Abstract
Tsetse flies (Glossina spp.) act as the sole vectors of the African trypanosome species that cause Human African Trypanosomiasis (HAT or African Sleeping Sickness) and Nagana in animals. These flies have undergone a variety of specializations during their evolution including an exclusive diet consisting solely of vertebrate blood for both sexes as well as an obligate viviparous reproductive biology. Alongside these adaptations, Glossina species have developed intricate relationships with specific microbes ranging from mutualistic to parasitic. These relationships provide fundamental support required to sustain the specializations associated with tsetse’s biology. This chapter provides an overview on the knowledge to date regarding the biology behind these relationships and focuses primarily on four bacterial species that are consistently associated with Glossina species. Here their interactions with the host are reviewed at the morphological, biochemical and genetic levels. This includes: the obligate symbiont Wigglesworthia, which is found in all tsetse species and is essential for nutritional supplementation to the blood-specific diet, immune system maturation and facilitation of viviparous reproduction; the commensal symbiont Sodalis, which is a frequently associated symbiont optimized for survival within the fly via nutritional adaptation, vertical transmission through mating and may alter vectorial capacity of Glossina for trypanosomes; the parasitic symbiont Wolbachia, which can manipulate Glossina via cytoplasmic incompatibility and shows unique interactions at the genetic level via horizontal transmission of its genetic material into the genome in two Glossina species; finally, knowledge on recently observed relations between Spiroplasma and Glossina is explored and potential interactions are discussed based on knowledge of interactions between this bacterial Genera and other insect species. These flies have a simple microbiome relative to that of other insects. However, these relationships are deep, well-studied and provide a window into the complexity and function of host/symbiont interactions in an important disease vector.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 Introduction
Human African Trypanosomiasis (HAT, sleeping sickness) and Animal African Trypanosomiasis (AAT, Nagana) are neglected tropical diseases affecting humans and animals throughout countries in sub-Saharan Africa. These diseases are inevitably fatal if left untreated. Recently, coordinated efforts by affected countries have brought case numbers down dramatically (Franco et al. 2018; Meyer et al. 2016). However, these efforts need to be maintained to prevent future resurgences as has occurred in the past when control efforts waned. The animal form of the disease remains a large problem and has tremendous impacts on the economy and people of affected areas (Isaac et al. 2017). The trypanosomes responsible for these diseases are vectored from a reservoir to host or from host to host by tsetse flies (Glossina spp.). Flies within the genus Glossina are the sole vectors of African trypanosomes in humans and the primary vectors of trypanosomes in animals. Vector control is a primary mechanism for control of trypanosomiasis transmission (Vreysen et al. 2013). This is due to the low natural population numbers of Glossina in the wild. This feature differentiates tsetse flies from more prolific vectors such as mosquitoes and is due to their unique biology and unusual adaptations.
Tsetse flies are specialized vectors. They differ from other Dipteran vectors in that both sexes feed on blood alone and derive no nutrition from alternative sources such as floral nectar as mosquitoes do (Buxton 1955; Magnarelli 1978). Tsetse flies also differ in their reproductive biology as females give birth to live fully developed 3rd instar larvae (Mellanby 1937; Tobe 1978). The female fly provides for all of the nutritional requirements of the larvae by secretion of a lactation product (or milk) into the uterus where the developing larvae live (Ma and Denlinger 1974). The evolutionary pressures behind these adaptations are unknown. However, these pressures resulted in extreme adaptations, which required or were facilitated by the presence of symbiotic bacteria. Obligate mutualistic bacterial relationships with insects are well documented and often function to compensate for dietary adaptations to exploit rich but nutrient-deficient dietary sources (Buchner 1965; Douglas 1989). Examples include homopteran insects such as aphids that feed exclusively on plant sap, which lacks essential amino acids. Other insect species with wood-based diets such as carpenter ants (Hymenoptera) (Schroder et al. 1996) and termites (Isoptera) (Jucci 1952) require microbial assistance with the digestion of cellulose. Hematophagous insects subsist entirely on blood, which lacks in B vitamin-associated compounds. In addition to tsetse, examples of other blood feeders with symbiotic relationships include bed bugs (Cimicidae) (Chang and Musgrave 1973) and lice (Anoplura) (Puchta 1955).
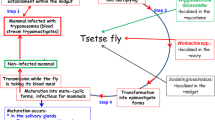
Symbiotic relationships can range from mutualistic where the insect and the bacteria are receiving positive effects from their relationship, to parasitic where the bacteria are exploiting the host resulting in negative impacts on fitness and/or reproduction and some relationships are commensal where the bacteria benefit at no apparent cost to the insect host. To date, research shows that tsetse flies have consistent relationships with a few bacterial species. In general, tsetse flies have a relatively simple microbiome consisting primarily of four characterized species. These include Wigglesworthia glossinidia, Sodalis glossinidius, Wolbachia pipientis, and Spiroplasma (Fig. 19.1). The presence or absence of all but one of these bacterial species is variable. The only bacterial species consistently found in all tsetse fly species is Wigglesworthia glossinidia, which functions as an obligate mutualist. The interactions between vectors, their microbiota, and the parasites they vector are intricate. This chapter will explore the biology behind these relationships and the implications for tsetse physiology, vector control, and vectorial capacity.
2 Wigglesworthia glossinidia
2.1 Discovery
The first documentations of bacteria (bacteroids) living within the digestive tract of tsetse flies were made in Glossina fuscipes and Glossina tachinoides by Robert Koch in 1907 (Stuhlmann 1907). This was followed by a paper reported by Roubaud who hypothesized that the bacteria were symbiotic and aided in the digestion of blood by the fly (Roubaud 1919). These papers documented the presence of the bacteria and observed they were living intracellularly in a tissue (at the time termed the mycetome, currently referred to as the bacteriome), made up of giant hypertrophic cells. These cells contained bacteria, which Roubaud hypothesized that they were released during blood feeding to aid in digestion. The symbionts were further characterized in detail by Wigglesworth in 1929 in his analysis of the physiology of the tsetse digestive tract as well as by Buxton in his extensive 1955 memoir on Glossina biology for the London School of Tropical Medicine and Hygiene (Buxton 1955; Wigglesworth 1929). Wigglesworth did not observe free forms of the bacteria and suspected the observed free bacteria may have been released from the bacteriome due to damage during dissection. These papers hypothesized that Wigglesworthia functioned to supplement or aid in the digestion of tsetse’s blood-specific diet; however, the lack of antibiotics made it difficult to target the bacteria for experimental purposes. Roubaud and Wigglesworth also hypothesized that these bacteria were vertically transmitted to the intrauterine larvae via the milk secretions generated by the female accessory (milk) glands.
The first functional evidence of Wigglesworthia’s role as a symbiont was described when flies were fed on antibiotic-treated rabbits. The antibiotic fed flies were observed to have morphological disturbances in the bacteriome and a lack of bacteria. These flies showed decreased blood-feeding rates and appeared incapable of developing larvae, which were aborted early in their development (Hill et al. 1973). This observation was repeated by Nogge by direct antibiotic treatment of tsetse flies via an artificial blood meal (Nogge 1976). It was also shown that flies feeding on rabbits inoculated with Wigglesworthia become sterile. The antisera against Wigglesworthia from the rabbits are specifically bound and killed the bacteria in the bacteriome of flies (Nogge 1978). Nogge also noted that the loss of fecundity in tsetse in which endosymbionts had been removed (aposymbiotic) could be partially complemented by supplementation of blood meals with B-vitamins (Nogge 1976). While the loss of symbionts has a negative impact on fecundity, it does not seem to influence lifespan (Nogge and Gerresheim 1982).
2.2 Localization and Transmission
The localization of Wigglesworthia in Glossina is specific to two locations. As mentioned previously, they are found living intracellularly within the giant bacteriocyte cells of the bacteriome tissue. The bacteriome is located in the anterior section of the Glossina midgut and from the exterior appears as a horseshoe-shaped structure that encircles the dorsal side of the midgut. The bacteriocyte giant cells protrude into the anterior midgut from the basal lamina of the gut and occupy a significant portion of the gut lumen (Wigglesworth 1929). Ultrastructural analysis of the bacteriome by transmission electron microscopy revealed many new observations about the relationship between tsetse and Wigglesworthia. Visualization of the bacteriome revealed that Wigglesworthia was gram-negative based on their membrane composition and rod-shaped up to 8 microns in length and 1–1.4 microns in width. The analysis also identified the presence of secretory vacuoles within Wigglesworthia connected to the plasma membrane (Reinhardt et al. 1972).
Analysis of genes enriched in the bacteriome of Glossina morsitans revealed tissue-specific gene expression patterns indicative of the specialization of these cells. The bacteriocytes are enriched in the expression of vesicular transport/exocytosis-related genes, sodium/potassium pumps, a lectin, and the peptidoglycan recognition protein LB (PGRP-LB) (Bing et al. 2017). The lectin is a carbohydrate-binding protein that in some systems is associated with symbiont uptake and localization (Bulgheresi et al. 2006; Chaston and Goodrich-Blair 2010; Kita et al. 2015; Wang et al. 2010). The ion pumps and secretion-associated genes suggest that the bacteriocytes are maintaining an ion gradient that may assist in the exocytosis and secretion of intracellular compounds and may function to export Wigglesworthia-derived cofactors. The presence of PGRP-LB is thought to have an immunosuppressive function by binding and cleaving free Wigglesworthia-derived peptidoglycan (Wang et al. 2009).
Initial hypotheses by Wigglesworth suggested that Wigglesworthia were transmitted vertically from mother to offspring via the glandular “milk” secretions produced by the female accessory gland (milk gland) that flow into the uterus and are imbibed by the developing larvae. Ultrastructural analysis of the milk gland tubules by Denlinger and Ma revealed the presence of bacteria living within the lumen of the gland and often located in close association with the openings of the secretory reservoirs from which milk is released (Ma and Denlinger 1974). The milk gland-associated bacteria showed differences from those in the bacteriome. The bacteria in the milk gland were extracellular, had a thicker cell wall, and were coated in filamentous fimbriae 5–7 nm in diameter and 2 μm in length. These bacteria were later confirmed as Wigglesworthia by in situ staining of milk gland tissues with a specific 16S ribosomal RNA probe (Attardo et al. 2008; Balmand et al. 2013). The morphological differences between the bacteriocyte and milk gland-derived Wigglesworthia appear to reflect differential morphological states required for intra and extracellular life within Glossina. Larval Glossina are exposed to Wigglesworthia only via their gut contents. During the larval stage, the gut is a closed system with larvae lacking an anal opening. A possible mechanism is if bacteria ingested during larvigenesis end up being partitioned, with some invading the cells of the bacteriome and others making their way to the milk gland possibly during metamorphosis. However, the route and mechanism of this colonization process remains an open question.
2.3 Phylogeny
Genetic analysis of Wigglesworthia by 16S ribosomal DNA analysis revealed that they fall into a discrete lineage close to members of the Enterobacteriaceae family within the class γ Proteobacteria. The Enterobacteriaceae are facultative anaerobes and include many species that function as gastrointestinal pathogens including Escherichia coli (E. coli) O157:H7, Salmonella, and Shigella. The γ Proteobacteria also contain a lineage of obligate symbionts of aphids in the genus Buchnera (Munson et al. 1991; Unterman et al. 1989). Analysis of Wigglesworthia derived from five species of Glossina representing three subgenera of tsetse flies revealed that these bacteria also form a specific lineage within the γ-3 subdivision of the Proteobacteria. Phylogenetic comparison of Wigglesworthia by 16S rDNA sequencing revealed them as close relatives of Buchnera, symbionts identified in other insects and E. coli (Aksoy 1995). The phylogeny of Wigglesworthia from the different species of Glossina closely matched the phylogeny of the flies themselves. This suggests the evolution of Wigglesworthia is parallel to that of Glossina reflecting an ancient relationship derived from a common ancestor (Aksoy et al. 1995; Chen et al. 1999; Symula et al. 2011). This relationship is estimated to have begun in an ancestral tsetse 50–100 million years ago.
2.4 Genetics, Functional Conservation, and Gene Loss
Sequencing of the Wigglesworthia genome revealed multiple insights into its biology and functional role as an obligate symbiont. The sequence was obtained from Wigglesworthia derived from the tsetse species Glossina brevipalpis. The genome is contained in a single chromosome and a plasmid with a total size of 697,742 base pairs containing 621 protein-coding sequences with 89.1% of the genome containing coding regions (Akman et al. 2002). The size of the genome is very small relative to most free-living bacteria and is similar in size to that of the obligate symbiont of the Pea aphid Buchnera aphidicola. The small size of these genomes likely reflects the symbiotic nature of these bacteria as they exist in a protected state within the bacteriome tissues of the host (Bennett and Moran 2013; Moran and Bennett 2014). The protective environment and the presence of host-derived nutrients allow for relaxed evolutionary selection on functions required for free-living and competitive environments. In addition, obligate endosymbionts also tend to undergo rapid evolution due to the potential bottlenecks between generations during vertical transmission. To date, attempts to culture Wigglesworthia have failed, suggesting that these reductions have compromised their ability to survive outside the parameters of the host environment. Wigglesworthia’s genome also has a very low guanine/cytosine content of 22%, which is also similar to that of other intracellular bacteria. This is thought to result from the loss of genes coding for repair and recombination enzymes associated with the SOS, base excision, and nucleotide excision repair systems. However, genes with high expression levels such as ribosomal proteins and chaperonins have a bias toward the use of GC-rich amino acid codons relative to the rest of the genome (Herbeck et al. 2003). In addition, Wigglesworthia lacks the DnaA and OriC genes, which are essential components of the chromosomal replication complex, suggesting they are accomplishing this by an alternative method. Another interesting observation is that Wigglesworthia also lacks the gene coding for phosphofructokinase (PfkA), which is required for energy production via glycolysis and genes required for amino acid biosynthesis (Zientz et al. 2004).
The genes retained by Wigglesworthia are informative as to the selective pressures and unique environmental requirements within Glossina. These illustrate the biological demands required for maintenance of the vitality and fecundity of the host. While Wigglesworthia lacks a key enzyme for glycolysis, it has retained the transketolase and transaldolase enzymes utilized by the nonoxidative branch of the pentose phosphate pathway. This pathway may be facilitating the oxidation of abundant blood meal-derived amino acids for energy. In addition, they have retained the fructose bisphosphatase (fbp) gene, which facilitates the synthesis of complex carbohydrates by gluconeogenesis as well as all the enzymatic components required for lipid, phospholipid, and nucleotide biosynthesis (Zientz et al. 2004).
The Wigglesworthia genome has maintained genes associated with biosynthetic pathways required for the synthesis of B-vitamins and cofactors such as pantothenate (B5), biotin (B7), riboflavin (B2), folate (B9), thiamine (B1), nicotinamide (B3), and pyridoxine (B6) (Akman et al. 2002). These compounds are deficient in blood and the retention of these biosynthetic pathways appears to be associated with nutritional supplementation of the tsetse host. This observation reinforces the original hypotheses of Roubaud, Wigglesworth, and Nogge. In addition, Wigglesworthia maintains all the genes required for the assembly and function of flagella. The function of the flagella in Wigglesworthia is unknown. The observation of fimbriae associated with extracellular milk gland Wigglesworthia suggests that the fimbriae are flagella and may be involved in motility and cellular invasion of the bacteriome during vertical transfer. Expression of the flagella-associated genes motA and fliC is specific to maternal milk glands, larvae and early pupal stages of development and their expression are not observed in bacteriome-associated Wigglesworthia (Rio et al. 2012).
High-throughput analysis of bacteriome-associated Wigglesworthia gene expression revealed enriched expression of functional classes of genes by the symbiont (Bing et al. 2017). The most abundant genes expressed by Wigglesworthia code for chaperonins, which aid in protein folding, as well as proteins associated with the degradation of misfolded proteins. These are hypothesized to compensate for the AT-rich nature of Wigglesworthia’s genome and the absence of rigorous DNA repair systems lost over evolutionary time. The second most highly expressed class of Wigglesworthia genes was associated with B-vitamin biosynthesis. Genes associated with thiamine (B1) biosynthesis were most highly expressed, followed by biotin (B7), riboflavin (B2), pantothenate and CoA (B5), nicotinamide (B3), pyridoxine (B6), and folate (B9) (Bing et al. 2017).
2.5 Roles in Glossina Digestion and Metabolism
The relationship between Glossina and Wigglesworthia is complex and is essential to multiple aspects of tsetse fly biology including digestion, metabolism, reproduction, and immunity. In the absence of Wigglesworthia, female Glossina becomes unable to develop intrauterine larval offspring, however, aposymbiotic males do not appear to suffer significant impacts to their fertility (Hill et al. 1973; Nogge 1976; Pais et al. 2008). Females in this state develop and ovulate oocytes. After fertilization, the oocytes appear to undergo embryogenesis, however, females abort their developing larvae early in intrauterine development. This suggests that either the larvae are dying due to malnutrition or that something is initiating premature parturition. Malnutrition could result from the lack of a Wigglesworthia-derived compound or the female’s inability to effectively transfer nutrients via the milk in the aposymbiotic state. The retention of genes required for B-vitamin compound biosynthesis in the context of Wigglesworthia’s extreme genomic reduction highlights their significance within the context of the host and their role in dietary supplementation.
Research on the function of these compounds in Glossina biology has revealed important insights into this relationship. Dietary supplementation of Glossina blood meals with thiamine results in decreased Wigglesworthia population density and reduced expression of the Wigglesworthia thiC gene. This suggests that Wigglesworthia can sense environmental thiamine levels and regulate the expression of its biosynthetic pathway accordingly. Ectopic treatment with thiamine also caused changes in Wigglesworthia density suggesting that there is a potential population-regulatory mechanism either via the Glossina immune system or a regulatory mechanism within Wigglesworthia (Snyder et al. 2012).
In Glossina, the amino acid proline is the primary source of ATP production via the tricarboxylic acid (TCA) cycle. The proline is catabolized to alanine to produce ATP. The alanine is then shuttled back to lipid storage tissues where it is restored to proline by the addition of lipid-derived acetyl-CoA. This differs from many other insects that utilize carbohydrates as their primary source of ATP (Bursell 1960, 1963, 1966). A key enzyme required for the conversion of alanine back to proline is alanine-glyoxylate aminotransferase (AGAT). This enzyme requires vitamin B6 (pyridoxal 5′-phosphate) as a cofactor to function. Aposymbiotic flies have significantly lower levels of vitamin B6 as well as lower levels of free proline in their hemolymph. The lack of a key energy-associated metabolite during pregnancy is likely a significant impediment during the energetically demanding process of lactation in females (Michalkova et al. 2014).
The production of folate (Vitamin B9) by Wigglesworthia is important for fitness and reproductive function in Glossina. Folate functions as a cofactor in many pathways/processes including DNA/RNA synthesis, repair, methylation, and production of the amino acid methionine. Wigglesworthia upregulates genes associated with the chorismite and folate biosynthesis in young and pregnant female flies relative to males and virgin females. This suggests that Wigglesworthia responds to the requirements of the host. In addition, Glossina expresses a folate transporter protein in proportion to the folate level in the bacteriome (Snyder and Rio 2015). There are significant molecular and biochemical interactions occurring at the interface of the symbiont and host that maintain the equilibrium of the system. Female flies fed on glyphosate, an inhibitor of the chorismate and folate biosynthetic pathways, show a number of pathologies associated with folate deprivation. Offspring from folate-deficient mothers had longer larval and pupal development times, weighed less, and had a smaller adult body size (Snyder and Rio 2015).
Comparison of tetracycline-treated aposymbiotic female flies with age-matched pregnant females by untargeted metabolomic analyses revealed dysfunction in multiple metabolic pathways. As predicted by previous work, aposymbiotic females show deficiencies in B-vitamins and associated compounds resulting from the loss of Wigglesworthia (Bing et al. 2017). Many pathways with altered metabolite profiles are those with enzymes dependent on B vitamins as enzymatic cofactors. Metabolism of glycogen appears disrupted as aposymbiotic flies show increased levels of unprocessed glycogen metabolites relative to symbiotic flies. This is likely a result of the deficiency in vitamin B6, which is required as a cofactor by the enzyme glycogen phosphorylase. Another disruption appears in a pathway downstream of the glycogen pathway, the pentose phosphate pathway. This pathway processes glucose-6-phosphate derived from glycogen catabolism and converts it into NADPH (utilized in reduction reactions and fatty acid biosynthesis) and 5 carbon sugars (pentoses), which are required for the synthesis of aromatic amino acids and nucleotides. A key enzyme in this pathway is transketolase, which is dependent on vitamin B1 (thiamine). The disruption of this pathway results in a severe deficiency in the metabolite phosphoribosyl pyrophosphate (PRPP). This compound is an essential precursor to nucleotide biosynthesis and aposymbiotic flies appear to be impacted by this as they show significant deficiencies in purine and pyrimidine nucleotide biosynthesis (Bing et al. 2017). The deficiencies in nucleotide biosynthesis in combination with the B-vitamin deficiencies have further impacts downstream in the methionine metabolism pathway. This pathway is responsible for the production of S-adenosyl-methionine (SAM), which functions as a universal methyl donor in methylation reactions. Levels of SAM in aposymbiotic flies relative to controls were the second-lowest among all the metabolites identified after PRPP. The biosynthesis of SAM requires adenosine, folate, and vitamin B6 all of which are deficient in aposymbiotic tsetse.
The exact cause behind why aposymbiotic female tsetse is unable to develop intrauterine larvae remains undetermined. However, the observed deficiencies indicate that lipid metabolism could be impacted. The reduced levels of NADPH could negatively impact fatty acid biosynthesis. In addition, the reduced levels of SAM could be impacting the synthesis of phospholipids, which play an important role in lipid storage, metabolism, and mobilization.
2.6 Roles in Immunity and Development
The interactions between Glossina and Wigglesworthia are ancient and complex affecting many aspects of the system. The role of Glossina species as vectors of trypanosomes makes the topic of immunity of particular interest. The obligate presence of symbionts requires fine tuning of immune responses such that potential pathogens are selectively targeted to avoid damage of symbiotic populations. Experimental treatment of female flies with the antibiotic ampicillin resulted in selective elimination of Wigglesworthia from the milk gland. However, the intracellular population in the bacteriome remained intact due to the inability of the antibiotic to penetrate the bacteriocytes (Pais et al. 2008). This resulted in females that maintain their fecundity; however, they do not pass on Wigglesworthia to their offspring. This finding allowed the study of the developmental impacts on tsetse physiology/biology associated with the loss of Wigglesworthia.
Glossina that develop in the absence of Wigglesworthia shows phenotypic effects that highlight the impacts of development in the absence of their symbionts. Aposymbiotic flies derived from ampicillin-treated mothers show a significant decrease in survival over time relative to symbiotic flies. This phenotype is exacerbated at higher temperatures. These flies also show deficiencies in digestion manifested as a reduced rate of blood meal digestion and the abundant presence of undigested hemoglobin in the gut 2 days after blood feeding. Flies lacking Wigglesworthia also appear more susceptible to infection by trypanosomes (Pais et al. 2008).
Analysis of immune gene expression associated with the presence or absence of Wigglesworthia showed that the peptidoglycan recognition protein PGRP-LB is upregulated in the bacteriome and milk gland tissues in the presence of Wigglesworthia and is downregulated in aposymbiotic flies (Dawadi et al. 2018; Wang and Aksoy 2012). PGRP-LB was first shown to moderate the immune system function in Drosophila through degradation of bacterially derived peptidoglycan and buffering activation of the Immune Deficient (IMD) immune pathway (Zaidman-Remy et al. 2006). Flies in which PGRP-LB is knocked down show higher levels of IMD-dependent antimicrobial gene expression. In Glossina, this protein is thought to regulate symbiont population numbers and protect the symbionts from the immune system in the bacteriome and the milk gland. A similar role for PGRP-LB in mediating the symbiont/host relationship is also documented in the weevil (Sitophilus zeamais) (Anselme et al. 2006). Progeny of female PGRP-LB knockdown flies shows lower densities of Wigglesworthia suggesting that this is an important mechanism that ensures safe transfer of the symbiont from mother to offspring (Wang and Aksoy 2012).
Aposymbiotic Glossina show higher levels of IMD pathway-associated innate immune gene expression. However, if larvae develop in the absence of symbionts, they display an immunocompromised phenotype resulting from incomplete cellular immune cell development. This manifests as an inability to melanize and clot cuticular wounds and lack of mature hemocytes in their hemolymph (Weiss et al. 2011). Adult flies with this phenotype are very susceptible to hemocelic infections by E. coli relative to wild flies. Aposymbiotic Glossina appear to be deficient in plasmatocyte and crystal cell type hemocytes. These types of cells are responsible for phagocytosis of foreign bodies (plasmatocytes) and secretion of chemical components required for melanization (crystal cells). The hemocyte deficiency results from an inability of hemocyte precursor cells to differentiate into active immune cells. These findings suggest that Wigglesworthia provides stimuli required for proper hemocyte and immune system maturation (Weiss et al. 2011). Deeper analysis of this phenotype revealed that the presence of Wigglesworthia in developing larvae stimulates the expression of a gene coding for an odorant-binding protein, obp6. Knockdown of obp6 expression in larval tsetse inhibited differentiation of precursor hemocytes into crystal cells specifically. The lack of crystal cells results in an inability of flies to melanize cuticular wounds or invading pathogens in a manner similar to aposymbiotic flies (Benoit et al. 2017). OBPs are thought to function as transporters for small hydrophobic molecules typically associated with olfaction (Zhou 2010). In this case, OBP6 is hypothesized to mediate the activity of a Wigglesworthia-derived compound required for crystal cell hemocyte differentiation. This finding expands the functional role of OBPs (previously thought to primarily function in olfaction) into the realm of development and immunity.
2.7 Summary
The relationship between Wigglesworthia and Glossina is intricate and essential for the survival of both organisms. The dependence of Glossina development, immunity, metabolic function, and reproduction on Wigglesworthia highlights the intricacy of this partnership. While this partnership is likely unique in some respects, the dependence of other blood-feeding insects suggests that there may be similarities in the relationships between obligate blood feeders and their associated obligate symbiotic relationships.
3 Sodalis glossinidius
3.1 Discovery and Genetic Characterization
Sodalis was first described in 1987 as a Rickettsia-like-organism isolated from the hemolymph of Glossina by culturing it in a cell line from the Asian Tiger mosquito Aedes albopictus (Welburn et al. 1987). The bacterium was classified as a member of the family Enterobacteriaceae within the γ-3 subdivision of the Proteobacteria. This family contains multiple species found as symbionts in other insects (Aksoy et al. 1997; Hosokawa et al. 2015; Novakova et al. 2015). The bacterium was cultured and characterized outside of the fly under microaerobic conditions using a solid-phase culture technique. A new genus, Sodalis, was derived from this work to contain secondary symbionts of other tsetse and insect species (Dale and Maudlin 1999). Initial analyses of the Sodalis genome by pulse field gel electrophoresis determined it to be ~2 Megabases (Mb) (Akman et al. 2001); however, follow up efforts with more advanced sequencing technologies revealed the genome to be 4.1 Mb (Toh et al. 2006). Sodalis also carries multiple extrachromosomal plasmids (Akman et al. 2001). The extrachromosomal material contains genes coding for pilus proteins required for bacterial conjugation and horizontal gene transfer as well as siderophores required for the binding and transport of iron (Darby et al. 2005).
Comparison of the genome relative to E. coli (another member of the Enterobacteriaceae) via microarray revealed that the two organisms are ~85% orthologous in terms of gene composition. The genome contains three regions encoding Type III secretion systems (SSR-1, SSR-2, and SSR-3), which appear to have independent ancestries and differences in their constitution (Toh et al. 2006). The SSR-1 region is similar in composition to the Type III secretion system ysa in the bacteria Yersinia enterolitica, while SSR-2+3 bear similarity to SPI-1+2 in Salmonella. Many pseudogenes were found in the Sodalis genome relative to that of free-living Salmonella typhi and Yersinia pestis. Genes lacking functional orthologs in Sodalis are associated with anaerobic metabolism and carbohydrate transport/metabolism. In addition, there are reductions in genes coding for membrane-associated proteins as well as those involved in cell structure (Rio et al. 2003). Further analysis of the Sodalis genome revealed the loss of the arginine biosynthesis pathway in Sodalis suggesting that it scavenges this amino acid from Glossina (Belda et al. 2010). Another feature of the Sodalis genome was the loss of the pathway for synthesis of the B-vitamin thiamine, yet they have retained a thiamine transporter gene. Wigglesworthia is capable of thiamine production suggesting that Sodalis may scavange Wigglesworthia-derived thiamine (Snyder et al. 2010). While Sodalis does not show the same level of genomic reduction as Wigglesworthia, the loss of these genes suggests it could be at an intermediate stage in the transition from a free-living facultative relationship to that of a symbiont.
An updated annotation of the Sodalis genome and development of a new culture system utilizing media with defined ingredients facilitated in-depth analysis of the nutritional and growth requirements of Sodalis in vitro. This work determined that Sodalis does encode an arginine biosynthesis system and is not completely auxotrophic. Growth on media lacking arginine is reduced, but not eliminated. Addition of excess L-glutamate to this system rescued the reduced growth rate associated with arginine depletion suggesting that Sodalis can compensate for low environmental arginine with L-glutamate and that this compound functions as an important source of carbon and nitrogen. Another interesting finding is that the carbohydrate N-acetyl-D-glucosamine, a component of insect chitin, is an important dietary factor for Sodalis. This suggests that Sodalis may have adapted to utilize an abundant carbohydrate associated with the Glossina physiological environment (Hall et al. 2019).
3.2 Biology, Localization, and Transmission
As opposed to Wigglesworthia, the Sodalis’s range is not limited to specific tissues and are found throughout the fly. During intrauterine larval development, Sodalis migrates into the developing larvae via the milk secretions. Upon eclosion from the pupa, a significant increase in the numbers of Sodalis relative to Glossina host cells is observed over a two-week period. The numbers of Sodalis then appear to fluctuate over time in adults (Rio et al. 2006). Sodalis is found intra and extracellularly in tissues throughout larval and adult Glossina including the midgut, fat body, milk gland, uterus, and oviduct. The ovaries and developing oocytes remain uninfected (Attardo et al. 2008; Balmand et al. 2013). Sodalis are also capable of transfer from infected males to females via seminal secretions during mating (De Vooght et al. 2015). Work by Dale and Welburn revealed that treatment of Glossina with the antibiotic streptozotocin selectively eliminates Sodalis while leaving bacteriome-based Wigglesworthia intact. This treatment did not impact Glossina fecundity, as observed with the elimination of Wigglesworthia. However, flies lacking Sodalis showed a significant reduction in lifespan and increased susceptibility to trypanosome infection in laboratory settings (Dale and Welburn 2001).
Investigations into the role of Type III secretion systems in the relationship between Sodalis and Glossina revealed differences relative to orthologous loci in free-living and parasitic bacterial species. These systems are often associated with pathogenicity due to their role in the secretion of toxins and inflammatory agents as well as invasion of host cells. Disruption of this system by transposon-mediated mutagenesis of the invasion protein C (invC) gene (a component of the type III secretion system) prevented an invasion of insect cells cultured in vitro (Dale et al. 2001). SSR-2 seems to have lost gene functionality for the proteins coding for the needle structure of the secretion system. Functional analysis of these pathways suggests that SSR-1 is required for cell invasion while SSR-2 is required for intracellular division (Dale et al. 2005).
Iron acquisition is essential for bacterial survival and proliferation and Sodalis has retained protein-coding genes required for iron chelation, transmembrane transport, heme metabolism, and iron storage. These include an inner membrane heme ABC transporter system (hemTUV), an outer membrane heme transporter (hemR), an iron/manganese transporter (sitABCD), ferritin-like proteins, and an iron-responsive negative transcriptional regulator (Fur). Analysis of the regulatory regions from two of these Sodalis genes in E. coli revealed that the Fur regulatory protein is required for the correct expression of these genes. Under iron-rich environmental conditions, these genes are repressed; however, in the absence of the Fur regulator, they are constitutively expressed. In Glossina, these genes appear to be responsive to environmental iron as in unfed teneral flies these genes are upregulated. However, at 48 hours post blood meal, the genes are significantly downregulated (Runyen-Janecky et al. 2010; Smith et al. 2013). The hemR outer membrane heme transporter is essential for the survival of Sodalis in Glossina as strains with mutations in this protein do not establish. The tonB gene codes for the protein that supplies energy to hemR and is also essential to this system. Strains with mutations in tonB show a similar phenotype to that of hemR mutants in that they are unable to colonize Glossina in its absence (Hrusa et al. 2015).
Analysis of quorum-sensing mechanisms in Sodalis revealed that the compound N-(3-oxohexanoyl) homoserine lactone regulates population numbers. The presence of this compound activates the transcription of genes coding for oxidative stress-response proteins that may function to reduce the oxidative burden associated with symbiosis (Pontes et al. 2008). The changes in these systems relative to free-living bacteria may reflect a reduction in mechanisms associated with pathogenesis and adaptations required for invasion by and survival of Sodalis in Glossina tissues. Transfer of Sodalis strains between Glossina species revealed that Sodalis originating in one species of Glossina are capable of colonization and survival in another species indicating that these adaptations are not species-specific. Rather, they allow Sodalis to be compatible across the Glossina genus (Weiss et al. 2006). Sodalis has a truncated lipopolysaccharide (LPS) structure that lacks the O-antigen, which may facilitate its ability to live within Glossina with the induction of an immune response (Toh et al. 2006). In depth analysis of outer membrane proteins revealed that the OmpA (Outer membrane protein A) gene in Sodalis contains polymorphisms not found in other pathogenic bacteria such as E. coli. The infection of Glossina with E. coli is fatal under normal conditions. However, infection with E. coli containing an OmpA mutation is nonpathogenic while infection with Sodalis containing the native E. coli OmpA gene results in a lethal infection (Weiss et al. 2008). In Sodalis, the OmpA surface protein is essential for the establishment of gut infections within Glossina. Mutation of the native Sodalis OmpA gene prevents biofilm formation that is required for the protection of bacteria from the flies’ immune response (Maltz et al. 2012).
3.3 Distribution in Wild Populations and Relationship with Vector Competence
Analysis of wild populations of flies revealed that the presence of Sodalis is heterogeneous in the field. The field-collected samples of Glossina austeni and Glossina pallidipes from Kenya and South Africa reveal Sodalis infection rates to be around 3.7% in G. austeni and 16% in G. pallidipes (Wamwiri et al. 2013). Analysis of Glossina populations from Luambe National Park in Zambia also revealed high levels of variability in the proportions of individuals infected with Sodalis. The species with the highest infection level was Glossina brevipalpis with 93.7% infection rate followed by Glossina morsitans and G. pallidipes with 17.5% and 1.4%, respectively (Dennis et al. 2014). llumina sequencing-based analyses of Glossina microbiota of fly populations in Uganda have revealed a broader diversity of microbial taxa than previously identified across multiple species of Glossina including Glossina fuscipes fuscipes (from five distinct populations), Glossina morsitans morsitans, and Glossina pallidipes. All samples were predominantly occupied by Wigglesworthia with it constituting the majority of bacterial sequences. A comparison of the profiles of the remaining bacterial taxa revealed the microbiomes from the different G. fuscipes populations showed significant diversity between their microbial constitution. The survey also identified a high prevalence of low-intensity Sodalis infections across all the groups tested (Aksoy et al. 2014). Another high-throughput microbiome analysis of Glossina palpalis palpalis from Cameroon revealed similar results with Wigglesworthia being the predominant species in the flies and the investigators also found low-level infections of Sodalis throughout the samples (Tsagmo Ngoune et al. 2019).
Comparison of the Trypanosoma infection rates in Sodalis infected versus uninfected field collected flies identified a significant positive correlation between the two suggesting that the presence of Sodalis may facilitate the establishment of blood meal-derived Trypanosoma infections (Farikou et al. 2010; Soumana et al. 2013; Wamwiri et al. 2013). Work on changes in Sodalis gene expression in permissive versus nonpermissive Glossina pallidipes revealed significant changes in expression profiles between the two groups. A large proportion of the changes associated with the refractory flies is associated with a viral prophage carried by Sodalis, which suggests that activation of this phage may be associated with an antitrypanosomal response (Hamidou Soumana et al. 2014a, b). However, recent work studying the correlation between Sodalis and Trypanosoma coinfections in wild-caught flies suggests that other factors such as geographic location, trypanosomal species, Glossina species, and the age and sex of the flies analyzed can be confounding factors (Channumsin et al. 2018).
Selective clearance of Sodalis using the antibiotic streptozotocin allowed researchers to generate a Sodalis-free line of Glossina (Sod-) in the lab for comparison with infected individuals (Sod+) by high-throughput gene expression analysis. An interesting finding from this research is that there were no significant changes in immune-responsive genes between Sod- and Sod+ flies. However, the challenge of these flies with E. coli or Sodalis praecaptivus, a free-living relative of S. glossinidius, resulted in a robust immune response. They also showed that activation of Glossina’s innate immune response did not have a significant impact on endogenous Sodalis numbers suggesting that the bacterium is resistant to host immune factors. Comparisons of susceptibility to Trypanosoma brucei brucei infection between Sod- and Sod+ flies showed no significant difference between the two groups.
3.4 Potential for Use in Paratransgenesis
The ability to isolate and culture Sodalis outside of Glossina provides the opportunity for a reduction of Glossina’s vectorial capacity via manipulation of its symbionts. This strategy is called paratransgenesis and is a promising alternative to direct genetic manipulation of the vector species (Coutinho-Abreu et al. 2010). Current genetic transformation technologies, functional in insects such as mosquitoes and Drosophila, are not a viable option in Glossina due to their low reproductive rate and viviparous physiology. Paratransgenesis works through the genetic transformation of cultured Sodalis with a gene encoding an antitrypanosomal factor. The engineered Sodalis are then reintroduced into Glossina where, in principle, the presence of the modified symbiont would create a hostile environment for invading trypanosomes resulting in reduced or eliminated vectorial capacity. Paratransgenesis was demonstrated in laboratory studies to be an effective strategy in kissing bugs (Order Hemiptera, Family Triatomidae) as a way to reduce their vectorial capacity for the Chagas pathogen, Trypanosoma cruzi (Beard et al. 2001, 2002).
Research into the use of Sodalis as a paratransgenic agent shows promise. Transformation of Sodalis with 9 different cationic antimicrobial peptides toxic to African trypanosomes revealed that Sodalis is resistant to 7 of them. This finding opened the door for the utilization of Sodalis as a paratrangenesis agent for the delivery of antitrypanosomal compounds in vivo (Haines et al. 2003). A study demonstrated that Sodalis transformed with a gene for a trypanolytic nanobody can secrete this factor and that they are capable of invading tissues throughout the fly. The effective establishment of this infection was dependent on the prior treatment of the flies with the antibiotic streptozotocin to deplete native Sodalis numbers and reduce competition. These bacteria express the nanobody throughout the tissues of the fly; however, vertical transmission of these bacteria only occurred at a low level (De Vooght et al. 2014). Follow up work shows that the establishment of a stable infection and reliable vertical transmission with engineered bacteria is dependent upon the route of introduction. Flies given intrathoracic injections with recombinant Sodalis as adults were able to establish infections, but those infections are not vertically transmitted to offspring. However, the investigators found that injection of larval Glossina facilitated disseminated bacterial infection as well as vertical transmission to the resulting offspring (De Vooght et al. 2018).
Sterile Insect Technique (SIT) is another important strategy for population control of Glossina. An issue associated with SIT in Glossina is that radiation-sterilized males are still capable of functioning as vectors, which is an ethical issue associated with this approach. An alternative approach that minimizes this risk is the release of sterile males infected with engineered Sodalis to reduce or eliminate their vectorial capacity. Sterilization of paratransgenic males with gamma radiation revealed that while the treatment caused an initial decline in the number of recombinant Sodalis, the bacterial numbers recovered over time making this a potentially practical approach (Demirbas-Uzel et al. 2018).
3.5 Summary
The relationship between Glossina and Sodalis is very different from that of Glossina and Wigglesworthia. Sodalis appears to be in an evolutionary transition between a free-living organism and a symbiont. Its genomic reductions are not at the level of that observed in Wigglesworthia; however, Sodalis has accumulated a significant number of pseudogenes associated with functions required for a free-living existence. Analysis of retained features suggests the development of specializations for life within the host. These include alterations to its outer membrane proteins and secretory systems to reduce its pathogenicity and immunogenicity. Sodalis has also made nutritional adaptations that optimize it for survival in the fly including the use of N-acetyl-D-glucosamine as a carbon source and the retention of a transporter to scavenge Wigglesworthia-derived thiamine. Glossina does not require the presence of Sodalis for essential functions such as reproduction. However, selective elimination of the bacteria does have a negative impact on Glossina lifespan suggesting a beneficial aspect to the relationship. The understanding of the relationship between Sodalis and the different Trypanosoma types remains uncertain given the conflicting results from field and lab-based studies. The ability to culture and genetically manipulate Sodalis makes it an ideal agent for use in paratransgenesis studies. Research on this method of control is ongoing and could be a valuable tool for integration into SIT strategies or on its own to reduce the vectorial capacity of wild Glossina populations.
4 Wolbachia
4.1 Discovery
Wolbachia is a genus of obligate intracellular gram-negative bacteria belonging to the Order Rickettsiales and were first identified from Culex pipiens in 1924 (Hertig and Wolbach 1924). In 1936, this finding was confirmed and described in detail (Hertig 1936). Since these early discoveries, research has shown that Wolbachia is widespread in arthropods (reviewed in (Serbus et al. 2008)), and is probably the most prevalent endosymbiont found in insect germlines. It is found in every insect order (Harris and Braig 2003), and is estimated to infect >65% insect species (de Oliveira et al. 2015; Hilgenboecker et al. 2008).
4.2 Basic Biology and Features
The widespread presence of Wolbachia is thought to be due to its efficient transmission and its capacity to manipulate host reproduction to favor infected females. Wolbachia bacteria are indeed able to colonize female germline cells, through which they are transovarially transmitted to the progeny. Wolbachia interspecific horizontal transmission was initially described as a rare phenomenon (O’Neill et al. 1992; Rousset et al. 1992; Turelli et al. 2018; Werren et al. 1995), but studies are increasingly showing that, in a variety of insect species, Wolbachia genes have been horizontally transferred to host chromosomes (Aikawa et al. 2009; Dunning Hotopp et al. 2007; Fenn et al. 2006; Klasson et al. 2009; Kondo et al. 2002; Nikoh and Nakabachi 2009; Nikoh et al. 2008; Woolfit et al. 2009).
In different arthropod hosts, Wolbachia infection is responsible for several mechanisms that enhance female fertility. These reproductive alterations induce different host phenotypes such as cytoplasmic incompatibility, the feminization of genetically male offspring, male-killing of infected males, and parthenogenesis by infected females (Harris and Braig 2003; Stouthamer et al. 1999; Tram et al. 2003).
Cytoplasmic Incompatibility (CI), the most prevalent and most widely investigated phenomenon, was first observed in the 1970s (Yen and Barr 1973) and results in embryonic mortality in the progeny derived from matings between insects with different Wolbachia infection status (Bourtzis et al. 1998; Clark et al. 2003). Unidirectional CI occurs when an infected male mates with an uninfected female; whereas the reciprocal crossing is compatible. Bidirectional CI occurs in crossings between individuals infected with different Wolbachia strains (Werren 1997). Embryo lethality has been related to the modifications induced by Wolbachia to the paternal chromosomes during spermatogenesis in a way that mitotic synchrony is lost (O’Neill and Karr 1990; Tram and Sullivan 2002). The genetic basis of CI remained unknown for a long time, and only recently LePage and colleagues identified two genes in the eukaryotic association module of prophage WO (Bordenstein and Bordenstein 2016) from Wolbachia strain wMel that acts as CI factors (Beckmann et al. 2017; LePage et al. 2017). These studies revealed that the mitotic defect is due to a deubiquitinating enzyme encoded by one of the two genes (i.e. CidB/CifB) and neutralized by the CidA/CifA product.
In different species, Wolbachia has also been proposed to play a key role in sex determination by enhancing female germline development (Cordaux and Gilbert 2017; Kageyama et al. 2017; Kageyama and Traut 2004; Sugimoto et al. 2015). For example, in the parasitoid wasp Asobara tabida, Wolbachia is essential for oogenesis as its elimination induces apoptosis in the ovaries thus impeding egg maturation (Dedeine et al. 2001; Pannebakker et al. 2007). The role of Wolbachia in supporting female germline development in Drosophila melanogaster began to be clarified when females carrying mutant alleles of the master gene in female sex determination, sex-lethal, rescued their fertility when infected with Wolbachia (Starr and Cline 2002). Following these findings, more recent work showed that Wolbachia interacts with RNAs encoding proteins involved in the support of germline stem cell maintenance and oocyte polarization (Ote and Yamamoto 2020). These data further support the idea that this bacterium is a genetic manipulator of the infected arthropod hosts (Kozek and Rao 2007).
Wolbachia-mediated manipulations were shown in recent work to affect several other reproduction-related functions. For example, in Drosophila, Wolbachia affects gene transcription in larval testes (Zheng et al. 2011) as well as the expression of seminal fluid proteins (Yuan et al. 2015). Moreover, Wolbachia was shown to impact the expression of immunity genes in a parasitoid wasp (Kremer et al. 2012). The functions Wolbachia exerts in mosquitoes are achieved through manipulations of host microRNAs and the production of small RNAs regulating host gene expression (Hussain et al. 2011; Mayoral et al. 2014).
Recent studies expanded knowledge on other effects exerted by Wolbachia on the behavior of its hosts. These include influences on sleep, learning, memory, feeding, mating, locomotion, and aggression [for a review see (Bi and Wang 2019)]. For example, it has been suggested that Wolbachia is able to affect sleep in Drosophila by interactions with the juvenile hormone/sex-determination genes/dopamine pathway. In particular, this bacterium appears to contribute to increased sleep time in order to favor the conservation of resources and energies to support reproductive outputs, thus supporting both the host and its transmission and indicating that the coevolution between Wolbachia and its hosts is even more multifaced than so far discovered (Bi and Wang 2019).
4.3 Localization in Insect Tissues
Wolbachia primarily resides in the germline tissues of both male and female insects (Dobson et al. 1999); in males, this bacterium is found in the spermatocytes but there is no transmission through the sperm (Bressac and Rousset 1993; Clark et al. 2002; Ijichi et al. 2002). In addition, since its early detection, this bacterium was described to be present also in somatic tissues [(Dobson et al. 1999; Hertig and Wolbach 1924) also see (Pietri et al. 2016) for a review].
In the germline tissue, some Wolbachia strains localize at the posterior end of mature oocytes (Ote and Yamamoto 2020) thanks to the presence of RNAs and proteins transported from the nurse cells along microtubules to form a pole plasm (Serbus and Sullivan 2007). Pole cells, together with somatic gonadal cells, form the embryonic gonads, becoming primordial germ cells. During the development of the female pupa, the primordial germ cells initiate their divisions resulting in the germ cell lineage in the ovaries of adult females. Wolbachia is found in the germline stem cells, which are localized at the anterior end of each ovariole within the ovaries (Ote et al. 2016; Serbus et al. 2008). In particular, Wolbachia tends to be located close to processing bodies in the cytoplasm of these cells that supply maternal factors to the oocyte (Ferree et al. 2005; Franks and Lykke-Andersen 2008; Ote et al. 2016; Serbus et al. 2011), resulting in an association that takes place from oogenesis to embryogenesis. In particular, Wolbachia produces a protein able to interact with RNAs encoding proteins that are involved in the support of germline stem cells and oocyte polarization (Ote and Yamamoto 2020).
As far as it concerns Wolbachia distribution in somatic tissues, this bacterium is known to be particularly abundant in the nervous system in Drosophila and several other insect species (Albertson et al. 2013; Casper-Lindley et al. 2011; Dobson et al. 1999; Mitsuhashi et al. 2002; Moreira et al. 2009; Osborne et al. 2009; Strunov and Kiseleva 2016), as well as in the fat body, gut, salivary glands, hemocytes, and Malphighian tubules, where it has been suggested to play roles related to the host immunity and metabolic regulation (see (Pietri et al. 2016) for a review). The age of the host also impacts Wolbachia infection levels and tropism, further suggesting that the physiological relationships between Wolbachia and its hosts are extremely complex and require a case-to-case analysis (Binnington and Hoffmann 1989; Bressac and Rousset 1993; Min and Benzer 1997).
4.4 Wolbachia in Glossina spp.
Early hybridization experiments conducted between different species belonging to the Glossina genus suggested the presence of incompatibilities resulting in females with reduced fecundity and sterile males. Both bidirectional and unidirectional incompatibility events were reported ((Curtis 1972; Rawlings 1985; Vanderplank 1948) see also (Gooding 1985, 1987, 1989, 1990) for a review). These data, together with light and electron microscopy studies showing gram-negative rods in tsetse ovaries and in the periphery of the yolk of early embryos (Huebner and Davey 1974; Pell and Southern 1975; Pinnock and Hess 1974) and 16s rRNA sequence analyses (Beard et al. 1993), prompted researchers to further investigate the identity of these bacteria. The exploration of 16S rRNA phylogenetic relationships and tissue distribution in tsetse resulted in the identification of Wolbachia in laboratory strains of Glossina species in 1993 (O’Neill et al. 1993). Initially, Wolbachia was detected in G. m. morsitans and G. m. centralis, while it was found to be absent in G. p. palpalis and G. p. gambiensis, supporting some of the reported events of reproductive incompatibilities in the genus (O’Neill et al. 1993).
4.5 Tissue Localization in Tsetse Tissues
The tissue tropism of Wolbachia to the ovarian tissues suggested that transovarial transmission is the primary mode of transmission for this bacterium (O’Neill et al. 1993), which supports previous studies (Pell and Southern 1975). Subsequent work on G. m. morsitans laboratory flies confirmed Wolbachia’s absence in the milk gland secretions, as well as any other somatic tissues surrounding the uterus, further supporting its transovarial transmission (Balmand et al., 2013). Wolbachia was not detected extracellularly, and it was shown to infect only the trophocytes and the oocytes in the ovaries, as well as embryos and larvae (Cheng et al. 2000; Balmand et al. 2013).
However, while in certain tsetse species Wolbachia appeared to be restricted to the reproductive tissues, in G. austeni, this bacterium was detected also in somatic tissues, in particular the head, salivary glands, milk glands, and fat body (Cheng et al. 2000). These findings stimulate novel studies aimed at understanding whether these differences are due to features of different Wolbachia strains infecting the different tsetse species or to insect-specific factors, such as immunity-regulatory mechanisms, controlling the infections. The application of a particularly effective hybridization approach (i.e. high-end Stellaris® RNA-FISH) surprisingly revealed that Wolbachia is also present in the lumen and secretory cells of the milk glands in G. m. morsitans (Schneider et al. 2018). This result opens up new pathways of investigation of a potentially yet undiscovered vertical transmission mechanism for this bacterium.
4.6 Wolbachia Role in Tsetse Physiology
Earlier studies described the existence of bidirectional CI between certain G. morsitans subspecies and between G. palpalis subspecies, while unidirectional CI has been described only in crossings of certain G. morsitans subspecies (Cheng et al. 2000).
The first study showing the functional role of Wolbachia in tsetse reared in the laboratory was performed by Alam and colleagues, who showed that the infection of this bacterium is able to support the expression of cytoplasmic incompatibility (Alam et al. 2011). The females expressing CI displayed loss of fecundity due to early embryogenic failure. Currently, there is no way to selectively cure Wolbachia infection through antibiotic administration. Aposymbiotic flies lack all endosymbionts, including obligate bacteria responsible for obligate nutritional dialogues, in the absence of which tsetse flies are sterile. Alam and coauthors set up a method to maintain Wolbachia-cured G. m. morsitans (symbiont-free, GmmApo) fertile through dietary provisioning of blood meals supplemented with tetracycline and yeast extract, thus rescuing tsetse fecundity, which is tightly dependent on Wigglesworthia. Moreover, cytoplasmic incompatibility in tsetse is particularly strong, differently from many other species that are characterized by incomplete CI (Sinkins and Gould 2006).
Whether Wolbachia plays a role in trypanosome infection is still under debate. Alam and colleagues reported the presence of a negative association between Wolbachia and trypanosome infections in G. f. fuscipes, suggesting that this bacterium could prevent trypanosome infections (Alam et al. 2012). However, the tripartite association between tsetse, trypanosomes, and Wolbachia remains unclear. In G. p. palpalis, Wolbachia infection appeared to have no impact on the establishment of trypanosomes (Kante et al. 2018). This is similar to what was found in G. tachinoides and G. m. submorsitans (Kame-Ngasse et al. 2018). To shed light on this relevant biological aspect, more extensive data on trypanosome and Wolbachia infections, as well as the other symbionts, in different tsetse species and populations are required. Moreover, the potential relationships between specific Wolbachia haplotypes and trypanosome infections will be essential to determine the presence and features of this dialogue.
4.7 Wolbachia Distribution in Tsetse Strains and Populations
Since its discovery, a number of studies have focused on expanding the analysis of the distribution of Wolbachia in tsetse species, in both laboratory strains and wild samples. The first extensive work aimed at understanding the presence and infection rates of Wolbachia in tsetse was performed by Cheng and colleagues, who analyzed the status of Wolbachia infections in laboratory colonies of G. brevipalpis and G. longipinnis (Fusca group), G. fuscipes, G. tachinoides, G. p. palpalis, and G. p. gambiensis (Palpalis group), as well as G. m. morsitans, G. m. centralis, G. swynnertoni, and G. pallidipes (Morsitans group) (Cheng et al. 2000). All individuals analyzed from the Morsitans and Fusca groups were positive for Wolbachia infection, while none of the flies belonging to the Palpalis group harbored Wolbachia, mirroring earlier findings (O’Neill et al. 1993). A 100% prevalence in laboratory strains was confirmed by a more recent study for G. m. morsitans and G. m. centralis. Similarly, the absence of Wolbachia was confirmed in laboratory strains of G. f. fuscipes and G. tachinoides, whereas in G. pallidipes, different colonies displayed different prevalences of Wolbachia ranging from the absence (Seibersdorf lab-colony) to low prevalence (KARI-TRC lab-colony, 3%) (Doudoumis et al. 2012). Also in this study, flies from G. p. palpalis and G. p. gambiensis laboratory colonies showed the absence of Wolbachia infection. Differences were reported for G. brevipalpis, where prevalence was not complete (41.2%) (Doudoumis et al. 2012).
Significant differences in Wolbachia infection frequencies are found between field populations and laboratory strains. In addition, differences are observed within wild populations, which may be dependent on ecological conditions (Mouton et al. 2007; Yun et al. 2011). For example, in the case of G. m. morsitans, the Wolbachia presence in wild populations ranges from 9.5 to 100% (Doudoumis et al. 2012). In G. brevipalpis sampled in South Africa Wolbachia appears absent, while, in Kenyan flies, infection levels are reported to be about 30% (Cheng et al. 2000). In G. austeni, infection rates ranged from 48 to 98%, in Kenya and South Africa, respectively (Cheng et al. 2000). More recent work also detected variations in Wolbachia prevalence in G. austeni samples in these two countries, but with the Kenyan population showing higher infection levels (Wamwiri et al. 2013). Low Wolbachia prevalence was detected in populations of G. pallidipes (below 8.5%) and in G. gambiensis (below 8.3%), and the absence of Wolbachia infection in G. p. palpalis and G. f. fuscipes populations was confirmed (Doudoumis et al. 2012). Alam and colleagues conversely showed the presence of Wolbachia infections in G. f. fuscipes from Uganda, although at low density, which may have influenced its detection in previous studies (Alam et al. 2012). A similar result was obtained by Schneider and colleagues, who used sensitive PCR-based methods that allowed the identification of Wolbachia in G. f. fuscipes (Schneider et al. 2013, 2018). Recent studies identified Wolbachia also in G. p. palpalis (Kante et al. 2018), as well as in G. tachinoides (68.1% prevalence) and in G. m. submorsitans (58.5%) (Kame-Ngasse et al. 2018), further supporting the idea that the choices of molecular markers and detection methods play a key role in the sensitivity of determining the infection rates of Wolbachia. Indeed, when present at low titers, Wolbachia infections can be detected only through the integration of different tools, such as high-sensitivity blot-PCR combined with hybridizations (Schneider et al. 2018).
Finally, in G. f. quanzensis (Palpalis group) from Congo, the analysis of midguts revealed that 85% of the analyzed samples were infected by Wolbachia, with infection rates varying according to sampling sites (Simo et al. 2019). Moreover, a low number of midguts were naturally coinfected by both Wolbachia and Sodalis, opening new questions on the potential interactions between these two tsetse symbionts.
4.8 Wolbachia Integrations in Tsetse Genomes
The genome of G. m. morsitans contains large segments of Wolbachia that were integrated via horizontal gene transfer (HGT) events. These integrated fragments contain a high degree of nucleotide polymorphisms, as well as insertions and deletions (Brelsfoard et al. 2014).
Subsequent comparative analysis of the genomes of six Glossina species, namely G. morsitans morsitans, G. pallidipes, G. austeni (Morsitans group), G. palpalis and G. fuscipes (Palpalis group), and G. brevipalpis (Fusca group), showed that all contain sequences with homology to Wolbachia, although the features of these integrations are different. Indeed, in G. pallidipes, G. fuscipes, G. palpalis, and G. brevipalpis, the homologous sequences consisted in short fragments and they were initially thought to be artifacts as PCR assays with Wolbachia-specific primers on these strains as well as on wild populations of these species resulted in negative results (Doudoumis et al. 2013). However, recent studies suggest that natural G. p. palpalis populations from Cameroon do carry Wolbachia symbionts (Kante et al. 2018).
However, G. austeni contains more extensive Wolbachia-derived chromosomal integrations. Both chromosomal and cytoplasmic Wolbachia sequences found in G. austeni were mapped against the reference Wolbachia genomes (i.e. wMel and wGmm), as well as the A and B chromosomal insertions found in G. m. morsitans (Attardo et al. 2019). The Wolbachia insertions in G. austeni range in size from 500 to 95,673 bps and display high-sequence homology not only to wMel and wGmm, but also to G. m. morsitans A and B insertions. The higher homology (98%) with A and B insertions from G. m. morsitans relative to cytoplasmic Wolbachia sequences suggests they could be derived from an event in a common ancestor, although the absence of comparable insertions in G. pallidipes (a closer relative to G. m. morsitans) requires further investigations on wild samples from Glossina species/subspecies to clarify the true origin of these events.
Whether these Wolbachia chromosomal integrations have functional roles in Glossina biology is still elusive. Indeed, gene expression analyses of Wolbachia insertions in G. morsitans found very limited evidence of expression (Brelsfoard et al. 2014), suggesting they may be accidental transfer events associated with the long-term symbiosis between Wolbachia and these tsetse species. However, since Wolbachia integrations in tsetse chromosomes include genes encoding proteins carrying ankyrin repeat domains, thought to be directly related to Wolbachia-host interactions, further research is required to determine their potential involvement in CI (Duron et al. 2007; Iturbe-Ormaetxe et al. 2005; Tram and Sullivan 2002).
4.9 Wolbachia Diversity
Wolbachia displays a high level of diversity in arthropods and nematodes and it currently comprises 17 phylogenetic clades (or supergroups), named from A to Q (Baldo et al. 2006; Bordenstein and Rosengaus 2005; Bordenstein et al. 2009; Casiraghi et al. 2005; Glowska et al. 2015; Gorham et al. 2003; Lo et al. 2002; Paraskevopoulos et al. 2006; Ros et al. 2009; Rowley et al. 2004). Each supergroup collects strains, most frequently named after their host species (e.g. wPip in Culex pipiens, wGff in G. f. fuscipes). The most common strategy adopted for strain genotyping relies on multilocus sequence typing (MLST), which includes the sequences of the five conserved genes fbpA, coxA, ftsZ, gatB, coxA, and hcpA and the amino acid sequences of the four hypervariable regions of the WSP protein (Baldo et al. 2006). The diversity displayed in this genus is visible at different levels, including the presence of variants within the same individual host, variation among Wolbachia sequences sampled from different individuals belonging to the same species, as well as the molecular changes occurring in the same Wolbachia infection in the case of transfer to different host species (i.e. in the case of horizontal gene transfer) (Hoffmann et al. 2015). In the case of Glossina, Wolbachia identified in tsetse species has been regarded to belong to the supergroup A, as determined based on the Wolbachia surface protein (wsp) gene (Cheng et al. 2000; Zhou et al. 1998). A recent study investigated Wolbachia genetic variability in G. f. fuscipes from Uganda (Symula et al. 2013). Two Wolbachia lineages were identified, suggesting the presence of superinfection in this species, and a high diversity within and between individuals. These data suggest that different Wolbachia strains infected this tsetse species multiple times independently.
4.10 Wolbachia as a Tool for Vector Population Control
Wolbachia has been proposed as a tool for controlling insect disease vectors and agricultural pests through multiple approaches, which are not mutually exclusive. These methods include the release of (1) Wolbachia-infected males that are incompatible with females by exploiting cytoplasmic incompatibility phenotypes (O’Connor et al. 2012), (2) Wolbachia strains able to induce deleterious fitness effects, in particular under seasonally variable environments (Rasić et al. 2014), and (3) Wolbachia strains interfering with pathogen transmission and thus decreasing the ability of vectors to transmit diseases (Kambris et al. 2009; Moreira et al. 2009; Teixeira et al. 2008; Walker et al. 2011). The capacity of rapidly invading insect populations that drive maternally inherited elements into wild insects has made Wolbachia a promising tool for inducing genetic manipulations of detrimental species (Beard et al. 1993; Sinkins et al. 1997). The data obtained by Alam and colleagues about CI in tsetse were incorporated into a mathematical model and suggest that Wolbachia has the potential for use as a gene-drive mechanism. This could be used to introduce desirable phenotypes, such as resistance to trypanosome infection, into wild Glossina populations (Alam et al. 2011).
The exploitation of Wolbachia-induced CI was proposed for use in the reduction of population sizes of several insect disease vectors and agricultural pests (Apostolaki et al. 2011; Bourtzis 2008; Stouthamer et al. 1999; Xi et al. 2005; Zabalou et al. 2009). Moreover, Wolbachia is able to protect their hosts against viral pathogens (Cook and McGraw 2010). The initial characterization of this phenomenon was performed in Drosophila (Hedges et al. 2008; Osborne et al. 2009; Teixeira et al. 2008) and mosquitoes (Bian et al. 2010; Glaser and Meola 2010; Moreira et al. 2009). More recent work demonstrates that wAlbB infection in the C6/36 Ae. albopictus cell line resulted in reduced titers of several Flaviviruses and Alphaviruses, suggesting a role of Wolbachia in reducing transmission of pathogenic RNA viruses (Ekwudu et al. 2020). Moreover, a triple-infected Ae. albopictus line carrying, in addition to the two natural symbiotic strains wAlbA and wAlbB, the wAu from D. simulans, showed complete resistance to Zika and dengue infections, with moderate fitness costs (Mancini et al. 2020). Pathogen-blocking function has been associated with the upregulation of antimicrobial peptides (e.g. DEFC, defensin c) (Caragata et al. 2019; Pan et al. 2018; Pan et al. 2012).
In the case of tsetse species, as mentioned above, contrasting results have been obtained. In wild populations of G. f. fuscipes, the presence of Wolbachia has been suggested to be able to prevent trypanosome infection (Alam et al. 2012), while in G. p. palpalis, G. tachinoides, and G. m. submorsitans, Wolbachia seems not to impact establishment of trypanosome infection (Kame-Ngasse et al. 2018; Kante et al. 2018). Further studies are thus necessary to understand whether Wolbachia may be used in Incompatible Insect Technique (IIT)-based approaches for population control. This strategy is based on the release of Wolbachia-infected males that both induce CI when mated with Wolbachia-free wild females and refractoriness to trypanosome infection and transmission. This concept is similar to strategies recently designed for mosquitoes (Bourtzis et al. 2016; Zhang et al. 2015a, b, 2016).
4.11 Summary
The Wolbachia symbiont is present predominantly in tsetse gonadal tissues, and it is transovarially transmitted from females to their progeny. An increasing number of studies now show that this bacterium can colonize also somatic tissues, with still unclarified functions. Horizontal transfer events have been detected in G. m. morsitans and G. austeni, but whether these integrated sequences play functional roles in tsetse biology is still unclear. Wolbachia induces strong cytoplasmic incompatibility in tsetse, supporting the idea that it may be exploited as a tool to control tsetse populations in the field.
5 Spiroplasma
The innovation of high-throughput 16S ribosomal RNA sequencing has allowed for deeper exploration of the microbiome of Glossina species. These studies have identified many other bacterial species in Glossina from laboratory colonies and from the field. From these analyses, bacteria from the Genus Spiroplasma were found in field-caught and lab colonies of Glossina f. fuscipes and G. tachinoides. The recent analysis of the genomes of six Glossina species also revealed evidence for a significant relationship between Spiroplasma and G. f. fuscipes. Analysis of the genomic scaffolds from this species revealed the presence of Spiroplasma genomic sequences. None of these sequences appear to be the result of a genomic integration and are likely derived from free-living Spiroplasma present in the flies used for sequencing (Attardo et al. 2019). Developmental stage and tissue specificity analyses revealed the Spiroplasma infections to be highest in the larval gut and male reproductive tissues. In addition, the analysis of flies from a collapsing colony of G. f. fuscipes showed that levels of Spiroplasma were higher in surviving flies versus recently deceased flies (Doudoumis et al. 2017). Another study found that Trypanosoma infection in field-collected G. f. fuscipes is negatively correlated with Spiroplasma coinfection. This finding was also demonstrated in the laboratory by comparing experimental infection rates between Spiroplasma infected and uninfected G. f. fuscipes (Schneider et al. 2019).
Spiroplasma species are found to be living in many insect and invertebrate species with relationships ranging from pathological to beneficial. Pathogenic Spiroplasma infections leading to death have been observed in aquatic invertebrates, such as shrimps and crabs (Nunan et al. 2005; Wang et al. 2004). In some Drosophila melanogaster, infection with Spiroplasma results in reproductive manipulation in the form of a male-killing phenotype (Montenegro et al. 2005; Paredes et al. 2015). However, Spiroplasma infections in Drosophila can also protect against infection by the parasitic nematode Howardula aoronymphium. The nematode infection results in loss of fecundity in female Drosophila. However, the coinfection of females with Spiroplasma results in the restoration of fecundity and inhibition of nematode development (Haselkorn et al. 2013; Jaenike et al. 2010). The early observations of the relationship between Spiroplasma and Glossina suggest that this relationship is beneficial in terms of its ability to extend lifespan and reduce the vectorial capacity for Trypanosoma. However, relative to the other Glossina symbionts, little is known about the details of the interactions between these two species and more work will be required to develop a more comprehensive understanding of this relationship.
6 Conclusions
The role bacteria play in the biology of tsetse flies spans the gamut, ranging from parasitic to obligate mutualists. This system is also unique in that the microbiome of species within Glossina is limited relative to other insects. This is primarily due to their restricted diet and the protected nature of intrauterine larval development. These limitations simplify the system and provide opportunities to study microbe-host interactions in a way that is impossible in insects with more diverse microbiomes. The relationship between Wigglesworthia and Glossina is ancient and provides a clear example of obligate symbiosis. It also demonstrates the evolutionary mechanisms and biology behind how an organism can evolve to survive and thrive on a nutrient-rich yet limited diet. The commensal yet nonessential relationship between Sodalis/Glossina provides a snapshot of two organisms in the early or intermediate stages of a symbiotic relationship and can provide insights as to the adaptations made by both parties to establish a harmonious relationship. In addition, the ability to culture and manipulate Sodalis provides opportunities to take advantage of this relationship for purposes of controlling trypanosome transmission by Glossina. The nature of the relationship between Glossina and Wolbachia is more complex and shows aspects of parasitism and manipulation by Wolbachia similar to that observed in other insects. However, a deeper understanding of the protective nature of Wolbachia infections against viral infections in other insects and the inherent reproductive manipulations driving the spread of these infections may provide opportunities to utilize this relationship to reduce Glossina vectorial capacity. The implementation of new technologies such as high-throughput metagenomics, advanced microbial imaging, and novel in vitro culture techniques has opened opportunities to investigate these well-studied relationships in more depth. They also provide the ability to identify previously undescribed microbial interactions that were overshadowed by the dominant microbial fauna. Analysis of the diversity of these lesser-explored interactions could provide valuable insights into the ecology, population dynamics, and biology of this unique system.
References
Aikawa T, Anbutsu H, Nikoh N, Kikuchi T, Shibata F, Fukatsu T (2009) Longicorn beetle that vectors pinewood nematode carries many Wolbachia genes on an autosome. Proc Biol Sci 276:3791–3798. https://doi.org/10.1098/rspb.2009.1022
Akman L, Rio RV, Beard CB, Aksoy S (2001) Genome size determination and coding capacity of Sodalis glossinidius, an enteric symbiont of tsetse flies, as revealed by hybridization to Escherichia coli gene arrays. J Bacteriol 183:4517–4525. https://doi.org/10.1128/JB.183.15.4517-4525.2001
Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M, Aksoy S (2002) Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat Genet 32:402–407. https://doi.org/10.1038/ng986
Aksoy S (1995) Wigglesworthia gen. nov. and Wigglesworthia glossinidia sp. nov., taxa consisting of the mycetocyte-associated, primary endosymbionts of tsetse flies. Int J Syst Bacteriol 45:848–851. https://doi.org/10.1099/00207713-45-4-848
Aksoy S, Pourhosseini AA, Chow A (1995) Mycetome endosymbionts of tsetse flies constitute a distinct lineage related to Enterobacteriaceae. Insect Mol Biol 4:15–22
Aksoy S, Chen X, Hypsa V (1997) Phylogeny and potential transmission routes of midgut-associated endosymbionts of tsetse (Diptera: Glossinidae). Insect Mol Biol 6:183–190
Aksoy E et al (2014) Analysis of multiple tsetse fly populations in Uganda reveals limited diversity and species-specific gut microbiota. Appl Environ Microbiol 80:4301–4312. https://doi.org/10.1128/AEM.00079-14
Alam U et al (2011) Wolbachia symbiont infections induce strong cytoplasmic incompatibility in the tsetse fly Glossina morsitans. PLoS Pathog 7:e1002415. https://doi.org/10.1371/journal.ppat.1002415
Alam U et al (2012) Implications of microfauna-host interactions for trypanosome transmission dynamics in Glossina fuscipes fuscipes in Uganda. Appl Environ Microbiol 78:4627–4637. https://doi.org/10.1128/AEM.00806-12
Albertson R, Tan V, Leads RR, Reyes M, Sullivan W, Casper-Lindley C (2013) Mapping Wolbachia distributions in the adult Drosophila brain. Cell Microbiol 15:1527–1544. https://doi.org/10.1111/cmi.12136
Anselme C, Vallier A, Balmand S, Fauvarque MO, Heddi A (2006) Host PGRP gene expression and bacterial release in endosymbiosis of the weevil Sitophilus zeamais. Appl Environ Microbiol 72:6766–6772. https://doi.org/10.1128/AEM.00942-06
Apostolaki A, Livadaras I, Saridaki A, Chrysargyris A, Savakis C, Bourtzis K (2011) Transinfection of the olive fruit fly Bactrocera oleae with Wolbachia: towards a symbiont-based population control strategy. J Appl Entomol 135:546–553. https://doi.org/10.1111/j.1439-0418.2011.01614.x
Attardo GM, Lohs C, Heddi A, Alam UH, Yildirim S, Aksoy S (2008) Analysis of milk gland structure and function in Glossina morsitans: milk protein production, symbiont populations and fecundity. J Insect Physiol 54:1236–1242. https://doi.org/10.1016/j.jinsphys.2008.06.008
Attardo GM et al (2019) Comparative genomic analysis of six Glossina genomes, vectors of African trypanosomes. Genome Biol 20:187. https://doi.org/10.1186/s13059-019-1768-2
Baldo L et al (2006) Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol 72:7098–7110. https://doi.org/10.1128/AEM.00731-06
Balmand S, Lohs C, Aksoy S, Heddi A (2013) Tissue distribution and transmission routes for the tsetse fly endosymbionts. J Invertebr Pathol 112. Suppl:S116–S122. https://doi.org/10.1016/j.jip.2012.04.002
Beard CB, O’Neill SL, Tesh RB, Richards FF, Aksoy S (1993) Modification of arthropod vector competence via symbiotic bacteria. Parasitol Today 9:179–183. https://doi.org/10.1016/0169-4758(93)90142-3
Beard CB, Dotson EM, Pennington PM, Eichler S, Cordon-Rosales C, Durvasula RV (2001) Bacterial symbiosis and paratransgenic control of vector-borne Chagas disease. Int J Parasitol 31:621–627. https://doi.org/10.1016/s0020-7519(01)00165-5
Beard CB, Cordon-Rosales C, Durvasula RV (2002) Bacterial symbionts of the triatominae and their potential use in control of Chagas disease transmission. Annu Rev Entomol 47:123–141. https://doi.org/10.1146/annurev.ento.47.091201.145144
Beckmann JF, Ronau JA, Hochstrasser M (2017) A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat Microbiol 2:17007. https://doi.org/10.1038/nmicrobiol.2017.7
Belda E, Moya A, Bentley S, Silva FJ (2010) Mobile genetic element proliferation and gene inactivation impact over the genome structure and metabolic capabilities of Sodalis glossinidius, the secondary endosymbiont of tsetse flies. BMC Genomics 11:449. https://doi.org/10.1186/1471-2164-11-449
Bennett GM, Moran NA (2013) Small, smaller, smallest: the origins and evolution of ancient dual symbioses in a Phloem-feeding insect. Genome Biol Evol 5:1675–1688. https://doi.org/10.1093/gbe/evt118
Benoit JB et al (2017) Symbiont-induced odorant binding proteins mediate insect host hematopoiesis. Elife 6. https://doi.org/10.7554/eLife.19535
Bi J, Wang YF (2019) The effect of the endosymbiont Wolbachia on the behavior of insect hosts. Insect Sci. https://doi.org/10.1111/1744-7917.12731
Bian G, Xu Y, Lu P, Xie Y, Xi Z (2010) The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog 6:e1000833. https://doi.org/10.1371/journal.ppat.1000833
Bing X et al (2017) Unravelling the relationship between the tsetse fly and its obligate symbiont Wigglesworthia: transcriptomic and metabolomic landscapes reveal highly integrated physiological networks. Proc Biol Sci 284. https://doi.org/10.1098/rspb.2017.0360
Binnington K, Hoffmann A (1989) Wolbachia-like organisms and cytoplasmic incompatibility in Drosophila simulans. J Invertebr Pathol 54:344–352
Bordenstein SR, Bordenstein SR (2016) Eukaryotic association module in phage WO genomes from Wolbachia. Nat Commun 7:13155. https://doi.org/10.1038/ncomms13155
Bordenstein S, Rosengaus RB (2005) Discovery of a novel Wolbachia super group in Isoptera. Curr Microbiol 51:393–398. https://doi.org/10.1007/s00284-005-0084-0
Bordenstein SR et al (2009) Parasitism and mutualism in Wolbachia: what the phylogenomic trees can and cannot say. Mol Biol Evol 26:231–241. https://doi.org/10.1093/molbev/msn243
Bourtzis K (2008) Wolbachia-based technologies for insect pest population control. In: Aksoy S (ed) Transgenesis and the management of vector-borne disease. Springer, New York, NY, pp 104–113. https://doi.org/10.1007/978-0-387-78225-6_9
Bourtzis K, Dobson SL, Braig HR, O’Neill SL (1998) Rescuing Wolbachia have been overlooked. Nature 391:852–853. https://doi.org/10.1038/36017
Bourtzis K, Lees RS, Hendrichs J, Vreysen MJ (2016) More than one rabbit out of the hat: radiation, transgenic and symbiont-based approaches for sustainable management of mosquito and tsetse fly populations. Acta Trop 157:115–130. https://doi.org/10.1016/j.actatropica.2016.01.009
Brelsfoard C et al (2014) Wolbachia symbiont genome sequence and extensive chromosomal insertions present in the host Glossina morsitans morsitans genome. PLoS Negl Trop Dis 8:e2728. https://doi.org/10.1371/journal.pntd.0002728
Bressac C, Rousset F (1993) The reproductive incompatibility system in Drosophila simulans: DAPI-staining analysis of the Wolbachia symbionts in sperm cysts. J Invertebr Pathol 61:226–230. https://doi.org/10.1006/jipa.1993.1044
Buchner P (1965) Endosymbiosis of animals with plant microorganisms. Interscience Publishers, New York
Bulgheresi S, Schabussova I, Chen T, Mullin NP, Maizels RM, Ott JA (2006) A new C-type lectin similar to the human immunoreceptor DC-SIGN mediates symbiont acquisition by a marine nematode. Appl Environ Microbiol 72:2950–2956. https://doi.org/10.1128/AEM.72.4.2950-2956.2006
Bursell E (1960) Free amino-acids of the tsetse fly (Glossina). Nature 187:778–778
Bursell E (1963) Aspects of the metabolism of amino acids in the tsetse fly, Glossina (Diptera). J Insect Physiol 9:439–452
Bursell E (1966) Aspects of flight metabolism of tsetse flies (Glossina). Comp Biochem Physiol 19:809–818
Buxton PA (1955) London school of hygiene and tropical medicine memoir 10: the natural history of tsetse flies. H. K. Lewis, London
Caragata EP, Rocha MN, Pereira TN, Mansur SB, Dutra HLC, Moreira LA (2019) Pathogen blocking in Wolbachia-infected Aedes aegypti is not affected by Zika and dengue virus co-infection. PLoS Negl Trop Dis 13:e0007443. https://doi.org/10.1371/journal.pntd.0007443
Casiraghi M et al (2005) Phylogeny of Wolbachia pipientis based on gltA, groEL and ftsZ gene sequences: clustering of arthropod and nematode symbionts in the F supergroup, and evidence for further diversity in the Wolbachia tree. Microbiology 151:4015–4022. https://doi.org/10.1099/mic.0.28313-0
Casper-Lindley C, Kimura S, Saxton DS, Essaw Y, Simpson I, Tan V, Sullivan W (2011) Rapid fluorescence-based screening for Wolbachia endosymbionts in Drosophila germ line and somatic tissues. Appl Environ Microbiol 77:4788–4794. https://doi.org/10.1128/AEM.00215-11
Chang KP, Musgrave AJ (1973) Morphology, histochemistry, and ultrastructure of mycetome and its rickettsial symbiotes in Cimex lectularius L. Can J Microbiol 19:1075–1081. https://doi.org/10.1139/m73-171
Channumsin M, Ciosi M, Masiga D, Turner CMR, Mable BK (2018) Sodalis glossinidius presence in wild tsetse is only associated with presence of trypanosomes in complex interactions with other tsetse-specific factors. BMC Microbiol 18:163. https://doi.org/10.1186/s12866-018-1285-6
Chaston J, Goodrich-Blair H (2010) Common trends in mutualism revealed by model associations between invertebrates and bacteria. FEMS Microbiol Rev 34:41–58. https://doi.org/10.1111/j.1574-6976.2009.00193.x
Chen X, Li S, Aksoy S (1999) Concordant evolution of a symbiont with its host insect species: molecular phylogeny of genus Glossina and its bacteriome-associated endosymbiont, Wigglesworthia glossinidia. J Mol Evol 48:49–58. https://doi.org/10.1007/pl00006444
Cheng Q, Ruel TD, Zhou W, Moloo SK, Majiwa P, O’Neill SL, Aksoy S (2000) Tissue distribution and prevalence of Wolbachia infections in tsetse flies, Glossina spp. Med Vet Entomol 14:44–50. https://doi.org/10.1046/j.1365-2915.2000.00202.x
Clark ME, Veneti Z, Bourtzis K, Karr TL (2002) The distribution and proliferation of the intracellular bacteria Wolbachia during spermatogenesis in Drosophila. Mech Dev 111:3–15. https://doi.org/10.1016/s0925-4773(01)00594-9
Clark ME, Veneti Z, Bourtzis K, Karr TL (2003) Wolbachia distribution and cytoplasmic incompatibility during sperm development: the cyst as the basic cellular unit of CI expression. Mech Dev 120:185–198. https://doi.org/10.1016/s0925-4773(02)00424-0
Cook PE, McGraw EA (2010) Wolbachia pipientis: an expanding bag of tricks to explore for disease control. Trends Parasitol 26:373–375. https://doi.org/10.1016/j.pt.2010.05.006
Cordaux R, Gilbert C (2017) Evolutionary significance of Wolbachia-to-animal horizontal gene transfer: female sex determination and the f element in the isopod Armadillidium vulgare. Genes 8. https://doi.org/10.3390/genes8070186
Coutinho-Abreu IV, Zhu KY, Ramalho-Ortigao M (2010) Transgenesis and paratransgenesis to control insect-borne diseases: current status and future challenges. Parasitol Int 59:1–8. https://doi.org/10.1016/j.parint.2009.10.002
Curtis CF (1972) Sterility from crosses between sub-species of the tsetse fly Glossina morsitans. Acta Trop 29:250–268
Dale C, Maudlin I (1999) Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int J Syst Bacteriol 49(Pt 1):267–275. https://doi.org/10.1099/00207713-49-1-267
Dale C, Welburn SC (2001) The endosymbionts of tsetse flies: manipulating host-parasite interactions. Int J Parasitol 31:628–631. https://doi.org/10.1016/s0020-7519(01)00151-5
Dale C, Young SA, Haydon DT, Welburn SC (2001) The insect endosymbiont Sodalis glossinidius utilizes a type III secretion system for cell invasion. Proc Natl Acad Sci U S A 98:1883–1888. https://doi.org/10.1073/pnas.021450998
Dale C, Jones T, Pontes M (2005) Degenerative evolution and functional diversification of type-III secretion systems in the insect endosymbiont Sodalis glossinidius. Mol Biol Evol 22:758–766. https://doi.org/10.1093/molbev/msi061
Darby AC, Lagnel J, Matthew CZ, Bourtzis K, Maudlin I, Welburn SC (2005) Extrachromosomal DNA of the symbiont Sodalis glossinidius. J Bacteriol 187:5003–5007. https://doi.org/10.1128/JB.187.14.5003-5007.2005
Dawadi B, Wang X, Xiao R, Muhammad A, Hou Y, Shi Z (2018) PGRP-LB homolog acts as a negative modulator of immunity in maintaining the gut-microbe symbiosis of red palm weevil, Rhynchophorus ferrugineus Olivier. Dev Comp Immunol 86:65–77. https://doi.org/10.1016/j.dci.2018.04.021
de Oliveira CD, Gonçalves DS, Baton LA, Shimabukuro PHF, Carvalho FD, Moreira LA (2015) Broader prevalence of Wolbachia in insects including potential human disease vectors. Bull Entomol Res 105:305–315. https://doi.org/10.1017/S0007485315000085
De Vooght L, Caljon G, De Ridder K, Van Den Abbeele J (2014) Delivery of a functional anti-trypanosome Nanobody in different tsetse fly tissues via a bacterial symbiont, Sodalis glossinidius. Microb Cell Fact 13:156. https://doi.org/10.1186/s12934-014-0156-6
De Vooght L, Caljon G, Van Hees J, Van Den Abbeele J (2015) Paternal transmission of a secondary symbiont during mating in the viviparous tsetse Fly. Mol Biol Evol 32:1977–1980. https://doi.org/10.1093/molbev/msv077
De Vooght L, Van Keer S, Van Den Abbeele J (2018) Towards improving tsetse fly paratransgenesis: stable colonization of Glossina morsitans morsitans with genetically modified Sodalis. BMC Microbiol 18:165. https://doi.org/10.1186/s12866-018-1282-9
Dedeine F, Vavre F, Fleury F, Loppin B, Hochberg ME, Bouletreau M (2001) Removing symbiotic Wolbachia bacteria specifically inhibits oogenesis in a parasitic wasp. Proc Natl Acad Sci U S A 98:6247–6252. https://doi.org/10.1073/pnas.101304298
Demirbas-Uzel G, De Vooght L, Parker AG, Vreysen MJB, Mach RL, Van Den Abbeele J, Abd-Alla AMM (2018) Combining paratransgenesis with SIT: impact of ionizing radiation on the DNA copy number of Sodalis glossinidius in tsetse flies. BMC Microbiol 18:160. https://doi.org/10.1186/s12866-018-1283-8
Dennis JW, Durkin SM, Horsley Downie JE, Hamill LC, Anderson NE, MacLeod ET (2014) Sodalis glossinidius prevalence and trypanosome presence in tsetse from Luambe National Park, Zambia. Parasit Vectors 7:378. https://doi.org/10.1186/1756-3305-7-378
Dobson SL, Bourtzis K, Braig HR, Jones BF, Zhou W, Rousset F, O’Neill SL (1999) Wolbachia infections are distributed throughout insect somatic and germ line tissues. Insect Biochem Mol Biol 29:153–160. https://doi.org/10.1016/s0965-1748(98)00119-2
Doudoumis V et al (2012) Detection and characterization of Wolbachia infections in laboratory and natural populations of different species of tsetse flies (genus Glossina). BMC Microbiol 12(Suppl 1):S3. https://doi.org/10.1186/1471-2180-12-S1-S3
Doudoumis V et al (2013) Tsetse-Wolbachia symbiosis: comes of age and has great potential for pest and disease control. J Invertebr Pathol 112. Suppl:S94–S103. https://doi.org/10.1016/j.jip.2012.05.010
Doudoumis V et al (2017) Challenging the Wigglesworthia, Sodalis, Wolbachia symbiosis dogma in tsetse flies: Spiroplasma is present in both laboratory and natural populations. Sci Rep 7:4699. https://doi.org/10.1038/s41598-017-04740-3
Douglas AE (1989) Mycetocyte symbiosis in insects. Biol Rev Camb Philos Soc 64:409–434. https://doi.org/10.1111/j.1469-185x.1989.tb00682.x
Dunning Hotopp JC et al (2007) Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317:1753–1756. https://doi.org/10.1126/science.1142490
Duron O, Boureux A, Echaubard P, Berthomieu A, Berticat C, Fort P, Weill M (2007) Variability and expression of ankyrin domain genes in Wolbachia variants infecting the mosquito Culex pipiens. J Bacteriol 189:4442–4448. https://doi.org/10.1128/JB.00142-07
Ekwudu O, Devine GJ, Aaskov JG, Frentiu FD (2020) Wolbachia strain wAlbB blocks replication of flaviviruses and alphaviruses in mosquito cell culture. Parasit Vectors 13:54. https://doi.org/10.1186/s13071-020-3936-3
Farikou O et al (2010) Tripartite interactions between tsetse flies, Sodalis glossinidius and trypanosomes – an epidemiological approach in two historical human African trypanosomiasis foci in Cameroon. Infect Genet Evol 10:115–121. https://doi.org/10.1016/j.meegid.2009.10.008
Fenn K, Conlon C, Jones M, Quail MA, Holroyd NE, Parkhill J, Blaxter M (2006) Phylogenetic relationships of the Wolbachia of nematodes and arthropods. PLoS Pathog 2:e94. https://doi.org/10.1371/journal.ppat.0020094
Ferree PM, Frydman HM, Li JM, Cao J, Wieschaus E, Sullivan W (2005) Wolbachia utilizes host microtubules and Dynein for anterior localization in the Drosophila oocyte. PLoS Pathog 1:e14. https://doi.org/10.1371/journal.ppat.0010014
Franco JR et al (2018) Monitoring the elimination of human African trypanosomiasis: Update to 2016. PLoS Negl Trop Dis 12:e0006890. https://doi.org/10.1371/journal.pntd.0006890
Franks TM, Lykke-Andersen J (2008) The control of mRNA decapping and P-body formation. Mol Cell 32:605–615. https://doi.org/10.1016/j.molcel.2008.11.001
Glaser RL, Meola MA (2010) The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS One 5:e11977. https://doi.org/10.1371/journal.pone.0011977
Glowska E, Dragun-Damian A, Dabert M, Gerth M (2015) New Wolbachia supergroups detected in quill mites (Acari: Syringophilidae). Infect Genet Evol 30:140–146. https://doi.org/10.1016/j.meegid.2014.12.019
Gooding R (1985) Electrophoretic and hybridization comparisons of Glossina morsitans morsitans, G. m. centralis and G. m. submorsitans (Diptera:Glossinidae). Can J Zool 63:2694–2702
Gooding R (1987) Genetic basis of sterility in hybrid males from crosses of Glossina morsitans morsitans and Glossina morsitans centralis. Can J Zool 65:640–646
Gooding R (1989) Genetic basis of sterility in hybrids from crosses of Glossina morsitans submorsitans and Glossina morsitans morsitans (Diptera:Glossinidae). Genome 32:479–485
Gooding RH (1990) Postmating barriers to gene flow among species and subspecies of tsetse flies (Diptera: Glossinidae). Can J Zool 68:1727–1734
Gorham CH, Fang QQ, Durden LA (2003) Wolbachia endosymbionts in fleas (Siphonaptera). J Parasitol 89:283–289. https://doi.org/10.1645/0022-3395(2003)089[0283:WEIFS]2.0.CO;2
Haines LR, Hancock RE, Pearson TW (2003) Cationic antimicrobial peptide killing of African trypanosomes and Sodalis glossinidius, a bacterial symbiont of the insect vector of sleeping sickness. Vector Borne Zoonotic Dis 3:175–186. https://doi.org/10.1089/153036603322662165
Hall RJ et al (2019) A tale of three species: adaptation of Sodalis glossinidius to tsetse biology, Wigglesworthia metabolism, and host Diet. mBio 10:e02106–e02118. https://doi.org/10.1128/mBio.02106-18
Hamidou Soumana I, Loriod B, Ravel S, Tchicaya B, Simo G, Rihet P, Geiger A (2014a) The transcriptional signatures of Sodalis glossinidius in the Glossina palpalis gambiensis flies negative for Trypanosoma brucei gambiense contrast with those of this symbiont in tsetse flies positive for the parasite: possible involvement of a Sodalis-hosted prophage in fly Trypanosoma refractoriness? Infect Genet Evol 24:41–56. https://doi.org/10.1016/j.meegid.2014.03.005
Hamidou Soumana I, Tchicaya B, Loriod B, Rihet P, Geiger A (2014b) Identification of overexpressed genes in Sodalis glossinidius inhabiting trypanosome-infected self-cured tsetse flies. Front Microbiol 5:255. https://doi.org/10.3389/fmicb.2014.00255
Harris HL, Braig HR (2003) Sperm chromatin remodelling and Wolbachia-induced cytoplasmic incompatibility in Drosophila. Biochem Cell Biol 81:229–240. https://doi.org/10.1139/o03-053
Haselkorn TS, Cockburn SN, Hamilton PT, Perlman SJ, Jaenike J (2013) Infectious adaptation: potential host range of a defensive endosymbiont in Drosophila. Evolution 67:934–945. https://doi.org/10.1111/evo.12020
Hedges LM, Brownlie JC, O’Neill SL, Johnson KN (2008) Wolbachia and virus protection in insects. Science 322:702. https://doi.org/10.1126/science.1162418
Herbeck JT, Wall DP, Wernegreen JJ (2003) Gene expression level influences amino acid usage, but not codon usage, in the tsetse fly endosymbiont Wigglesworthia. Microbiology 149:2585–2596. https://doi.org/10.1099/mic.0.26381-0
Hertig M (1936) The Rickettsia, Wolbachia pipientis (gen. et sp.n.) and associated inclusions of the mosquito, Culex pipiens. Parasitology 28:453–486. https://doi.org/10.1017/S0031182000022666
Hertig M, Wolbach SB (1924) Studies on Rickettsia-like micro-organisms in insects. J Med Res 44:329–374.327
Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH (2008) How many species are infected with Wolbachia? – a statistical analysis of current data. FEMS Microbiol Lett 281:215–220. https://doi.org/10.1111/j.1574-6968.2008.01110.x
Hill P, Saunders DS, Campbell JA (1973) Letter: The production of “symbiont-free” Glossina morsitans and an associated loss of female fertility. Trans R Soc Trop Med Hyg 67:727–728. https://doi.org/10.1016/0035-9203(73)90051-5
Hoffmann AA, Ross PA, Rašić G (2015) Wolbachia strains for disease control: ecological and evolutionary considerations. Evol Appl 8:751–768. https://doi.org/10.1111/eva.12286
Hosokawa T, Kaiwa N, Matsuura Y, Kikuchi Y, Fukatsu T (2015) Infection prevalence of Sodalis symbionts among stinkbugs. Zoological Lett 1:5. https://doi.org/10.1186/s40851-014-0009-5
Hrusa G et al (2015) TonB-dependent heme iron acquisition in the tsetse fly symbiont Sodalis glossinidius. Appl Environ Microbiol 81:2900–2909. https://doi.org/10.1128/AEM.04166-14
Huebner E, Davey KG (1974) Bacteroids in the ovaries of a tsetse fly. Nature 249:260–261
Hussain M, Frentiu FD, Moreira LA, O’Neill SL, Asgari S (2011) Wolbachia uses host microRNAs to manipulate host gene expression and facilitate colonization of the dengue vector Aedes aegypti. Proc Natl Acad Sci U S A 108:9250–9255. https://doi.org/10.1073/pnas.1105469108
Ijichi N, Kondo N, Matsumoto R, Shimada M, Ishikawa H, Fukatsu T (2002) Internal spatiotemporal population dynamics of infection with three Wolbachia strains in the adzuki bean beetle, Callosobruchus chinensis (Coleoptera: Bruchidae). Appl Environ Microbiol 68:4074–4080. https://doi.org/10.1128/aem.68.8.4074-4080.2002
Isaac C, Ohiolei JA, Ebhodaghe F, Igbinosa IB, Eze AA (2017) Animal African Trypanosomiasis in Nigeria: A long way from elimination/eradication. Acta Trop 176:323–331. https://doi.org/10.1016/j.actatropica.2017.08.032
Iturbe-Ormaetxe I, Burke GR, Riegler M, O’Neill SL (2005) Distribution, expression, and motif variability of ankyrin domain genes in Wolbachia pipientis. J Bacteriol 187:5136–5145. https://doi.org/10.1128/JB.187.15.5136-5145.2005
Jaenike J, Unckless R, Cockburn SN, Boelio LM, Perlman SJ (2010) Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science 329:212–215. https://doi.org/10.1126/science.1188235
Jucci C (1952) Symbiosis and phylogenesis in the isoptera. Nature 169:837. https://doi.org/10.1038/169837a0
Kageyama D, Traut W (2004) Opposite sex-specific effects of Wolbachia and interference with the sex determination of its host Ostrinia scapulalis. Proc Biol Sci 271:251–258. https://doi.org/10.1098/rspb.2003.2604
Kageyama D et al (2017) Feminizing Wolbachia endosymbiont disrupts maternal sex chromosome inheritance in a butterfly species. Evol Lett 1:232–244. https://doi.org/10.1002/evl3.28
Kambris Z, Cook PE, Phuc HK, Sinkins SP (2009) Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science 326:134–136. https://doi.org/10.1126/science.1177531
Kame-Ngasse GI, Njiokou F, Melachio-Tanekou TT, Farikou O, Simo G, Geiger A (2018) Prevalence of symbionts and trypanosome infections in tsetse flies of two villages of the “Faro and Deo” division of the Adamawa region of Cameroon. BMC Microbiol 18:159. https://doi.org/10.1186/s12866-018-1286-5
Kante ST, Melachio T, Ofon E, Njiokou F, Simo G (2018) Detection of Wolbachia and different trypanosome species in Glossina palpalis palpalis populations from three sleeping sickness foci of southern Cameroon. Parasit Vectors 11:630. https://doi.org/10.1186/s13071-018-3229-2
Kita A, Jimbo M, Sakai R, Morimoto Y, Miki K (2015) Crystal structure of a symbiosis-related lectin from octocoral. Glycobiology 25:1016–1023. https://doi.org/10.1093/glycob/cwv033
Klasson L, Kambris Z, Cook PE, Walker T, Sinkins SP (2009) Horizontal gene transfer between Wolbachia and the mosquito Aedes aegypti. BMC Genomics 10:33. https://doi.org/10.1186/1471-2164-10-33
Kondo N, Nikoh N, Ijichi N, Shimada M, Fukatsu T (2002) Genome fragment of Wolbachia endosymbiont transferred to X chromosome of host insect. Proc Natl Acad Sci U S A 99:14280–14285. https://doi.org/10.1073/pnas.222228199
Kozek W, Rao R (2007) The Discovery of Wolbachia in arthropods and nematodes – a historical perspective. In: Hoerauf AaR RU (ed) Wolbachia: A Bug’s Life in another Bug, vol 5. Wolbachia. Issues Infect Dis. Karger, Basel, pp 1–14. https://doi.org/10.1159/000104228
Kremer N, Charif D, Henri H, Gavory F, Wincker P, Mavingui P, Vavre F (2012) Influence of Wolbachia on host gene expression in an obligatory symbiosis. BMC Microbiol 12(Suppl 1):S7. https://doi.org/10.1186/1471-2180-12-S1-S7
LePage DP et al (2017) Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature 543:243–247. https://doi.org/10.1038/nature21391
Lo N, Casiraghi M, Salati E, Bazzocchi C, Bandi C (2002) How many Wolbachia supergroups exist? Mol Biol Evol 19:341–346. https://doi.org/10.1093/oxfordjournals.molbev.a004087
Ma WC, Denlinger DL (1974) Secretory discharge and microflora of milk gland in tsetse flies. Nature 247:301–303
Magnarelli LA (1978) Nectar-feeding by female mosquitoes and its relation to follicular development and parity. J Med Entomol 14:527–530. https://doi.org/10.1093/jmedent/14.5.527
Maltz MA, Weiss BL, O’Neill M, Wu Y, Aksoy S (2012) OmpA-mediated biofilm formation is essential for the commensal bacterium Sodalis glossinidius to colonize the tsetse fly gut. Appl Environ Microbiol 78:7760–7768. https://doi.org/10.1128/AEM.01858-12
Mancini MV, Herd CS, Ant TH, Murdochy SM, Sinkins SP (2020) Wolbachia strain wAu efficiently blocks arbovirus transmission in Aedes albopictus. PLoS Negl Trop Dis 14:e0007926. https://doi.org/10.1371/journal.pntd.0007926
Mayoral JG, Hussain M, Joubert DA, Iturbe-Ormaetxe I, O’Neill SL, Asgari S (2014) Wolbachia small noncoding RNAs and their role in cross-kingdom communications. Proc Natl Acad Sci U S A 111:18721–18726. https://doi.org/10.1073/pnas.1420131112
Mellanby H (1937) Experimental work on reproduction in the tsetse fly, Glossina palpalis. Parasitology 1:131–141
Meyer A, Holt HR, Selby R, Guitian J (2016) Past and ongoing tsetse and animal trypanosomiasis control operations in five African Countries: a systematic review. PLoS Negl Trop Dis 10:e0005247. https://doi.org/10.1371/journal.pntd.0005247
Michalkova V, Benoit JB, Weiss BL, Attardo GM, Aksoy S (2014) Vitamin B6 generated by obligate symbionts is critical for maintaining proline homeostasis and fecundity in tsetse flies. Appl Environ Microbiol 80:5844–5853. https://doi.org/10.1128/AEM.01150-14
Min K-T, Benzer S (1997) Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and death. Proc Natl Acad Sci U S A 94:10792–10796
Mitsuhashi W, Saiki T, Wei W, Kawakita H, Sato M (2002) Two novel strains of Wolbachia coexisting in both species of mulberry leafhoppers. Insect Mol Biol 11:577–584. https://doi.org/10.1046/j.1365-2583.2002.00368.x
Montenegro H, Solferini VN, Klaczko LB, Hurst GDD (2005) Male-killing Spiroplasma naturally infecting Drosophila melanogaster. Insect Mol Biol 14:281–287. https://doi.org/10.1111/j.1365-2583.2005.00558.x
Moran NA, Bennett GM (2014) The tiniest tiny genomes. Annu Rev Microbiol 68:195–215. https://doi.org/10.1146/annurev-micro-091213-112901
Moreira LA et al (2009) A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 139:1268–1278. https://doi.org/10.1016/j.cell.2009.11.042
Mouton L, Henri H, Charif D, Boulétreau M, Vavre F (2007) Interaction between host genotype and environmental conditions affects bacterial density in Wolbachia symbiosis. Biol Lett 3:210–213. https://doi.org/10.1098/rsbl.2006.0590
Munson MA, Baumann P, Kinsey MG (1991) Buchnera gen. nov. and Buchnera aphidicola sp. nov., a taxon consisting of the mycetocyte-associated, primary endosymbionts of aphids. Int J Syst Evol Micr 41:566–568
Nikoh N, Nakabachi A (2009) Aphids acquired symbiotic genes via lateral gene transfer. BMC Biol 7:12. https://doi.org/10.1186/1741-7007-7-12
Nikoh N, Tanaka K, Shibata F, Kondo N, Hizume M, Shimada M, Fukatsu T (2008) Wolbachia genome integrated in an insect chromosome: evolution and fate of laterally transferred endosymbiont genes. Genome Res 18:272–280. https://doi.org/10.1101/gr.7144908
Nogge G (1976) Sterility in tsetse flies (Glossina morsitans Westwood) caused by loss of symbionts. Experientia 32:995–996
Nogge G (1978) Aposymbiotic tsetse flies, Glossina morsitans morsitans obtained by feeding on rabbits immunized specifically with symbionts. J Insect Physiol 24:299–304
Nogge G, Gerresheim A (1982) Experiments on the elimination of symbionts from the tsetse-fly, Glossina morsitans morsitans (Diptera, Glossinidae), by antibiotics and lysozyme. J Invertebr Pathol 40:166–179
Novakova E, Husnik F, Sochova E, Hypsa V (2015) Arsenophonus and Sodalis symbionts in louse flies: an analogy to the Wigglesworthia and Sodalis system in tsetse flies. Appl Environ Microbiol 81:6189–6199. https://doi.org/10.1128/AEM.01487-15
Nunan LM, Lightner DV, Oduori MA, Gasparich GE (2005) Spiroplasma penaei sp. nov., associated with mortalities in Penaeus vannamei, Pacific white shrimp. Int J Syst Evol Micr 55:2317–2322
O’Connor L, Plichart C, Sang AC, Brelsfoard CL, Bossin HC, Dobson SL (2012) Open release of male mosquitoes infected with a Wolbachia biopesticide: field performance and infection containment. PLoS Negl Trop Dis 6:e1797. https://doi.org/10.1371/journal.pntd.0001797
O’Neill SL, Karr TL (1990) Bidirectional incompatibility between conspecific populations of Drosophila simulans. Nature 348:178–180. https://doi.org/10.1038/348178a0
O’Neill SL, Giordano R, Colbert AM, Karr TL, Robertson HM (1992) 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc Natl Acad Sci U S A 89:2699–2702. https://doi.org/10.1073/pnas.89.7.2699
O’Neill SL, Gooding RH, Aksoy S (1993) Phylogenetically distant symbiotic microorganisms reside in Glossina midgut and ovary tissues. Med Vet Entomol 7:377–383. https://doi.org/10.1111/j.1365-2915.1993.tb00709.x
Osborne SE, Leong YS, O’Neill SL, Johnson KN (2009) Variation in antiviral protection mediated by different Wolbachia strains in Drosophila simulans. PLoS Pathog 5:e1000656. https://doi.org/10.1371/journal.ppat.1000656
Ote M, Yamamoto D (2020) Impact of Wolbachia infection on Drosophila female germline stem cells. Curr Opin Insect Sci 37:8–15. https://doi.org/10.1016/j.cois.2019.10.001
Ote M, Ueyama M, Yamamoto D (2016) Wolbachia protein TomO targets nanos mRNA and restores germ stem cells in Drosophila sex-lethal mutants. Curr Biol 26:2223–2232. https://doi.org/10.1016/j.cub.2016.06.054
Pais R, Lohs C, Wu Y, Wang J, Aksoy S (2008) The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl Environ Microbiol 74:5965–5974. https://doi.org/10.1128/AEM.00741-08
Pan X, Zhou G, Wu J, Bian G, Lu P, Raikhel AS, Xi Z (2012) Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc Natl Acad Sci U S A 109:E23–E31. https://doi.org/10.1073/pnas.1116932108
Pan X et al (2018) The bacterium Wolbachia exploits host innate immunity to establish a symbiotic relationship with the dengue vector mosquito Aedes aegypti. ISME J 12:277–288. https://doi.org/10.1038/ismej.2017.174
Pannebakker BA, Loppin B, Elemans CP, Humblot L, Vavre F (2007) Parasitic inhibition of cell death facilitates symbiosis. Proc Natl Acad Sci U S A 104:213–215. https://doi.org/10.1073/pnas.0607845104
Paraskevopoulos C, Bordenstein SR, Wernegreen JJ, Werren JH, Bourtzis K (2006) Toward a Wolbachia multilocus sequence typing system: discrimination of Wolbachia strains present in Drosophila species. Curr Microbiol 53:388–395. https://doi.org/10.1007/s00284-006-0054-1
Paredes JC et al (2015) Genome sequence of the Drosophila melanogaster male-killing Spiroplasma strain MSRO endosymbiont. mBio 6:e02437-14. https://doi.org/10.1128/mBio.02437-14
Pell PE, Southern DI (1975) Symbionts in the female tsetse fly Glossina morsitans morsitans. Experientia 31:650–651. https://doi.org/10.1007/BF01944608
Pietri JE, DeBruhl H, Sullivan W (2016) The rich somatic life of Wolbachia. Microbiologyopen 5:923–936. https://doi.org/10.1002/mbo3.390
Pinnock DE, Hess RT (1974) The occurrence of intracellular rickettsia-like organisms in the tsetse flies, Glossina morsitans, G. fuscipes, G. brevipalpis and G. pallidipes. Acta Trop 31:70–79
Pontes MH, Babst M, Lochhead R, Oakeson K, Smith K, Dale C (2008) Quorum sensing primes the oxidative stress response in the insect endosymbiont, Sodalis glossinidius. PLoS One 3:e3541. https://doi.org/10.1371/journal.pone.0003541
Puchta O (1955) Experimental studies on the significance of symbiosis in the clothes louse Pediculus vestimenti Burm. Z Parasitenkd 17:1–40. https://doi.org/10.1007/bf00260226
Rasić G, Endersby NM, Williams C, Hoffmann AA (2014) Using Wolbachia-based release for suppression of Aedes mosquitoes: insights from genetic data and population simulations. Ecol Appl 24:1226–1234. https://doi.org/10.1890/13-1305.1
Rawlings P (1985) The genetics of hybrid sterility between subspecies of the complex of Glossina morsitans Westwood (Diptera: Glossinidae). B Entomol Res 75:689–699
Reinhardt C, Steiger R, Hecker H (1972) Ultrastructural study of the midgut mycetome-bacteroids of the tsetse flies Glossina morsitans, G. fuscipes, and G. brevipalpis (Diptera, Brachycera). Acta Trop 29:280–288
Rio RV, Lefevre C, Heddi A, Aksoy S (2003) Comparative genomics of insect-symbiotic bacteria: influence of host environment on microbial genome composition. Appl Environ Microbiol 69:6825–6832. https://doi.org/10.1128/aem.69.11.6825-6832.2003
Rio RV, Wu YN, Filardo G, Aksoy S (2006) Dynamics of multiple symbiont density regulation during host development: tsetse fly and its microbial flora. Proc Biol Sci 273:805–814. https://doi.org/10.1098/rspb.2005.3399
Rio RV et al (2012) Insight into the transmission biology and species-specific functional capabilities of tsetse (Diptera: glossinidae) obligate symbiont Wigglesworthia. MBio 3:00240-11. https://doi.org/10.1128/mBio.00240-11
Ros VID, Fleming VM, Feil EJ, Breeuwer JAJ (2009) How diverse is the genus Wolbachia? Multiple-gene sequencing reveals a putatively new Wolbachia supergroup recovered from spider mites (Acari: Tetranychidae). Appl Environ Microbiol 75:1036–1043. https://doi.org/10.1128/AEM.01109-08
Roubaud B (1919) Les particularites de la nutrition et de la vie symbiotique chez les mouches tsetse. Ann Inst Pasteur 33:489–537
Rousset F, Bouchon D, Pintureau B, Juchault P, Solignac M (1992) Wolbachia endosymbionts responsible for various alterations of sexuality in arthropods. Proc Biol Sci 250:91–98. https://doi.org/10.1098/rspb.1992.0135
Rowley SM, Raven RJ, McGraw EA (2004) Wolbachia pipientis in Australian spiders. Curr Microbiol 49:208–214. https://doi.org/10.1007/s00284-004-4346-z
Runyen-Janecky LJ, Brown AN, Ott B, Tujuba HG, Rio RV (2010) Regulation of high-affinity iron acquisition homologues in the tsetse fly symbiont Sodalis glossinidius. J Bacteriol 192:3780–3787. https://doi.org/10.1128/JB.00161-10
Schneider DI, Garschall KI, Parker AG, Abd-Alla AMM, Miller WJ (2013) Global Wolbachia prevalence, titer fluctuations and their potential of causing cytoplasmic incompatibilities in tsetse flies and hybrids of Glossina morsitans subgroup species. J Invertebr Pathol 112. Suppl:S104–S115. https://doi.org/10.1016/j.jip.2012.03.024
Schneider DI, Parker AG, Abd-Alla AM, Miller WJ (2018) High-sensitivity detection of cryptic Wolbachia in the African tsetse fly (Glossina spp.). BMC Microbiol 18:140. https://doi.org/10.1186/s12866-018-1291-8
Schneider DI et al (2019) Spatio-temporal distribution of Spiroplasma infections in the tsetse fly (Glossina fuscipes fuscipes) in northern Uganda. PLoS Negl Trop Dis 13:e0007340. https://doi.org/10.1371/journal.pntd.0007340
Schroder D et al (1996) Intracellular endosymbiotic bacteria of Camponotus species (carpenter ants): systematics, evolution and ultrastructural characterization. Mol Microbiol 21:479–489. https://doi.org/10.1111/j.1365-2958.1996.tb02557.x
Serbus LR, Sullivan W (2007) A cellular basis for Wolbachia recruitment to the host germline. PLoS Pathog 3:e190. https://doi.org/10.1371/journal.ppat.0030190
Serbus LR, Casper-Lindley C, Landmann F, Sullivan W (2008) The genetics and cell biology of Wolbachia-host interactions. Annu Rev Genet 42:683–707. https://doi.org/10.1146/annurev.genet.41.110306.130354
Serbus LR, Ferreccio A, Zhukova M, McMorris CL, Kiseleva E, Sullivan W (2011) A feedback loop between Wolbachia and the Drosophila gurken mRNP complex influences Wolbachia titer. J Cell Sci 124:4299–4308. https://doi.org/10.1242/jcs.092510
Simo G et al (2019) Molecular identification of Wolbachia and Sodalis glossinidius in the midgut of Glossina fuscipes quanzensis from the Democratic Republic of Congo. Parasite 26:5. https://doi.org/10.1051/parasite/2019005
Sinkins SP, Gould F (2006) Gene drive systems for insect disease vectors. Nat Rev Genet 7:427–435. https://doi.org/10.1038/nrg1870
Sinkins S, Curtis C, O’Neill SL (1997) The potential application of inherited symbiont systems to pest control. In: Influential passengers: inherited microorganisms and arthropod reproduction. Oxford University Press, Oxford, pp 155–175
Smith CL, Weiss BL, Aksoy S, Runyen-Janecky LJ (2013) Characterization of the achromobactin iron acquisition operon in Sodalis glossinidius. Appl Environ Microbiol 79:2872–2881. https://doi.org/10.1128/AEM.03959-12
Snyder AK, Rio RV (2015) “Wigglesworthia morsitans” folate (Vitamin B9) biosynthesis contributes to tsetse host fitness. Appl Environ Microbiol 81:5375–5386. https://doi.org/10.1128/AEM.00553-15
Snyder AK, Deberry JW, Runyen-Janecky L, Rio RV (2010) Nutrient provisioning facilitates homeostasis between tsetse fly (Diptera: Glossinidae) symbionts. Proc Biol Sci 277:2389–2397. https://doi.org/10.1098/rspb.2010.0364
Snyder AK, McLain C, Rio RV (2012) The tsetse fly obligate mutualist Wigglesworthia morsitans alters gene expression and population density via exogenous nutrient provisioning. Appl Environ Microbiol 78:7792–7797. https://doi.org/10.1128/AEM.02052-12
Soumana IH, Simo G, Njiokou F, Tchicaya B, Abd-Alla AM, Cuny G, Geiger A (2013) The bacterial flora of tsetse fly midgut and its effect on trypanosome transmission. J Invertebr Pathol 112. Suppl:S89–S93. https://doi.org/10.1016/j.jip.2012.03.029
Starr DJ, Cline TW (2002) A host parasite interaction rescues Drosophila oogenesis defects. Nature 418:76–79. https://doi.org/10.1038/nature00843
Stouthamer R, Breeuwer JA, Hurst GD (1999) Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu Rev Microbiol 53:71–102. https://doi.org/10.1146/annurev.micro.53.1.71
Strunov A, Kiseleva E (2016) Drosophila melanogaster brain invasion: pathogenic Wolbachia in central nervous system of the fly. Insect Sci 23:253–264. https://doi.org/10.1111/1744-7917.12187
Stuhlmann F (1907) Beitrage zur Kenntnis der Tsetsefliege (G. fusca und G. tachinoides). Arb Gesundh Amte (Berlin) xxxiii:489–536
Sugimoto TN, Kayukawa T, Shinoda T, Ishikawa Y, Tsuchida T (2015) Misdirection of dosage compensation underlies bidirectional sex-specific death in Wolbachia-infected Ostrinia scapulalis. Insect Biochem Mol Biol 66:72–76. https://doi.org/10.1016/j.ibmb.2015.10.001
Symula RE et al (2011) Influence of host phylogeographic patterns and incomplete lineage sorting on within-species genetic variability in Wigglesworthia species, obligate symbionts of tsetse flies. Appl Environ Microbiol 77:8400–8408. https://doi.org/10.1128/AEM.05688-11
Symula RE et al (2013) Wolbachia association with the tsetse fly, Glossina fuscipes fuscipes, reveals high levels of genetic diversity and complex evolutionary dynamics. BMC Evol Biol 13:31. https://doi.org/10.1186/1471-2148-13-31
Teixeira L, Ferreira A, Ashburner M (2008) The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol 6:e2. https://doi.org/10.1371/journal.pbio.1000002
Tobe SS (1978) Reproductive physiology of Glossina. Annu Rev Entomol 23:283–307. https://doi.org/10.1146/annurev.en.23.010178.001435
Toh H, Weiss BL, Perkin SA, Yamashita A, Oshima K, Hattori M, Aksoy S (2006) Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res 16:149–156. https://doi.org/10.1101/gr.4106106
Tram U, Sullivan W (2002) Role of delayed nuclear envelope breakdown and mitosis in Wolbachia-induced cytoplasmic incompatibility. Science 296:1124–1126. https://doi.org/10.1126/science.1070536
Tram U, Ferree PM, Sullivan W (2003) Identification of Wolbachia-host interacting factors through cytological analysis. Microbes Infect 5:999–1011. https://doi.org/10.1016/s1286-4579(03)00192-8
Tsagmo Ngoune JM et al (2019) The composition and abundance of bacterial communities residing in the gut of Glossina palpalis palpalis captured in two sites of southern Cameroon. Parasit Vectors 12:151. https://doi.org/10.1186/s13071-019-3402-2
Turelli M et al (2018) Rapid global spread of wRi-like Wolbachia across multiple Drosophila. Curr Biol 28 e968:963–971. https://doi.org/10.1016/j.cub.2018.02.015
Unterman BM, Baumann P, McLean DL (1989) Pea aphid symbiont relationships established by analysis of 16S rRNAs. J Bacteriol 171:2970–2974. https://doi.org/10.1128/jb.171.6.2970-2974.1989
Vanderplank FL (1948) Experiments in crossbreeding tsetse-flies, Glossina species. Ann Trop Med Parasitol 42:131–152. https://doi.org/10.1080/00034983.1948.11685357
Vreysen MJ, Seck MT, Sall B, Bouyer J (2013) Tsetse flies: their biology and control using area-wide integrated pest management approaches. J Invertebr Pathol 112. Suppl:S15–S25. https://doi.org/10.1016/j.jip.2012.07.026
Walker T et al (2011) The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476:450–453. https://doi.org/10.1038/nature10355
Wamwiri FN et al (2013) Wolbachia, Sodalis and trypanosome co-infections in natural populations of Glossina austeni and Glossina pallidipes. Parasit Vectors 6:232. https://doi.org/10.1186/1756-3305-6-232
Wang J, Aksoy S (2012) PGRP-LB is a maternally transmitted immune milk protein that influences symbiosis and parasitism in tsetse’s offspring. Proc Natl Acad Sci U S A 109:10552–10557. https://doi.org/10.1073/pnas.1116431109
Wang W, Wen B, Gasparich GE, Zhu N, Rong L, Chen J, Xu Z (2004) A spiroplasma associated with tremor disease in the Chinese mitten crab (Eriocheir sinensis). Microbiology 150:3035–3040
Wang J, Wu Y, Yang G, Aksoy S (2009) Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc Natl Acad Sci U S A 106:12133–12138. https://doi.org/10.1073/pnas.0901226106
Wang Y, Carolan JC, Hao F, Nicholson JK, Wilkinson TL, Douglas AE (2010) Integrated metabonomic-proteomic analysis of an insect-bacterial symbiotic system. J Proteome Res 9:1257–1267. https://doi.org/10.1021/pr9007392
Weiss BL, Mouchotte R, Rio RV, Wu YN, Wu Z, Heddi A, Aksoy S (2006) Inter-specific transfer of bacterial endosymbionts between tsetse species: infection establishment and effect on host fitness. Appl Environ Microbiol 72:7013–7021
Weiss BL, Wu Y, Schwank JJ, Tolwinski NS, Aksoy S (2008) An insect symbiosis is influenced by bacterium-specific polymorphisms in outer-membrane protein A. Proc Natl Acad Sci U S A 105:15088–15093. https://doi.org/10.1073/pnas.0805666105
Weiss BL, Wang J, Aksoy S (2011) Tsetse immune system maturation requires the presence of obligate symbionts in larvae. PLoS Biol 9:e1000619. https://doi.org/10.1371/journal.pbio.1000619
Welburn SC, Maudlin I, Ellis DS (1987) In vitro cultivation of rickettsia-like-organisms from Glossina spp. Ann Trop Med Parasit 81:331–335
Werren JH (1997) Wolbachia run amok. Proc Natl Acad Sci U S A 94:11154–11155. https://doi.org/10.1073/pnas.94.21.11154
Werren JH, Zhang W, Guo LR (1995) Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc Biol Sci 261:55–63. https://doi.org/10.1098/rspb.1995.0117
Wigglesworth VB (1929) Digestion in the tsetse-fly: a study of structure and function. Parasitology 21:288–321
Woolfit M, Iturbe-Ormaetxe I, McGraw EA, O’Neill SL (2009) An ancient horizontal gene transfer between mosquito and the endosymbiotic bacterium Wolbachia pipientis. Mol Biol Evol 26:367–374. https://doi.org/10.1093/molbev/msn253
Xi Z, Khoo CCH, Dobson SL (2005) Wolbachia establishment and invasion in an Aedes aegypti laboratory population. Science 310:326–328. https://doi.org/10.1126/science.1117607
Yen JH, Barr AR (1973) The etiological agent of cytoplasmic incompatibility in Culex pipiens. J Invertebr Pathol 22:242–250. https://doi.org/10.1016/0022-2011(73)90141-9
Yuan L-L et al (2015) Quantitative proteomic analyses of molecular mechanisms associated with Cytoplasmic Incompatibility in Drosophila melanogaster induced by Wolbachia. J Proteome Res 14:3835–3847. https://doi.org/10.1021/acs.jproteome.5b00191
Yun Y, Lei C, Peng Y, Liu F, Chen J, Chen L (2011) Wolbachia strains typing in different geographic population spider, Hylyphantes graminicola (Linyphiidae). Curr Microbiol 62:139–145. https://doi.org/10.1007/s00284-010-9686-2
Zabalou S, Apostolaki A, Livadaras I, Franz G, Robinson AS, Savakis C, Bourtzis K (2009) Incompatible insect technique: incompatible males from a Ceratitis capitata genetic sexing strain. Entomol Exp Appl 132:232–240. https://doi.org/10.1111/j.1570-7458.2009.00886.x
Zaidman-Remy A et al (2006) The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity 24:463–473. https://doi.org/10.1016/j.immuni.2006.02.012
Zhang D, Lees RS, Xi Z, Gilles JRL, Bourtzis K (2015a) Combining the Sterile Insect Technique with Wolbachia-based approaches: II-A safer approach to Aedes albopictus population suppression programmes, designed to minimize the consequences of inadvertent female release. PLoS One 10:e0135194. https://doi.org/10.1371/journal.pone.0135194
Zhang D, Zheng X, Xi Z, Bourtzis K, Gilles JRL (2015b) Combining the Sterile Insect Technique with the Incompatible Insect Technique: I-Impact of Wolbachia infection on the fitness of triple- and double-infected strains of Aedes albopictus. PLoS One 10:e0121126. https://doi.org/10.1371/journal.pone.0121126
Zhang D, Lees RS, Xi Z, Bourtzis K, Gilles JRL (2016) Combining the Sterile Insect Technique with the Incompatible Insect Technique: III-Robust mating competitiveness of irradiated triple Wolbachia-infected Aedes albopictus males under semi-field conditions. PLoS One 11:e0151864. https://doi.org/10.1371/journal.pone.0151864
Zheng Y, Wang J-L, Liu C, Wang C-P, Walker T, Wang Y-F (2011) Differentially expressed profiles in the larval testes of Wolbachia infected and uninfected Drosophila. BMC Genomics 12:595. https://doi.org/10.1186/1471-2164-12-595
Zhou JJ (2010) Odorant-binding proteins in insects. Vitam Horm 83:241–272. https://doi.org/10.1016/S0083-6729(10)83010-9
Zhou W, Rousset F, O’Neil S (1998) Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc Biol Sci 265:509–515. https://doi.org/10.1098/rspb.1998.0324
Zientz E, Dandekar T, Gross R (2004) Metabolic interdependence of obligate intracellular bacteria and their insect hosts. Microbiol Mol Biol Rev 68:745–770. https://doi.org/10.1128/MMBR.68.4.745-770.2004
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Attardo, G.M., Scolari, F., Malacrida, A. (2020). Bacterial Symbionts of Tsetse Flies: Relationships and Functional Interactions Between Tsetse Flies and Their Symbionts. In: Kloc, M. (eds) Symbiosis: Cellular, Molecular, Medical and Evolutionary Aspects. Results and Problems in Cell Differentiation, vol 69. Springer, Cham. https://doi.org/10.1007/978-3-030-51849-3_19
Download citation
DOI: https://doi.org/10.1007/978-3-030-51849-3_19
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-51848-6
Online ISBN: 978-3-030-51849-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)