pierantonius was acquired by cereal weevils less than 1 million years ago (Clayton et al. 2012), likely as a result of the replacement of Candidatus Nardonella, the ancestor endosymbiont of the Dryophthoridae family (Lefèvre et al. 2004; Conord et al. 2008). The weevil-S. pierantonius association is therefore a fairly recent symbiosis involving a bacteriocyte-associated nutritional mutualist. By comparison, the symbiosis between aphids and Buchnera was established over 100 million years ago (Chong et al. 2019). This makes S. pierantonius a valuable model for examining the early stages of obligate nutritional mutualism (Heddi et al. 1998, 1999; Gil et al. 2008).
4.1.2 A model for understanding the dialogue host-symbiont
The weevil-S. pierantonius system has enabled major advances to be made in elucidating the key mechanisms by which the insect controls symbionts, notably via its bacteriocytes. In association with S. oryzae, S. pierantonius produces amino acids that the insect needs to manufacture its cuticle, and which it cannot find in sufficient quantities in the grain (Vigneron et al. 2014). Interestingly, although the symbiont is an obligate partner, it has only a temporary nutritional function, multiplying primarily in young adults whose protective exoskeleton is developing, before being rapidly eliminated within a few days. The multiplication of S. pierantonius corresponds to the host’s physiological need for the amino acids tyrosine (Tyr) and phenylalanine (Phe). Indeed, these amino acids are precursors of the dihydroxyphenylalanine (DOPA) molecule, an essential component in cuticle synthesis. Once the cuticle is formed, DOPA reaches high levels in insects, triggering the elimination of the endosymbiont through apoptosis and the activation of autophagy (Vigneron et al. 2014; Ferrarini et al. 2023).
The S. pierantonius symbionts are contained in large bacteriocytes which, at the larval stage, form a specialized organ (the bacteriome) at the junction of the foregut and the midgut. The molecular dialogue between S. oryzae and S. pierantonius has been extensively studied (Galambos et al. 2023). Maire et al. (2020) found that the symbiont uses its virulence factors, in particular a type III secretion system (T3SS), during metamorphosis: the stem cells are infected by the symbiont before bacteriocytes develop and assemble into a bacteriome (Maire et al. 2020). The S. pierantonius genome also encodes genes required for the synthesis of microbe-associated molecular patterns (MAMPs), including peptidoglycans (PGs), which can activate insect immune responses through their interaction with host pattern recognition receptors (Zaidman-Rémy et al. 2018). Injection of S. pierantonius into insect hemolymph triggers a plethora of antimicrobial peptides (AMPs), suggesting that the nutritional symbiont is perceived as a foe in the host body (Anselme et al. 2008). However, several studies indicate that the immune response within bacteriocytes differs markedly from that expressed outside these cells and is conducive to the multiplication of S. pierantonius (Anselme et al. 2008; Login et al. 2011; Maire et al. 2019). For instance, the antimicrobial peptide coleoptericin A (ColA) keeps S. pierantonius within the bacteriocytes by regulating their multiplication through the inhibition of cell division (Login et al. 2011). The expression of colA is dependent on relish and imd, two genes belonging to the immune deficiency (IMD) pathway. Maire et al. (2019) also found that the weevil peptidoglycan recognition protein LB (PGRP-LB) plays a key role in this homeostasis by cleaving tracheal cytotoxin (TCT), a monomeric form of DAP-type peptidoglycan constantly produced in the bacteriome (Maire et al. 2019). This cleavage prevents the outflow of TCT into the hemolymph and thus the activation of a chronic immune response at this level. Other work has shown that bacteriocytes play a central role in the insect’s general immune response, while providing a safe haven for the internalized symbionts during the period in which they are required for insect development, i.e. during the development of its cuticle (Ferrarini et al. 2022; Galambos et al. 2023). As S. pierantonius is a “young” obligate symbiont that still exhibits a wide range of virulence factors and may be perceived as a pathogen, its interaction with Sitophilus weevils gives the opportunity to decipher mechanisms by which an antagonistic interaction between a eukaryote and a prokaryote can be circumvented during the evolution of bacterial mutualism.
4.2 Other cases of obligate Sodalis symbionts
4.2.1 Sodalis symbionts in Hemiptera
Sodalis bacteria have been identified as obligate partners in many other insect species, particularly hemipterans, although these cases have not yet been studied as thoroughly as in Sitophilus weevils. An example is the long-tailed mealybug Pseudococcus longispinus (Hemiptera: Coccoidea: Pseudococcidae) where Sodalis endolongispinus is a recent obligate partner involved in an interdependent system based on four partners: the insect and its ancestral obligate symbiont Tremblaya princeps and two additional gammaproteobacterial (Symbiopectobacterium endolongispinus and S. endolongispinus). These two additional partners acquired more recently have rather large genomes (3.73 Mb) (Table 1), and exhibit an atypical localization as they live within the cytoplasm of the ancestral symbiont T. princeps (Gatehouse et al. 2012; Garber et al. 2021). The two gammaproteobacterial genomes complement those of T. princeps and the host for biosynthetic pathways of amino acids and vitamins (Garber et al. 2021). Similarly, the mealybug species Maconellicoccus hirsutus, Planococcus citri, Trionymus perrisii, Paracoccus marginatus and Ferrisia virgata (Hemiptera: Coccoidea: Pseudococcidae) are associated with obligate symbionts with highly reduced genomes and considered Sodalis-allied insect endosymbionts, but which are not officially part of the genus, although they form a unique clade with members of the genus Sodalis (Husnik and McCutcheon 2016).
Sodalis symbionts have also evolved with certain aphid species. Virtually all aphid species (Hemiptera: Aphidoidea: Aphididae) host the ancestral obligate symbiont Buchnera aphidicola. But some species host an additional, more recently acquired, co-obligate symbiont that complements B. aphidicola in the biosynthesis of certain essential amino acids and vitamins (Monnin et al. 2020; Renoz et al. 2022; Manzano-Marín et al. 2023). These nutritional di-symbiotic systems have notably evolved in aphids of the subfamilies Lachninae and Chaitophorinae. In the white pine aphid Cinara strobi, a recently acquired Sodalis symbiont having a genome size of 3.07 Mb (Table 1) metabolically complements B. aphidicola for vitamin biosynthesis (biotin and riboflavin), a function previously fulfilled by a co-obligate but now obsolete Serratia symbiotica symbiont (Manzano-Marín et al. 2018). Interestingly, Manzano-Marı́n et al. (2020) have shown that genes of Sodalis-related bacteria providing the basis for the synthesis of certain amino acids and vitamins are found in other co-obligate symbiont species as a result of horizontal transfers as is the case in Erwinia symbionts associated with Cinara aphids (Manzano-Marı́n et al. 2020). This suggests that circulating Sodalis strains can be integrated as nutritional partners in aphids and exchange genetic material with other bacteria, thus acting as a source of metabolic innovations.
In sap-feeding insects of hemipteran suborder Auchenorrhyncha, which feed exclusively on xylem sap, the production of essential nutrients is ensured by a consortium of obligate symbionts, originally Sulcia muelleri (Bacteroidetes) and Zinderia insecticola (Betaproteobacteria), each confined to distinct bacteriocytes (Koga et al. 2013). However, in the meadow spittlebug Philaenus spumarius (Hemiptera: Cercopoidea: Aphrophoridae), a third symbiont, more recently acquired and belonging to the genus Sodalis, has replaced the Zinderia symbiont (Koga and Moran 2014; Ankrah et al. 2018). This co-obligate Sodalis symbiont with a genome size of 2.24 Mb (Table 1) is located in syncytial bacteriocytes near the bacteriocytes specifically hosting Sulcia. Genes replacing its predecessor’s functions for amino acid biosynthesis have been selectively maintained in Sodalis genome. For instance, the genome retains genes supporting efficient energy production pathways, including a complete tricarboxylic acid (TCA) cycle, which could alleviate severe energy limitations in the host due to its xylem-based diet (Koga and Moran 2014).
A Sodalis bacterium is also an obligate partner of the seed bug species Henestaris halophilus (Hemiptera: Lygaeoidea: Geocoridae), where it is found in bacteriocytes forming tubular-shaped bacteriomes on the abdominal flanks (Santos-Garcia et al. 2017). In this insect, Sodalis baculum, is a mutualistic partner capable of providing the amino acids tyrosine, lysine, and some cofactors. The size of its genome (1.62 Mb) is small compared with other Sodalis genomes (Table 1). Interestingly, it still includes many pseudogenes but few mobile elements, indicating an intermediate stage in reductive evolution.
Other Sodalis symbionts are potential obligate insect partners, although they have been less studied to date. For this reason, they are referred to as putative obligate symbionts (Table 1). Among them the Sodalis symbiont strain TME1 (genome size: 3.42 Mb) associated with the wax cochineal Llaveia axin axin (Hemiptera: Coccoidea: Margarodidae), probably a co-obligate symbiont that metabolically complements the more ancient symbiont Walczuchella monophlebidarum endowed with a 0.3 Mb genome (Rosas-Pérez et al. 2017). However, this metabolic complementation has yet to be formally demonstrated. Another case is the Sodalis SPI-1 strain associated with the sucking lice Proechinophthirus fluctus (Phthiraptera: Echinophthiriidae) and featured by a genome of 2.19 Mb (Table 1). The role of this Sodalis-allied symbiont has yet to be elucidated. It appears to be a recently acquired obligate symbiont, capable of infecting a wide range of tissues, exhibiting both an intracellular and extracellular lifestyle. Indeed, it infects bacteriocytes, but also other tissue types such as fat bodies, oviduct and oocytes, a rather singular tissue tropism for an obligate symbiont. Another putative obligate Sodalis symbiont is the strain IL reported in the carrot psyllid Bactericera trigonica (Hemiptera: Psylloidea: Triozidae) (Ghosh et al. 2020). Despite the availability of its genome sequence (1.57 Mb) (Table 1), the function associated with this Sodalis symbiont has yet to be established. However, this strain offers a rare case of intranuclear symbiont since the Sodalis bacteria in this psyllid species are localized inside the nuclei of midgut cells, in addition to being internalized in bacteriocytes. This atypical localization raises the question of the influence that this Sodalis symbiont could have on biological processes taking place in the nuclei of the midgut cells, such as gene regulation and cell division. In the psyllid species Ctenarytaina eucalypti and Heteropsylla cubana (Hemiptera: Psylloidea: Psyllidae), a Sodalis-allied symbiont metabolically complements the ancient obligate symbiont Carsonella ruddii for the biosynthesis of essential amino acids such as tryptophan and arginine (Sloan and Moran 2012).
4.2.2 Sodalis symbionts in blood-feeding insects
The award for the smallest genome for a symbiont officially belonging to the genus Sodalis goes to the symbiont associated with the slender pigeon louse Columbicola wolffhuegeli (Psocodea: Philopteridae) that parasitizes the pied imperial pigeon (Ducula bicolor). Genome analysis reveals features similar to other obligate symbionts in insects, namely a sharp reduction in size and a GC content shift (0.8 Mb, 31.4% of GC) (Table 1), but provides no clear evidence of its function in the host (Alickovic et al. 2021).
4.2.3 Sodalis symbionts in ants
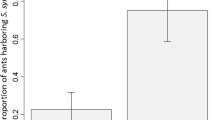
Until recently, the biological significance of Sodalis bacteria in ants was unknown. However, Jackson et al. (2022) recently discovered that Sodalis symbionts have evolved obligate symbioses with ant species of the genera Formica and Plagiolepis (Hymenoptera: Formicidae) (Jackson et al. 2022). All these Sodalis-allied insect symbionts exhibit genomes between 1.0 and 1.5 Mb (Table 1) and are obligate associates involved in the biosynthesis of tyrosine, an amino acid involved in cuticle synthesis. They are found in the bacteriocytes surrounding the midgut of adult queens, but also infect eggs and ovaries. Interestingly, the authors found that the prevalence of Sodalis is not 100% in different species of these ant genera. The retention of Sodalis symbionts in queens and workers of some species, but not in others, suggests that species differ in their dependence on symbiont-derived nutrients, or that Sodalis has lost the ability to produce nutrients in certain host lineages (Jackson et al. 2022). The authors hypothesize that these differences are linked to the feeding ecology of the species examined. Ant species with a predominantly plant-based diet would be more likely to have evolved a dependence on nutritional symbionts than species with an omnivorous diet.
5 The free-living Sodalis bacteria
5.1 Sodalis praecaptivus: a human pathogen
The genus Sodalis also include free-living members. Sodalis praecaptivus, originally known as the HS strain, was isolated from a human hand wound caused by impalement on a crabapple branch (Clayton et al. 2012; Chari et al. 2015). It is assumed, however, that the tree was the original source of the S. praecaptivus infection (Fig. 3) (Clayton et al. 2012). The bacterium is a prototroph capable of growing in minimal media at 37 °C, whose genome has been sequenced and annotated (Clayton et al. 2012; Chari et al. 2015). Comparative genomic analyses revealed that the free-living S. praecaptivus strain has a large and less altered genome (5.16 Mb) than S. glossinidius (4.3 Mb), the facultative symbiont of tsetse flies, and S. pierantonius (4.5 Mb), the obligate symbiont of cereal weevils Sitophilus sp. (Table 1) (Clayton et al. 2012; Oakeson et al. 2014). The free-living strain has preserved many genes inactivated and lost in Sodalis-allied insect symbionts due to rapid genome degeneration, consistent with the hypothesis that S. praecaptivus evolves under strong stabilizing selection in a free-living lifestyle (Clayton et al. 2012; Chari et al. 2015). For instance, compared with S. glossinidius, S. praecaptivus has a more extensive metabolic capability and can utilize carbon sources contained in plants and animals (Clayton et al. 2012). In contrast to the free-living strain genome, those of Sodalis-allied insect symbionts has undergone evolutionary adaptations during the transition to their symbiotic lifestyle: pseudogenes thus account for a significant fraction of their total genomic coding capacity, the result of relaxed selection on non-essential loci (Clayton et al. 2012; Chari et al. 2015). Importantly, comparative genomic and phylogenetic analyses have shown that S. praecaptivus and Sodalis-allied insect symbionts are closely related (Clayton et al. 2012; Oakeson et al. 2014), suggesting that Sodalis symbionts could evolve independently and repeatedly from S. praecaptivus-like environmental bacteria. This makes S. praecaptivus an ideal model for examining the early stages of bacterial mutualism in insects.
5.2 Sodalis members as ubiquitous environmental bacteria
The existence of free-living S. praecaptivus suggests a diverse spectrum of living strategies among Sodalis members that extend beyond associations with insects. Recently, Tláskal et al. (2021) described two free-living isolates from the Sodalis genus, Sodalis ligni sp. nov. strain dw23 and Sodalis sp. strain dw96, which inhabit decomposing deadwood and possess genome characteristics consistent with a non-symbiotic lifestyle (Tláskal et al. 2021). They can grow on laboratory media and have a larger genome relative to other Sodalis strains (6.44 and 5.93 Mb respectively, Table 1). These strains also have a higher number of longer genes and a lower coding density. A pangenome analysis revealed genomic differences between the various Sodalis genomes, showing that deadwood-associated strains encode genes for the active decomposition of biopolymers of plant and fungal origin and can use more diverse carbon sources than the symbiotic strains associated with insects (Tláskal et al. 2021). Accessory genes specific to deadwood-associated strains account for a significant portion of the total identified genes and are involved, for example, in amino acid and carbohydrate metabolism (Tláskal et al. 2021). It has also been suggested that deadwood-associated strains encode multiple nifHDK operons expressing nitrogenase, a key enzyme for metal cofactor-dependent nitrogen fixation (Tláskal et al. 2021). Their genetic potential for nitrogen fixation makes them one of the few members of the nitrogen-fixing Enterobacterales described so far.
Another novel free-living S. ligni strain, called Sodalis ligni strain 159R, has been isolated from an anaerobic enrichment culture of temperate forest soils (Tláskal et al. 2021; Chaput et al. 2022). Whole-genome sequencing revealed a genome size of 6.38 Mb, which appears similar in genome size to S. ligni strain dw23 (Table 1) (Tláskal et al. 2021). This strain is closely related phylogenetically to the deadwood-associated Sodalis strains (Chaput et al. 2022). The authors suggest that S. ligni strain 159R has a genetic potential for anaerobic lignin degradation and aromatic catabolism (Chaput et al. 2022). This strain has a potentially applied dimension, as microorganisms with the ability to depolymerize and catabolize lignin are relevant candidates for lignocellulosic biofuel applications. Removing lignin from lignocellulosic materials is a generally unsustainable and expensive process, but using microorganisms and their enzymes to break down lignin could be a more environmentally-friendly approach (Chaput et al. 2022). However, the process has its limitations, not least the need for constant aeration and mixing. Anaerobic bacteria could therefore be a substitute for lignin depolymerization and conversion into valuable by-products.
5.3 A potential reservoir for new symbiotic associations
The presence of Sodalis in phylogenetically distant insect taxa and the isolation of free-living strains from non-insect material suggest that associations involving the bacterial genus arose during multiple independent infectious events, such as through horizontal transfers or direct acquisition of free-living bacteria from the environment (Clayton et al. 2012; Tláskal et al. 2021). The close relationship between the free-living S. praecaptivus bacterium and Sodalis-allied insect symbionts suggests that this free-living bacterium may be a close relative and putative environmental progenitor of Sodalis-allied insect symbionts (Clayton et al. 2012; Tláskal et al. 2021) (Fig. 2). All this information constitutes a rich data set from which to examine the origin of the insect-Sodalis symbiosis.
Over the past decade, studies have examined how Sodalis-allied insect symbionts have evolved in their transition to symbiosis, using the free-living S. praecaptivus bacterium as a model. One study computationally examined the pathway by which the S. glossinidius symbiont may have taken in its transition to symbiosis with tsetse flies (Hall et al. 2020). The authors used the free-living S. praecaptivus and metabolic modeling in combination with a multi-objective evolutionary algorithm to investigate the evolutionary trajectories of the bacterial symbiont after being internalized (Hall et al. 2020). They found that the order in which key genes are lost influence the evolved populations. Although S. praecaptivus has a free-living lifestyle, it can colonize insects known to be associated with Sodalis-allied insect symbionts by suppressing its virulence through quorum sensing (QS) (Enomoto et al. 2017; Munoz et al. 2020). QS can play a pivotal role in the regulation of symbiotic relationships, including the coordination of group behavior, as demonstrated by the symbiont Vibrio fischeri, which uses this mechanism to regulate bioluminescence in the light organ of its squid host, Euprymna scolopes (Visick et al. 2000). QS can also control the production of extracellular factors, such as antibiotic production in the plant-associated pathogen Erwinia carotovorum (McGowan et al. 1995). Enomoto et al. (2017) identified three genic components of a QS system within the S. praecaptivus genome: a luxI-like synthase homolog (ypeI), which produces N-(3-oxohexanoyl) homoserine lactone, and two luxR-like response regulators. They created mutant strains of S. praecaptivus lacking these genetic mechanisms and showed that the bacterium then expresses an extremely virulent (“killing”) phenotype in the cereal weevil Sitophilus zeamais (Enomoto et al. 2017). The authors thus established the importance of QS in promoting the symbiotic domestication process by repressing virulence factors after infection. Attenuation of virulence by QS occurs when the bacterial population reaches a certain density (Enomoto et al. 2017). Munoz et al. (2020) performed similar experiments with tsetse flies and showed that QS virulence suppression also facilitates the establishment of S. praecaptivus infection in tsetse flies (Munoz et al. 2020). In addition, S. praecaptivus can infect the reproductive tissues of these insects, enabling vertical transmission within a single host generation (Munoz et al. 2020). The modulation of virulence by QS could be a mechanism enabling Sodalis bacteria to establish themselves in insect hosts with attenuated virulence and could explain their propensity to establish symbiotic associations with a wide range of insect hosts. These results therefore suggest that QS modulation may represent a crucial evolutionary step in the early stage of symbiosis between insects and bacteria.
In continuation of these studies, researchers have set up a protocol for microinjecting S. praecaptivus into grain weevil eggs to study its transmission and maintenance through host generation (Su et al. 2022). They showed that the free-living strain can colonize prototypical somatic and germinal bacteriomes after microinjection, leading to its maternal transmission over several generations. The finding that S. praecaptivus can be found in weevil bacteriocytes along with the native symbiont S. pierantonius and can be co-transmitted to offspring (albeit at a low rate) offers the possibility of exploring the evolution of the interaction between an established symbiont and a new bacterial associate (Su et al. 2022). Indeed, studies have suggested that S. pierantonius has recently replaced Nardonella, the ancestral endosymbiont of the Dryophthoridae family to which cereal weevils belong (Charles et al. 2001; Lefèvre et al. 2004; Conord et al. 2008). The ancestor of S. pierantonius could be functionally close to S. praecaptivus, given their phylogenetic proximity. The replacement of S. pierantonius by S. praecaptivus could mimic the way in which Nardonella was replaced by S. pierantonius (Vigneron and Kaltenpoth 2022).
The close relationship of the free-living S. praecaptivus bacterium with Sodalis-allied insect symbionts, as well as its ability to colonize and associate with insect hosts, raise questions about its principal habitat and ecological functions (Clayton et al. 2012; Enomoto et al. 2017; Munoz et al. 2020). Based on virulence factors in its genome and its metabolic capability to use animal and plant hosts, S. praecaptivus could be an opportunistic plant or animal pathogen that could develop associations with insects to achieve vector-based transmission (Su et al. 2022). That is consistent with the fact that S. praecaptivus uses quorum sensing to reduce its virulence towards insects and, in general, natural selection favors bacteria that minimize the fitness costs associated with their transmission by vectors.
Unlike the S. praecaptivus strain, free-living strains associated with deadwood and temperate forest soils do not belong to the same phylogenetic clade as Sodalis-allied insect symbionts. However, a study identified a Sodalis bacterium massively infecting the epithelial cells of the larva’s proximal midgut of the longicorn beetle T. castaneum (Grünwald et al. 2010). No genomic or phylogenetic analysis has been carried out, making it impossible to position this Sodalis member phylogenetically and to study its biological role. It is possible that the ancestor of this bacterium came from the wood on which the beetle’s lava feeds, and resembles the strains living in dead wood (Tláskal et al. 2021; Chaput et al. 2022). It would be interesting to test this hypothesis to confirm that Sodalis-allied insect symbionts originate from different sources and multiple, independent infectious events.
Overall, the data provide evidence that the Sodalis genus is predisposed to evolve associations with insects due to its ability to maintain minor infections, colonize many different insect tissues, and undergo vertical transmission in diverse insect hosts (Clayton et al. 2012; Tláskal et al. 2021). They also corroborate that mutualism can be initiated in insects by the rapid acquisition of environmental bacteria, echoing a recent study showing the quick establishment of a novel symbiosis in the stinkbug Plautia stali from an Escherichia coli strain (Koga et al. 2022). This study demonstrated that a non-symbiotic bacterial strain can be converted into a mutualistic insect associate via a single mutation that disrupts the carbon catabolite repression global transcriptional regulator system.
6 Concluding remarks and future directions
The Sodalis genus is highly diverse and undeniably a powerful tool for studying the many facets of bacterial symbiosis in eukaryotes, as its members embody the different degrees of host dependence and anatomical integration that bacteria can experience during their evolution with insects. Members of the Sodalis genus have repeatedly succeeded in entering new hosts and forming heritable symbioses in a variety of insects. This is reflected in the incongruence between Sodalis phylogeny and insect host taxonomy. Associations between insects and Sodalis symbionts have been studied mainly in tsetse flies and weevils, but their study has been extended to a range of other insect taxa (e.g. aphids, spittlebugs, mealybugs). This repeated success for an endosymbiotic lifestyle reflects a pre-adaptation of Sodalis bacteria to associate with insects, as demonstrated by the use of free-living members in the context of groundbreaking experiments aimed at understanding the initial stages of bacterial endosymbiosis in insects (Munoz et al. 2020; Su et al. 2022). In particular, the Sodalis genus offers an exciting playground for understanding how genes encoding virulence factors are shaped to foster the entry of a pathogenic bacterium into a stabilized symbiotic lifestyle. However, many questions remain unanswered about the interactions that the Sodalis members may have with insects (and beyond). Here, we develop a few of them by way of perspective. One key issue is the exact nature of the relationship between S. glossinidius and tsetse flies. Although this is undoubtedly the most extensively studied Sodalis species, it remains unclear whether the symbiont maintains a mutualistic relationship with its host, or whether it is an opportunistic partner. Nor is it known whether environmental conditions can influence the nature of the interaction between the two partners on the parasitism-mutualism continuum, as has been demonstrated in aphids (Oliver et al. 2010). These are important issues because S. glossinidius could influence the transmission of pathogenic trypanosomes and is used in emerging paratransgenic approaches to counter their transmission. Further experimental efforts are therefore essential to clarify the symbiotic nature of S. glossinidius. Omics approaches could, for example, be used to understand how S. glossinidius populations are regulated in the various infected host tissues, and to determine the impact of the symbiont on host physiology. Furthermore, the diversity of S. glossinidius has been little studied in the different species of tsetse fly that host the symbiont. One hypothesis is that the strains associated with these insects are the result of independent acquisitions and are associated with different phenotypic effects. Genome sequencing and analysis of S. glossinidius strains associated with different tsetse species should be undertaken in the future to address these aspects.
The puzzling tissue localization of Sodalis associates in certain insects also raises questions about their associated phenotypic effects. This is the case of Sodalis bacteria that massively infect the digestive tract of certain Cerambycid beetles (Grünwald et al. 2010). Are these Sodalis bacteria opportunistic associates or do they play a potential role in wood digestion in host insects? Are these Sodalis bacteria systematically associated with these insects? The existence of free-living strains capable of degrading lignin raises the question of the ability of certain wood-eating insects to associate with strains of this type to benefit from their remarkable metabolic properties. Future work will need to examine the associations between wood-eating insects and Sodalis bacteria. Once again, sequencing the genome of these strains is an essential step towards understanding their metabolic capacities and the nature of their relationship with their host. In a similar vein, the atypical intranuclear localization of Sodalis symbionts in the cell nuclei of the midgut of the carrot psyllid B. trigonica also raises the question of the influence the symbiont may have on the molecular and cellular processes taking place in this organelle (Ghosh et al. 2020).
Interactions between Sodalis bacteria and plants are another aspect worth investigating. Indeed, the Sodalis genus is part of the Pectobacteriaceae family, which largely consists of plant pathogens (Adeolu et al. 2016; Motyka et al. 2017). Symbioses with Sodalis bacteria in different insect lineages are the result of multiple independent infections, and it is likely that Sodalis bacteria circulate in plants that likely mediate horizontal transfers to insects. This raises the question of the nature of interactions between Sodalis bacteria and plants. Do Sodalis bacteria have phytopathogenic properties? Are they beneficial endophytes or commensal associates? Can they circulate in plant sap like other insect symbionts (Caspi-Fluger et al. 2011; Pons et al. 2019)? Does interaction with the plant pre-adapt Sodalis bacteria to becoming insect symbionts? These questions need to be addressed, as they will provide a better understanding of the origin of bacterial symbioses in insects and, in the case of Sodalis, how independently established symbioses arose. The possibility of culturing certain Sodalis strains (especially the free-living members S. praecaptivus and S. ligni strains dw23, dw96, and 159R) is conducive to infection experiments on plants to examine these questions. At the same time, it is also essential to decipher in depth the metabolic capabilities of the free-living strains to determine whether they can be used for industrial and public health purposes.
An important technical point to consider is that several Sodalis-allied insect symbionts (e.g. S. glossinidius and S. praecaptivus) have been isolated in pure culture (Dale and Maudlin 1999; Matthew et al. 2005; Chari et al. 2015), opening the door to genetic engineering approaches applied to insect symbionts for fundamental and applied purposes. Indeed, most heritable insect symbionts are recalcitrant to culture approaches, given the dependence they have evolved towards their insect host. Despite these advantages, cultivable Sodalis bacteria, especially the facultative symbiont S. glossinidius, are reluctant to DNA transformation by the standard techniques (heat shock and electroporation) usually used on E. coli and other model bacteria (Pontes and Dale 2006, 2011; Kendra et al. 2020). However, it has recently been discovered that phage-mediated DNA transduction can be used to deliver exogenous DNA to S. glossinidius and S. praecaptivus (Keller et al. 2021). This is an important technical step, as it bypasses the culture requirement for genetically manipulating insect endosymbionts. In future research, this approach could greatly facilitate analyses aimed at identifying the genetic component and pathways governing interactions between bacteria and their eukaryotic hosts (Elston et al. 2022). This approach could also be used for paratransgenesis to control insect-borne microbes that pose economic and health concerns (Elston et al. 2022; Ratcliffe et al. 2022).
Finally, like Santos-Garcia et al. (2017), we recommend a taxonomic re-evaluation of the genus Sodalis (Santos-Garcia et al. 2017). Indeed, phylogenomic analyses suggest that insect symbionts affiliated with other genera actually form a single clade with members of the genus Sodalis (Sloan and Moran 2012; Husnik and McCutcheon 2016; Santos-Garcia et al. 2017). The unification of Sodalis-allied species into the same genus would undoubtedly facilitate further work on the genus and shed light on its tremendous diversity.
References
Adeolu M, Alnajar S, Naushad S, Gupta S R (2016) Genome-based phylogeny and taxonomy of the Enterobacteriales: proposal for Enterobacterales Ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. Nov. Int J Syst Evol Microbiol 66:5575–5599. https://doi.org/10.1099/ijsem.0.001485
Akman L, Yamashita A, Watanabe H et al (2002) Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat Genet 32:402–407. https://doi.org/10.1038/ng986
Aksoy S (1995) Wigglesworthia gen. nov. and Wigglesworthia glossinidia sp. nov., Taxa Consisting of the Mycetocyte-Associated, primary endosymbionts of tsetse flies. Int J Syst Bacteriol 45:848–851. https://doi.org/10.1099/00207713-45-4-848
Aksoy S, Chen X, Hypsa V (1997) Phylogeny and potential transmission routes of midgut-associated endosymbionts of tsetse (Diptera:Glossinidae). Insect Mol Biol 6:183–190. https://doi.org/10.1111/j.1365-2583.1997.tb00086.x
Aksoy S, Weiss B, Attardo G (2008) Paratransgenesis Applied for Control of Tsetse transmitted sleeping sickness. In: Aksoy S (ed) Transgenesis and the management of Vector-Borne Disease. Springer, New York, NY, pp 35–48
Aksoy E, Telleria EL, Echodu R et al (2014) Analysis of multiple tsetse fly populations in Uganda reveals limited diversity and species-specific gut microbiota. Appl Environ Microbiol 80:4301–4312. https://doi.org/10.1128/AEM.00079-14
Alam U, Medlock J, Brelsfoard C et al (2011) Wolbachia Symbiont Infections induce strong cytoplasmic incompatibility in the tsetse fly Glossina morsitans. PLOS Pathog 7:e1002415. https://doi.org/10.1371/journal.ppat.1002415
Alam U, Hyseni C, Symula RE et al (2012) Implications of Microfauna-Host Interactions for Trypanosome Transmission Dynamics in Glossina fuscipes fuscipes in Uganda. Appl Environ Microbiol. https://doi.org/10.1128/AEM.00806-12
Alickovic L, Johnson KP, Boyd BM (2021) The reduced genome of a heritable symbiont from an ectoparasitic feather feeding louse. BMC Ecol Evol 21:108. https://doi.org/10.1186/s12862-021-01840-7
Ankrah N, Chouaia B, Douglas A (2018) The cost of metabolic interactions in Symbioses between insects and Bacteria with reduced genomes. https://doi.org/10.1128/mBio.01433-18. mBio 9:
Anselme C, Pérez-Brocal V, Vallier A et al (2008) Identification of the Weevil immune genes and their expression in the bacteriome tissue. BMC Biol 6:43. https://doi.org/10.1186/1741-7007-6-43
Arp A, Munyaneza JE, Crosslin JM et al (2014) A global comparison of Bactericera cockerelli (Hemiptera: Triozidae) microbial communities. Environ Entomol 43:344–352. https://doi.org/10.1603/EN13256
Attardo GM, Lohs C, Heddi A et al (2008) Analysis of milk gland structure and function in Glossina morsitans: milk protein production, symbiont populations and fecundity. J Insect Physiol 54:1236–1242. https://doi.org/10.1016/j.jinsphys.2008.06.008
Attardo GM, Scolari F, Malacrida A (2020) Bacterial symbionts of Tsetse flies: relationships and functional interactions between tsetse flies and their symbionts. In: Kloc M (ed) Symbiosis: Cellular, Molecular, Medical and Evolutionary aspects. Springer International Publishing, Cham, pp 497–536
Balmand S, Lohs C, Aksoy S, Heddi A (2013) Tissue distribution and transmission routes for the tsetse fly endosymbionts. J Invertebr Pathol 112:S116–S122. https://doi.org/10.1016/j.jip.2012.04.002
Baumann P (2005) Biology of Bacteriocyte-Associated endosymbionts of Plant Sap-Sucking insects. Annu Rev Microbiol 59:155–189. https://doi.org/10.1146/annurev.micro.59.030804.121041
Beard CB, O’Neill SL, Mason P et al (1993) Genetic transformation and phylogeny of bacterial symbionts from tsetse. Insect Mol Biol 1:123–131. https://doi.org/10.1111/j.1365-2583.1993.tb00113.x
Belda E, Moya A, Bentley S, Silva FJ (2010) Mobile genetic element proliferation and gene inactivation impact over the genome structure and metabolic capabilities of Sodalis glossinidius, the secondary endosymbiont of tsetse flies. BMC Genomics 11:449. https://doi.org/10.1186/1471-2164-11-449
Boyd BM, Allen JM, Koga R et al (2016) Two bacterial Genera, Sodalis and Rickettsia, Associated with the seal louse Proechinophthirus fluctus (Phthiraptera: Anoplura). Appl Environ Microbiol 82:3185–3197. https://doi.org/10.1128/AEM.00282-16
Bruner-Montero G, Wood M, Horn HA et al (2021) Symbiont-mediated Protection of Acromyrmex Leaf-Cutter ants from the Entomopathogenic Fungus Metarhizium anisopliae. https://doi.org/10.1128/mBio.01885-21. mBio
Burke G, Fiehn O, Moran N (2010) Effects of facultative symbionts and heat stress on the metabolome of pea aphids. ISME J 4:242–252. https://doi.org/10.1038/ismej.2009.114
Caspi-Fluger A, Inbar M, Mozes-Daube N et al (2011) Horizontal transmission of the insect symbiont Rickettsia is plant-mediated. Proc R Soc B Biol Sci 279:1791–1796. https://doi.org/10.1098/rspb.2011.2095
Channumsin M, Ciosi M, Masiga D et al (2018) Sodalis glossinidius presence in wild tsetse is only associated with presence of trypanosomes in complex interactions with other tsetse-specific factors. BMC Microbiol 18:163. https://doi.org/10.1186/s12866-018-1285-6
Chaput G, Ford J, DeDiego L et al (2022) Sodalis Ligni strain 159R isolated from an anaerobic lignin-degrading Consortium. Microbiol Spectr 10:e0234621. https://doi.org/10.1128/spectrum.02346-21
Chari A, Oakeson KF, Enomoto S et al (2015) Phenotypic characterization of Sodalis praecaptivus sp. nov., a close non-insect-associated member of the Sodalis-allied lineage of insect endosymbionts. Int J Syst Evol Microbiol 65:1400–1405. https://doi.org/10.1099/ijs.0.000091
Charles H, Heddi A, Rahbe Y (2001) A putative insect intracellular endosymbiont stem clade, within the Enterobacteriaceae, infered from phylogenetic analysis based on a heterogeneous model of DNA evolution. Comptes Rendus Académie Sci - Ser III - Sci Vie 324:489–494. https://doi.org/10.1016/S0764-4469(01)01328-2
Chellappan M, Ranjith MT (2021) Metagenomic approaches for insect symbionts. In: Omkar (ed) Microbial approaches for Insect Pest Management. Springer, Singapore, pp 271–313
Chong RA, Park H, Moran NA (2019) Genome evolution of the Obligate Endosymbiont Buchnera aphidicola. Mol Biol Evol 36:1481–1489. https://doi.org/10.1093/molbev/msz082
Chrudimský T, Husník F, Nováková E, Hypša V (2012) Candidatus Sodalis melophagi sp. nov.: phylogenetically Independent comparative model to the tsetse fly Symbiont Sodalis glossinidius. PLoS ONE 7:e40354. https://doi.org/10.1371/journal.pone.0040354
Clayton AL, Oakeson KF, Gutin M et al (2012) A Novel Human-infection-derived bacterium provides insights into the Evolutionary origins of Mutualistic insect–bacterial Symbioses. PLOS Genet 8:e1002990. https://doi.org/10.1371/journal.pgen.1002990
Conord C, Despres L, Vallier A et al (2008) Long-term Evolutionary Stability of Bacterial endosymbiosis in Curculionoidea: additional evidence of Symbiont replacement in the Dryophthoridae Family. Mol Biol Evol 25:859–868. https://doi.org/10.1093/molbev/msn027
Cooper WR, Horton DR, Swisher-Grimm K et al (2022) Bacterial endosymbionts of Bactericera maculipennis and three mitochondrial haplotypes of B. cockerelli (Hemiptera: Psylloidea: Triozidae). Environ Entomol 51:94–107. https://doi.org/10.1093/ee/nvab133
Dale C, Maudlin I (1999) Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int J Syst Bacteriol 49 Pt 1:267–275. https://doi.org/10.1099/00207713-49-1-267
Dale C, Welburn SC (2001) The endosymbionts of tsetse flies: manipulating host-parasite interactions. Int J Parasitol 31:628–631. https://doi.org/10.1016/s0020-7519(01)00151-5
Dale C, Plague GR, Wang B et al (2002) Type III secretion systems and the evolution of mutualistic endosymbiosis. Proc Natl Acad Sci 99:12397–12402. https://doi.org/10.1073/pnas.182213299
Dale C, Jones T, Pontes M (2005) Degenerative evolution and functional diversification of type-III secretion systems in the insect endosymbiont Sodalis glossinidius. Mol Biol Evol 22:758–766. https://doi.org/10.1093/molbev/msi061
De Vooght L, Caljon G, Stijlemans B et al (2012) Expression and extracellular release of a functional anti-trypanosome Nanobody® in Sodalis glossinidius, a bacterial symbiont of the tsetse fly. Microb Cell Factories 11:23. https://doi.org/10.1186/1475-2859-11-23
De Vooght L, Caljon G, De Ridder K, Van Den Abbeele J (2014) Delivery of a functional anti-trypanosome Nanobody in different tsetse fly tissues via a bacterial symbiont, Sodalis glossinidius. Microb Cell Factories 13:156. https://doi.org/10.1186/s12934-014-0156-6
De Vooght L, Caljon G, Van Hees J, Van Den Abbeele J (2015) Paternal transmission of a secondary symbiont during mating in the Viviparous Tsetse fly. Mol Biol Evol 32:1977–1980. https://doi.org/10.1093/molbev/msv077
De Vooght L, Van Keer S, Van Den Abbeele J (2018) Towards improving tsetse fly paratransgenesis: stable colonization of Glossina morsitans morsitans with genetically modified Sodalis. BMC Microbiol 18:165. https://doi.org/10.1186/s12866-018-1282-9
Dedeine F, Vavre F, Fleury F et al (2001) Removing symbiotic Wolbachia bacteria specifically inhibits oogenesis in a parasitic wasp. Proc Natl Acad Sci 98:6247–6252. https://doi.org/10.1073/pnas.101304298
Dennis JW, Durkin SM, Horsley Downie JE et al (2014) Sodalis glossinidius prevalence and trypanosome presence in tsetse from Luambe National Park, Zambia. Parasit Vectors 7:378. https://doi.org/10.1186/1756-3305-7-378
Dhami MK, Buckley TR, Beggs JR, Taylor MW (2013) Primary symbiont of the ancient scale insect family Coelostomidiidae exhibits strict cophylogenetic patterns. Symbiosis 61:77–91. https://doi.org/10.1007/s13199-013-0257-8
Douglas AE (2014) Molecular dissection of nutrient exchange at the insect-microbial interface. Curr Opin Insect Sci 4:23–28. https://doi.org/10.1016/j.cois.2014.08.007
Drew GC, Stevens EJ, King KC (2021) Microbial evolution and transitions along the parasite–mutualist continuum. Nat Rev Microbiol 19:623–638. https://doi.org/10.1038/s41579-021-00550-7
Dyer NA, Lawton SP, Ravel S et al (2008) Molecular phylogenetics of tsetse flies (Diptera: Glossinidae) based on mitochondrial (COI, 16S, ND2) and nuclear ribosomal DNA sequences, with an emphasis on the palpalis group. Mol Phylogenet Evol 49:227–239. https://doi.org/10.1016/j.ympev.2008.07.011
El Yamlahi Y, Bel Mokhtar N, Maurady A et al (2023) Characterization of the Bacterial Profile from Natural and Laboratory Glossina populations. Insects 14:840. https://doi.org/10.3390/insects14110840
Elston KM, Leonard SP, Geng P et al (2022) Engineering insects from the endosymbiont out. Trends Microbiol 30:79–96. https://doi.org/10.1016/j.tim.2021.05.004
Emms DM, Kelly S (2019) OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20:238. https://doi.org/10.1186/s13059-019-1832-y
Engel P, Moran NA (2013) The gut microbiota of insects – diversity in structure and function. FEMS Microbiol Rev 37:699–735. https://doi.org/10.1111/1574-6976.12025
Enomoto S, Chari A, Clayton AL, Dale C (2017) Quorum sensing attenuates virulence in Sodalis praecaptivus. Cell Host Microbe 21:629–636e5. https://doi.org/10.1016/j.chom.2017.04.003
Farikou O, Njiokou F, Mbida Mbida JA et al (2010) Tripartite interactions between tsetse flies, Sodalis glossinidius and trypanosomes–an epidemiological approach in two historical human African Trypanosomiasis foci in Cameroon. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis 10:115–121. https://doi.org/10.1016/j.meegid.2009.10.008
Feldhaar H (2011) Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol Entomol 36:533–543. https://doi.org/10.1111/j.1365-2311.2011.01318.x
Ferrarini MG, Dell’Aglio E, Vallier A et al (2022) Efficient compartmentalization in insect bacteriomes protects symbiotic bacteria from host immune system. Microbiome 10:156. https://doi.org/10.1186/s40168-022-01334-8
Ferrarini MG, Vallier A, Vincent-Monégat C et al (2023) Coordination of host and endosymbiont gene expression governs endosymbiont growth and elimination in the cereal weevil Sitophilus spp. 2023.04.03.535335
Fourie A, Venter SN, Slippers B, Fourie G (2023) Pantoea bathycoeliae sp. nov and Sodalis sp. are core gut microbiome symbionts of the two-spotted stink bug. Front Microbiol 14
Frago E, Zytynska SE, Fatouros NE (2020) Chapter Four - Microbial symbionts of herbivorous species across the insect tree. In: Oliver KM, Russell JA (eds) Advances in Insect Physiology. Academic Press, pp 111–159
Fukatsu T, Koga R, Smith WA et al (2007) Bacterial endosymbiont of the Slender Pigeon Louse, Columbicola columbae, Allied to Endosymbionts of Grain Weevils and Tsetse flies. https://doi.org/10.1128/AEM.01131-07. Appl Environ Microbiol
Galambos N, Vincent-Monegat C, Vallier A et al (2023) Cereal Weevil’s antimicrobial peptides. At the Crosstalk between Development, Endosymbiosis and Immune Response, Immunology
Ganesan R, Janke RS, Kaltenpoth M, Flórez LV (2023) Colonization dynamics of a defensive insect ectosymbiont. Biol Lett 19:20230100. https://doi.org/10.1098/rsbl.2023.0100
Garber AI, Kupper M, Laetsch DR et al (2021) The evolution of interdependence in a four-way mealybug symbiosis. Genome Biol Evol 13:evab123. https://doi.org/10.1093/gbe/evab123
Gatehouse LN, Sutherland P, Forgie SA et al (2012) Molecular and histological characterization of primary (Betaproteobacteria) and secondary (Gammaproteobacteria) endosymbionts of three mealybug species. Appl Environ Microbiol. https://doi.org/10.1128/AEM.06340-11
Geiger A, Cuny G, Frutos R (2005) Two tsetse fly species, Glossina palpalis gambiensis and glossina morsitans morsitans, carry genetically distinct populations of the secondary Symbiont Sodalis glossinidius. Appl Environ Microbiol 71:8941–8943. https://doi.org/10.1128/AEM.71.12.8941-8943.2005
Geiger A, Ponton F, Simo G (2015) Adult blood-feeding tsetse flies, trypanosomes, microbiota and the fluctuating environment in sub-saharan Africa. ISME J 9:1496–1507. https://doi.org/10.1038/ismej.2014.236
Ghosh S, Sela N, Kontsedalov S et al (2020) An Intranuclear Sodalis-Like Symbiont and Spiroplasma Coinfect the Carrot psyllid, Bactericera Trigonica. Psylloidea) Microorganisms 8:692. https://doi.org/10.3390/microorganisms8050692. Hemiptera
Gil R, Belda E, Gosalbes MJ et al (2008) Massive presence of insertion sequences in the genome of SOPE, the primary endosymbiont of the rice weevil Sitophilus oryzae. Int Microbiol Off J Span Soc Microbiol 11:41–48
Goodhead I, Blow F, Brownridge P et al (2020) Large-scale and significant expression from pseudogenes in Sodalis glossinidius – a facultative bacterial endosymbiont. Microb Genomics 6:e000285. https://doi.org/10.1099/mgen.0.000285
Grossi AA, Tian C, Ren M et al (2024) Co-phylogeny of a hyper-symbiotic system: endosymbiotic bacteria (Gammaproteobacteria), chewing lice (Insecta: Phthiraptera) and birds (Passeriformes). Mol Phylogenet Evol 190:107957. https://doi.org/10.1016/j.ympev.2023.107957
Grünwald S, Pilhofer M, Höll W (2010) Microbial associations in gut systems of wood- and bark-inhabiting longhorned beetles [Coleoptera: Cerambycidae]. Syst Appl Microbiol 33:25–34. https://doi.org/10.1016/j.syapm.2009.10.002
Gruwell ME, Hardy NB, Gullan PJ, Dittmar K (2010) Evolutionary relationships among primary endosymbionts of the Mealybug Subfamily Phenacoccinae (Hemiptera: Coccoidea: Pseudococcidae). Appl Environ Microbiol. https://doi.org/10.1128/AEM.01354-10
Hall RJ, Flanagan LA, Bottery MJ et al (2019) A tale of three species: adaptation of Sodalis glossinidius to Tsetse Biology, Wigglesworthia Metabolism, and Host Diet. https://doi.org/10.1128/mBio.02106-18. mBio
Hall RJ, Thorpe S, Thomas GH, Wood AJ (2020) Simulating the evolutionary trajectories of metabolic pathways for insect symbionts in the genus Sodalis. Microb Genomics 6:mgen000378. https://doi.org/10.1099/mgen.0.000378
Hamidou Soumana I, Berthier D, Tchicaya B et al (2013) Population dynamics of Glossina palpalis gambiensis symbionts, Sodalis glossinidius, and Wigglesworthia glossinidia, throughout host-fly development. Infect Genet Evol 13:41–48. https://doi.org/10.1016/j.meegid.2012.10.003
Hao Z, Kasumba I, Lehane MJ et al (2001) Tsetse immune responses and trypanosome transmission: implications for the development of tsetse-based strategies to reduce Trypanosomiasis. Proc Natl Acad Sci U S A 98:12648–12653. https://doi.org/10.1073/pnas.221363798
Heddi A, Charles H, Khatchadourian C et al (1998) Molecular characterization of the principal symbiotic Bacteria of the Weevil Sitophilus oryzae: a peculiar G + C content of an endocytobiotic DNA. J Mol Evol 47:52–61. https://doi.org/10.1007/PL00006362
Heddi A, Grenier A-M, Khatchadourian C et al (1999) Four intracellular genomes direct weevil biology: Nuclear, mitochondrial, principal endosymbiont, and Wolbachia. Proc Natl Acad Sci 96:6814–6819. https://doi.org/10.1073/pnas.96.12.6814
Heyworth ER, Smee MR, Ferrari J (2020) Aphid Facultative symbionts Aid Recovery of their Obligate Symbiont and their host after heat stress. Front Ecol Evol 8
Hosokawa T, Kaiwa N, Matsuura Y et al (2015) Infection prevalence of Sodalis symbionts among stinkbugs. Zool Lett 1:5. https://doi.org/10.1186/s40851-014-0009-5
Hrusa G, Farmer W, Weiss BL et al (2015) TonB-Dependent Heme Iron Acquisition in the tsetse fly Symbiont Sodalis glossinidius. Appl Environ Microbiol. https://doi.org/10.1128/AEM.04166-14
Husnik F, McCutcheon JP (2016) Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. Proc Natl Acad Sci 113:E5416–E5424. https://doi.org/10.1073/pnas.1603910113
Jackson R, Monnin D, Patapiou PA et al (2022) Convergent evolution of a labile nutritional symbiosis in ants. ISME J 16:2114–2122. https://doi.org/10.1038/s41396-022-01256-1
Jousselin E, Cœur d’Acier A, Vanlerberghe-Masutti F, Duron O (2013) Evolution and diversity of Arsenophonus endosymbionts in aphids. Mol Ecol 22:260–270. https://doi.org/10.1111/mec.12092
Kaiwa N, Hosokawa T, Kikuchi Y et al (2010) Primary gut symbiont and secondary, Sodalis-Allied Symbiont of the Scutellerid Stinkbug Cantao Ocellatus. Appl Environ Microbiol. https://doi.org/10.1128/AEM.00421-10
Kaiwa N, Hosokawa T, Kikuchi Y et al (2011) Bacterial symbionts of the Giant Jewel Stinkbug Eucorysses Grandis (Hemiptera: Scutelleridae). Zoolog Sci 28:169–174. https://doi.org/10.2108/zsj.28.169
Kaiwa N, Hosokawa T, Nikoh N et al (2014) Symbiont-supplemented maternal investment underpinning host’s ecological adaptation. Curr Biol 24:2465–2470. https://doi.org/10.1016/j.cub.2014.08.065
Kallu SA, Ndebe J, Qiu Y et al (2023) Prevalence and Association of Trypanosomes and Sodalis glossinidius in Tsetse flies from the Kafue National Park in Zambia. Trop Med Infect Dis 8:80. https://doi.org/10.3390/tropicalmed8020080
Kanté Tagueu S, Farikou O, Njiokou F, Simo G (2018) Prevalence of Sodalis glossinidius and different trypanosome species in Glossina palpalis palpalis caught in the Fontem sleeping sickness focus of the southern Cameroon. Parasite Paris Fr 25:44. https://doi.org/10.1051/parasite/2018044
Kanyile SN, Engl T, Heddi A, Kaltenpoth M (2023) Endosymbiosis allows Sitophilus oryzae to persist in dry conditions. Front Microbiol 14
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kaur R, Shropshire JD, Cross KL et al (2021) Living in the endosymbiotic world of Wolbachia: a centennial review. Cell Host Microbe 29:879–893. https://doi.org/10.1016/j.chom.2021.03.006
Keller CM, Kendra CG, Bruna RE et al (2021) Genetic modification of Sodalis species by DNA transduction. mSphere 6. https://doi.org/10.1128/msphere.01331-20
Kendra CG, Keller CM, Bruna RE, Pontes MH (2020) Conjugal DNA transfer in Sodalis glossinidius, a maternally inherited Symbiont of Tsetse flies. mSphere 5:e00864–e00820. https://doi.org/10.1128/mSphere.00864-20
Kikuchi Y, Tada A, Musolin DL et al (2016) Collapse of insect gut symbiosis under simulated climate change. mBio 7:e01578–e01516. https://doi.org/10.1128/mBio.01578-16
Koga R, Moran NA (2014) Swapping symbionts in spittlebugs: evolutionary replacement of a reduced genome symbiont. ISME J 8:1237–1246. https://doi.org/10.1038/ismej.2013.235
Koga R, Bennett GM, Cryan JR, Moran NA (2013) Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ Microbiol 15:2073–2081. https://doi.org/10.1111/1462-2920.12121
Koga R, Moriyama M, Onodera-Tanifuji N et al (2022) Single mutation makes Escherichia coli an insect mutualist. Nat Microbiol 7:1141–1150. https://doi.org/10.1038/s41564-022-01179-9
Landmann F (2019) The Wolbachia endosymbionts. Microbiol Spectr. https://doi.org/10.1128/microbiolspec.BAI-0018-2019
Lefèvre C, Charles H, Vallier A et al (2004) Endosymbiont phylogenesis in the dryophthoridae weevils: evidence for bacterial replacement. Mol Biol Evol 21:965–973. https://doi.org/10.1093/molbev/msh063
Lindh JM, Lehane MJ (2011) The tsetse fly Glossina fuscipes fuscipes (Diptera: Glossina) harbours a surprising diversity of bacteria other than symbionts. Antonie Van Leeuwenhoek 99:711–720. https://doi.org/10.1007/s10482-010-9546-x
Lo W-S, Huang Y-Y, Kuo C-H (2016) Winding paths to simplicity: genome evolution in facultative insect symbionts. FEMS Microbiol Rev 40:855–874. https://doi.org/10.1093/femsre/fuw028
Login FH, Balmand S, Vallier A et al (2011) Antimicrobial peptides keep insect endosymbionts under control. Science 334:362–365. https://doi.org/10.1126/science.1209728
Maire J, Vincent-Monégat C, Balmand S et al (2019) Weevil pgrp-lb prevents endosymbiont TCT dissemination and chronic host systemic immune activation. Proc Natl Acad Sci U S A 116:5623–5632. https://doi.org/10.1073/pnas.1821806116
Maire J, Parisot N, Galvao Ferrarini M et al (2020) Spatial and morphological reorganization of endosymbiosis during metamorphosis accommodates adult metabolic requirements in a weevil. Proc Natl Acad Sci 117:19347–19358
Maltz MA, Weiss BL, O’Neill M et al (2012) OmpA-mediated biofilm formation is essential for the commensal bacterium Sodalis glossinidius to colonize the tsetse fly gut. Appl Environ Microbiol 78:7760–7768. https://doi.org/10.1128/AEM.01858-12
Malulu DJ, Nyingilili HS, Edward D et al (2023) Interactions among Sodalis, Glossina pallidipes salivary gland hypertrophy virus and trypanosomes in wild Glossina pallidipes. Int J Trop Insect Sci 43:1649–1657. https://doi.org/10.1007/s42690-023-01062-y
Manzano-Marín A, Coeur d’acier A, Clamens A-L et al (2018) A Freeloader? The highly eroded yet large genome of the Serratia symbiotica Symbiont of Cinara Strobi. Genome Biol Evol 10:2178–2189. https://doi.org/10.1093/gbe/evy173
Manzano-Marín A, Coeur d’acier A, Clamens A-L et al (2023) Co-obligate symbioses have repeatedly evolved across aphids, but partner identity and nutritional contributions vary across lineages. Peer Community J 3. https://doi.org/10.24072/pcjournal.278
Manzano-Marı́n A, D’acier AC, Clamens A-L et al (2020) Serial horizontal transfer of vitamin-biosynthetic genes enables the establishment of new nutritional symbionts in aphids’ di-symbiotic systems. ISME J 14:259–273. https://doi.org/10.1038/s41396-019-0533-6
Masson F, Zaidman-Rémy A, Heddi A (2016) Antimicrobial peptides and cell processes tracking endosymbiont dynamics. Philos Trans R Soc B Biol Sci 371:20150298. https://doi.org/10.1098/rstb.2015.0298
Matsuura Y, Hosokawa T, Serracin M et al (2014) Bacterial symbionts of a devastating coffee plant pest, the stinkbug Antestiopsis Thunbergii (Hemiptera: Pentatomidae). Appl Environ Microbiol 80:3769–3775. https://doi.org/10.1128/AEM.00554-14
Matsuura Y, Moriyama M, Łukasik P et al (2018) Recurrent symbiont recruitment from fungal parasites in cicadas. Proc Natl Acad Sci 115:E5970–E5979. https://doi.org/10.1073/pnas.1803245115
Matthew CZ, Darby AC, Young SA et al (2005) The rapid isolation and growth dynamics of the tsetse symbiont Sodalis glossinidius. FEMS Microbiol Lett 248:69–74. https://doi.org/10.1016/j.femsle.2005.05.024
Maudlin I, Welburn SC, Mehlitz D (1990) The relationship between rickettsia-like-organisms and trypanosome Infections in natural populations of tsetse in Liberia. Trop Med Parasitol off Organ Dtsch Tropenmedizinische Ges Dtsch Ges Tech Zusammenarbeit GTZ 41:265–267
McCutcheon JP, Moran NA (2012) Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10:13–26. https://doi.org/10.1038/nrmicro2670
McGowan S, Sebaihia M, Jones S et al (1995) Carbapenem antibiotic production in Erwinia carotovora is regulated by CarR, a homologue of the LuxR transcriptional activator. Microbiology 141:541–550. https://doi.org/10.1099/13500872-141-3-541
Medlock J, Atkins KE, Thomas DN et al (2013) Evaluating paratransgenesis as a potential control strategy for African Trypanosomiasis. PLoS Negl Trop Dis 7:e2374. https://doi.org/10.1371/journal.pntd.0002374
Mfopit YM, Engel JS, Chechet GD et al (2023) Molecular detection of Sodalis glossinidius, Spiroplasma species and Wolbachia endosymbionts in wild population of tsetse flies collected in Cameroon, Chad and Nigeria. BMC Microbiol 23:260. https://doi.org/10.1186/s12866-023-03005-6
Michalkova V, Benoit JB, Weiss BL et al (2014) Vitamin B6 generated by Obligate Symbionts is critical for maintaining Proline Homeostasis and Fecundity in Tsetse flies. Appl Environ Microbiol 80:5844–5853. https://doi.org/10.1128/AEM.01150-14
Moloo SK (1993) The distribution of Glossina species in Africa and their natural hosts. Int J Trop Insect Sci 14:511–527. https://doi.org/10.1017/S1742758400014211
Monnin D, Jackson R, Kiers ET et al (2020) Parallel evolution in the integration of a co-obligate aphid symbiosis. Curr Biol. https://doi.org/10.1016/j.cub.2020.03.011
Moran NA, McCutcheon JP, Nakabachi A (2008) Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet, 42, 165–190. https://doi.org/10.1146/annurev.genet.41.110306.130119
Moran NA, Bennett GM (2014) The tiniest tiny genomes. Annu Rev Microbiol 68:195–215. https://doi.org/10.1146/annurev-micro-091213-112901
Motyka A, Zoledowska S, Sledz W, Lojkowska E (2017) Molecular methods as tools to control plant Diseases caused by Dickeya and Pectobacterium spp: a minireview. New Biotechnol 39:181–189. https://doi.org/10.1016/j.nbt.2017.08.010
Munoz MM, Spencer N, Enomoto S et al (2020) Quorum sensing sets the stage for the establishment and vertical transmission of Sodalis praecaptivus in tsetse flies. PLOS Genet 16:e1008992. https://doi.org/10.1371/journal.pgen.1008992
Niepoth N, Ellers J, Henry LM (2018) Symbiont interactions with non-native hosts limit the formation of new symbioses. BMC Evol Biol 18:27. https://doi.org/10.1186/s12862-018-1143-z
Nováková E, Hypša V (2007) A new Sodalis lineage from bloodsucking fly Craterina melbae (Diptera, Hippoboscoidea) originated independently of the tsetse flies symbiont Sodalis glossinidius. FEMS Microbiol Lett 269:131–135. https://doi.org/10.1111/j.1574-6968.2006.00620.x
Nováková E, Hypša V, Klein J et al (2013) Reconstructing the phylogeny of aphids (Hemiptera: Aphididae) using DNA of the obligate symbiont Buchnera aphidicola. Mol Phylogenet Evol 68:42–54. https://doi.org/10.1016/j.ympev.2013.03.016
Nováková E, Husník F, Šochová E, Hypša V (2015) Arsenophonus and Sodalis symbionts in Louse flies: an analogy to the Wigglesworthia and Sodalis System in Tsetse flies. Appl Environ Microbiol 81:6189–6199. https://doi.org/10.1128/AEM.01487-15
Oakeson KF, Gil R, Clayton AL et al (2014) Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol Evol 6:76–93. https://doi.org/10.1093/gbe/evt210
Oliver KM, Degnan PH, Burke GR, Moran NA (2010) Facultative symbionts in Aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol 55:247–266. https://doi.org/10.1146/annurev-ento-112408-085305
Pais R, Lohs C, Wu Y et al (2008) The Obligate Mutualist Wigglesworthia glossinidia influences Reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl Environ Microbiol. https://doi.org/10.1128/AEM.00741-08
Perreau J, Moran NA (2022) Genetic innovations in animal–microbe symbioses. Nat Rev Genet 23:23–39. https://doi.org/10.1038/s41576-021-00395-z
Plarre R (2013) An attempt to reconstruct the natural and cultural history of the granary weevil, Sitophilus granarius (Coleoptera: Curculionidae). EJE 107:1–11. https://doi.org/10.14411/eje.2010.001
Pons I, Renoz F, Noël C, Hance T (2019) Circulation of the Cultivable Symbiont Serratia symbiotica in Aphids is mediated by plants. Front Microbiol 10
Pons I, González Porras MÁ, Breitenbach N et al (2022a) For the road: calibrated maternal investment in light of extracellular symbiont transmission. Proc R Soc B Biol Sci 289:20220386. https://doi.org/10.1098/rspb.2022.0386
Pons I, Scieur N, Dhondt L et al (2022b) Pervasiveness of the symbiont Serratia symbiotica in the aphid natural environment: distribution, diversity and evolution at a multitrophic level. FEMS Microbiol Ecol 98:fiac012. https://doi.org/10.1093/femsec/fiac012
Pontes MH, Dale C (2006) Culture and manipulation of insect facultative symbionts. Trends Microbiol 14:406–412. https://doi.org/10.1016/j.tim.2006.07.004
Pontes MH, Dale C (2011) Lambda red-mediated genetic modification of the insect endosymbiont Sodalis glossinidius. Appl Environ Microbiol 77:1918–1920. https://doi.org/10.1128/AEM.02166-10
Pontes MH, Babst M, Lochhead R et al (2008) Quorum sensing primes the oxidative stress response in the Insect Endosymbiont, Sodalis glossinidius. PLoS ONE 3:e3541. https://doi.org/10.1371/journal.pone.0003541
Ratcliffe NA, Furtado Pacheco JP, Dyson P et al (2022) Overview of paratransgenesis as a strategy to control pathogen transmission by insect vectors. Parasit Vectors 15:112. https://doi.org/10.1186/s13071-021-05132-3
Ratledge C, Dover LG (2000) Iron Metabolism in pathogenic Bacteria. Annu Rev Microbiol 54:881–941. https://doi.org/10.1146/annurev.micro.54.1.881
Renoz F, Ambroise J, Bearzatto B et al (2022) The Di-Symbiotic systems in the aphids Sipha maydis and periphyllus lyropictus provide a contrasting picture of recent Co-obligate Nutritional endosymbiosis in Aphids. Microorganisms 10:1360. https://doi.org/10.3390/microorganisms10071360
Rio RVM, Lefevre C, Heddi A, Aksoy S (2003) Comparative Genomics of insect-symbiotic Bacteria: influence of host environment on Microbial Genome Composition. Appl Environ Microbiol 69:6825–6832. https://doi.org/10.1128/AEM.69.11.6825-6832.2003
Rio RVM, Wu Y, Filardo G, Aksoy S (2006) Dynamics of multiple symbiont density regulation during host development: tsetse fly and its microbial flora. Proc R Soc B Biol Sci 273:805–814. https://doi.org/10.1098/rspb.2005.3399
Rio RVM, Symula RE, Wang J et al (2012) Insight into the transmission biology and species-specific functional capabilities of tsetse (Diptera: glossinidae) obligate symbiont Wigglesworthia. mBio 3:e00240–e00211. https://doi.org/10.1128/mBio.00240-11
Roma JS, D’Souza S, Somers PJ et al (2019) Thermal stress responses of Sodalis glossinidius, an indigenous bacterial symbiont of hematophagous tsetse flies. PLoS Negl Trop Dis 13:e0007464. https://doi.org/10.1371/journal.pntd.0007464
Rosas-Pérez T, de León AV-P, Rosenblueth M et al (2017) The Symbiome of Llaveia Cochineals (Hemiptera: Coccoidea: Monophlebidae) Includes a Gammaproteobacterial Cosymbiont Sodalis TME1 and the Known Candidatus Walczuchella monophlebidarum. IntechOpen
Rubin BER, Sanders JG, Turner KM et al (2018) Social behaviour in bees influences the abundance of Sodalis (Enterobacteriaceae) symbionts. R Soc Open Sci 5:180369. https://doi.org/10.1098/rsos.180369
Runyen-Janecky LJ, Brown AN, Ott B et al (2010) Regulation of High-Affinity Iron Acquisition homologues in the tsetse fly Symbiont Sodalis glossinidius. J Bacteriol 192:3780–3787. https://doi.org/10.1128/JB.00161-10
Runyen-Janecky LJ, Scheutzow JD, Farsin R et al (2022) Heme-induced genes facilitate endosymbiont (Sodalis glossinidius) colonization of the tsetse fly (Glossina Morsitans) midgut. PLoS Negl Trop Dis 16:e0010833. https://doi.org/10.1371/journal.pntd.0010833
Salem H, Bauer E, Kirsch R et al (2017) Drastic genome reduction in an Herbivore’s Pectinolytic Symbiont. Cell 171:1520–1531e13. https://doi.org/10.1016/j.cell.2017.10.029
Santos-Garcia D, Silva FJ, Morin S et al (2017) The All-Rounder Sodalis: a New Bacteriome-Associated Endosymbiont of the lygaeoid bug Henestaris Halophilus (Heteroptera: Henestarinae) and a critical examination of its evolution. Genome Biol Evol 9:2893–2910. https://doi.org/10.1093/gbe/evx202
Shaida SS, Weber JS, Gbem TT et al (2018) Diversity and phylogenetic relationships of Glossina populations in Nigeria and the Cameroonian border region. BMC Microbiol 18:180. https://doi.org/10.1186/s12866-018-1293-6
Simonet P, Duport G, Gaget K et al (2016) Direct flow cytometry measurements reveal a fine-tuning of symbiotic cell dynamics according to the host developmental needs in aphid symbiosis. Sci Rep 6:1–13. https://doi.org/10.1038/srep19967
Simonet P, Gaget K, Balmand S et al (2018) Bacteriocyte cell death in the pea aphid/Buchnera symbiotic system. Proc Natl Acad Sci 115:E1819–E1828. https://doi.org/10.1073/pnas.1720237115
Sloan DB, Moran NA (2012) Genome reduction and co-evolution between the primary and secondary bacterial symbionts of psyllids. Mol Biol Evol 29:3781–3792. https://doi.org/10.1093/molbev/mss180
Smith CL, Weiss BL, Aksoy S, Runyen-Janecky LJ (2013a) Characterization of the Achromobactin Iron Acquisition Operon in Sodalis glossinidius. Appl Environ Microbiol 79:2872–2881. https://doi.org/10.1128/AEM.03959-12
Smith WA, Oakeson KF, Johnson KP et al (2013b) Phylogenetic analysis of symbionts in feather-feeding lice of the genus Columbicola: evidence for repeated symbiont replacements. BMC Evol Biol 13:109. https://doi.org/10.1186/1471-2148-13-109
Snyder AK, Rio RVM (2015) Wigglesworthia morsitans Folate (vitamin B9) biosynthesis contributes to tsetse host fitness. Appl Environ Microbiol 81:5375–5386. https://doi.org/10.1128/AEM.00553-15
Snyder AK, Deberry JW, Runyen-Janecky L, Rio RVM (2010) Nutrient provisioning facilitates homeostasis between tsetse fly (Diptera: Glossinidae) symbionts. Proc Biol Sci 277:2389–2397. https://doi.org/10.1098/rspb.2010.0364
Snyder AK, McMillen CM, Wallenhorst P, Rio RVM (2011) The phylogeny of Sodalis-like symbionts as reconstructed using surface-encoding loci. FEMS Microbiol Lett 317:143–151. https://doi.org/10.1111/j.1574-6968.2011.02221.x
Šochová E, Husník F, Nováková E et al (2017) Arsenophonus and Sodalis replacements shape evolution of symbiosis in louse flies. PeerJ 5:e4099. https://doi.org/10.7717/peerj.4099
Stoy KS, Gibson AK, Gerardo NM, Morran LT (2020) A need to consider the evolutionary genetics of host–symbiont mutualisms. J Evol Biol 33:1656–1668. https://doi.org/10.1111/jeb.13715
Su Y, Lin H-C, Teh LS et al (2022) Rational engineering of a synthetic insect-bacterial mutualism. Curr Biol 32:3925–3938e6. https://doi.org/10.1016/j.cub.2022.07.036
Sudakaran S, Kost C, Kaltenpoth M (2017) Symbiont Acquisition and replacement as a source of Ecological Innovation. Trends Microbiol 25:375–390. https://doi.org/10.1016/j.tim.2017.02.014
Tláskal V, Pylro VS, Žifčáková L, Baldrian P (2021) Ecological divergence within the Enterobacterial Genus Sodalis: from insect symbionts to inhabitants of decomposing Deadwood. Front Microbiol 12
Toh H, Weiss BL, Perkin SAH et al (2006) Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res 16:149–156. https://doi.org/10.1101/gr.4106106
Toju H, Fukatsu T (2011) Diversity and Infection prevalence of endosymbionts in natural populations of the chestnut weevil: relevance of local climate and host plants. Mol Ecol 20:853–868. https://doi.org/10.1111/j.1365-294X.2010.04980.x
Toju H, Hosokawa T, Koga R et al (2010) Candidatus Curculioniphilus Buchneri, a novel clade of bacterial endocellular symbionts from weevils of the Genus Curculio. Appl Environ Microbiol 76:275–282. https://doi.org/10.1128/AEM.02154-09
Toju H, Tanabe AS, Notsu Y et al (2013) Diversification of endosymbiosis: replacements, co-speciation and promiscuity of bacteriocyte symbionts in weevils. ISME J 7:1378–1390. https://doi.org/10.1038/ismej.2013.27
Trappeniers K, Matetovici I, Van Den Abbeele J, De Vooght L (2019) The tsetse fly displays an attenuated Immune response to its secondary Symbiont, Sodalis glossinidius. Front Microbiol 10
Tsagmo Ngoune JM, Reveillaud J, Sempere G et al (2019) The composition and abundance of bacterial communities residing in the gut of Glossina palpalis palpalis captured in two sites of southern Cameroon. Parasit Vectors 12:151. https://doi.org/10.1186/s13071-019-3402-2
Vigneron A, Kaltenpoth M (2022) Symbiosis: creating a tractable intracellular insect-microbe association. Curr Biol CB 32:R943–R946. https://doi.org/10.1016/j.cub.2022.08.011
Vigneron A, Masson F, Vallier A et al (2014) Insects recycle endosymbionts when the benefit is over. Curr Biol 24:2267–2273. https://doi.org/10.1016/j.cub.2014.07.065
Visick KL, Foster J, Doino J et al (2000) Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J Bacteriol 182:4578–4586. https://doi.org/10.1128/jb.182.16.4578-4586.2000
Vooght LD, Ridder KD, Hussain S et al (2022) Targeting the tsetse-trypanosome interplay using genetically engineered Sodalis glossinidius. PLOS Pathog 18:e1010376. https://doi.org/10.1371/journal.ppat.1010376
Wamwiri FN, Alam U, Thande PC et al (2013) Wolbachia, Sodalis and trypanosome co-infections in natural populations of Glossina Austeni and Glossina pallidipes. Parasit Vectors 6:232. https://doi.org/10.1186/1756-3305-6-232
Wang J, Brelsfoard C, Wu Y, Aksoy S (2013) Intercommunity effects on microbiome and GpSGHV density regulation in tsetse flies. J Invertebr Pathol 112:S32–S39. https://doi.org/10.1016/j.jip.2012.03.028
Weeks AR, Turelli M, Harcombe WR et al (2007) From parasite to Mutualist: Rapid Evolution of Wolbachia in Natural populations of Drosophila. PLOS Biol 5:e114. https://doi.org/10.1371/journal.pbio.0050114
Weiss BL, Mouchotte R, Rio RVM et al (2006) Interspecific transfer of bacterial endosymbionts between tsetse fly species: Infection establishment and effect on host fitness. Appl Environ Microbiol 72:7013–7021. https://doi.org/10.1128/AEM.01507-06
Weiss BL, Wu Y, Schwank JJ et al (2008) An insect symbiosis is influenced by bacterium-specific polymorphisms in outer-membrane protein A. Proc Natl Acad Sci 105:15088–15093. https://doi.org/10.1073/pnas.0805666105
Welburn SC, Maudlin I (1999) Tsetse–trypanosome interactions: rites of passage. Parasitol Today 15:399–403. https://doi.org/10.1016/S0169-4758(99)01512-4
Welburn SC, Maudlin I, Ellis DS (1987) In vitro cultivation of rickettsia-like-organisms from Glossina spp. Ann Trop Med Parasitol 81:331–335. https://doi.org/10.1080/00034983.1987.11812127
Welburn SC, Arnold K, Maudlin I, Gooday GW (1993) Rickettsia-like organisms and chitinase production in relation to transmission of trypanosomes by tsetse flies. Parasitology 107(Pt 2):141–145. https://doi.org/10.1017/s003118200006724x
Whittle M, Barreaux AMG, Bonsall MB et al (2021) Insect-host control of obligate, intracellular symbiont density. Proc R Soc B Biol Sci 288:20211993. https://doi.org/10.1098/rspb.2021.1993
Wicker C (1983) Differential vitamin and choline requirements of symbiotic and aposymbiotic s. oryzae (coleoptera: curculionidae). https://doi.org/10.1016/0300-9629(83)90311-0
Wicker C, Nardon P (1982) Development responses of symbiotic and aposymbiotic weevils Sitophilus oryzae L. (Coleoptera, curculionidae) to a diet supplemented with aromatic amino acids. https://doi.org/10.1016/0022-1910(82)90008-7
Zaidman-Rémy A, Vigneron A, Weiss BL, Heddi A (2018) What can a weevil teach a fly, and reciprocally? Interaction of host immune systems with endosymbionts in Glossina and Sitophilus. BMC Microbiol 18:150. https://doi.org/10.1186/s12866-018-1278-5
Zhang W, Wang J, Huang Z et al (2023) Symbionts in Hodgkinia-free cicadas and their implications for co-evolution between endosymbionts and host insects. Appl Environ Microbiol 0:e01373–e01323. https://doi.org/10.1128/aem.01373-23
Zientz E, Dandekar T, Gross R (2004) Metabolic Interdependence of Obligate Intracellular Bacteria and their insect hosts. Microbiol Mol Biol Rev 68:745–770. https://doi.org/10.1128/MMBR.68.4.745-770.2004
Zug R, Hammerstein P (2015) Bad guys turned nice? A critical assessment of Wolbachia mutualisms in arthropod hosts. Biol Rev 90:89–111. https://doi.org/10.1111/brv.12098
Zytynska SE, Tighiouart K, Frago E (2021) Benefits and costs of hosting facultative symbionts in plant-sucking insects: a meta-analysis. Mol Ecol 30:2483–2494. https://doi.org/10.1111/mec.15897
Acknowledgements
This study was financially supported by the National Fund for Scientific Research, FNRS grant no. 1B374.21. François Renoz acknowledges support from the Japan Society for the Promotion of Science (JSPS) - Postdoctoral Fellowships for Research in Japan [Fellowship Number PE22052]. The authors thank Prof. Thierry Hance for his valuable comments. This paper is publication BRC411 of the Biodiversity Research Centre (Université Catholique de Louvain).
Author information
Authors and Affiliations
Contributions
FR and IP wrote the paper. HA performed the phylogenomic analyses and made manuscript revisions. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Renoz, F., Arai, H. & Pons, I. The genus Sodalis as a resource for understanding the multifaceted evolution of bacterial symbiosis in insects. Symbiosis 92, 187–208 (2024). https://doi.org/10.1007/s13199-023-00966-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13199-023-00966-0