Abstract
The importance of plant genetic resources has received increasing attention in crop improvement and plant variation in breeding procedures. Here, plant variation and the structures of plant populations were examined among 93 flax accessions using SCoT and URP molecular markers. Polymorphic-fragments by URP and SCoT primers were, on average, 9.3 and 10.9, respectively, and the mean of polymorphism information content for primers revealed a good efficiency of both marker techniques (0.34 and 0.35 for URP and SCoT, respectively). The neighbor-joining (NJ)-based clustering, via combined data, classified all investigated accessions into four main groups that were further confirmed by principal coordinate analysis (PCoA). Moreover, molecular diversity (88%) was mostly observed inside the populations. STRUCTURE-analysis confirmed the cluster analysis and all samples were separated into four subpopulations (∆K = 4). The results implied that the sub-populations consisted of diverse accessions, suggesting that this gene pool has the potential for flax breeding programs. Furthermore, our findings indicated that URP and SCoT markers are reliable techniques in assessing plant variation, especially in elucidating the structures of flax populations. However, gene-targeting markers like SCoT are preferable because of their roots in functional genome-based regions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Flax (Linum usitatissimum L.), the common type of linseed, belongs to the Linaceae family and is grown for different purposes. Its high amount of linolenic acid (45–60%) has made linseed oil popular with several applications in the industry, such as the manufacture of paints, printing inks, varnishes, and stains (Kurt and Bozkurt 2006; Patial et al. 2019). Linseed has high linolenic acid (ALA, omega 3 fatty acid) and high lignans, making it nutritious with anti-cancer properties. The essential polyunsaturated fatty acids in the seed oil of linseed also make it very effective to reduce coronary heart diseases and add to the health benefits of this valuable herb. In addition, this plant is primarily grown for the extraction of fiber (Bassett et al. 2009; Goyal et al. 2014; Patial et al. 2019; Thakur et al. 2021).
Since plants play a key role in agriculture, their variation seems essential in plant breeding. Having genetic diversity in crop germplasm can guarantee a successful selection of parents in providing populations with superior plant varieties. In fact, enough variation is a precursor to new, enhanced plant cultivars (Govindaraj et al. 2015). The commercialization of optimal cultivars has put less competitive variations at risk of permanent loss. The evaluation of plant variation not only assists in generating new varieties but also helps with the efficient management and conservation of germplasms (Vinu et al. 2013). Genetic diversity is also substantially crucial when maximizing breeding for efficiency. Genetic mapping with an evaluation of plant diversity helps with the maintenance of the genetic well-being of plant populations. It assists in discovering new valuable alleles for breeding programs (Diederichsen and Richards 2003; Kumari et al. 2017; Patial et al. 2019). Although the investigation of a genetic construct, with a focus on variation, among plant accessions can be carried out via molecular, morphological, and biochemical markers, the use of new, molecular-based methods can be more efficient than traditional phenotypic and biochemical markers. Molecular markers, in particular, DNA markers, are more reliable, and reproducible, with a larger capacity to be detected among the plant tissue. Moreover, molecular-based markers can be less influenced by environmental factors and offer higher polymorphism than the other types of markers (Mohammadi and Prasanna 2003; Collard and Mackill 2009; Govindaraj et al. 2015).
Many applications are known for molecular-based markers in plant breeding, such as assessment of genetic diversity, analysis of population structure, marker-assisted selection, cultivar identification and fingerprinting, Association mapping, and QTL mapping (Govindaraj et al. 2015). More recently, many different DNA-based marker(s) are introduced because of developments in the area of genomics. Among different types of DNA markers, gene-targeted markers are a preferable choice to random markers for the assessment of genetic diversity because of their concentration on gene loci, rendering them efficiently operative in QTL-mapping, gene-targeting, and marker-assisted breeding. Furthermore, gene-targeted markers work with lengthier primers that have hotter annealing temperatures, causing a greater level of reproducibility in them, more stability and reliability, compared to other types of non-efficient constructs (Collard and Mackill 2009; de Lucena et al. 2013; Ding et al. 2015).
Collard and Mackill (2009) reported a simple and novel DNA marker technique Start Codon Targeted (SCoT) Polymorphism. SCoT can be regarded as a reliable, new gene-based marker technique, with a wide range of applications in plants. The markers can be made with single-primer constructs, while relying on short-conserved regions in genes in the vicinity of the ATGs translation initiation codon, occurring as a conserved format in the majority of genes (Collard and Mackill 2009; Feng et al. 2018). This marker system does not require any sequence information for primer designing and SCoT markers usually correspond with functional modes of genetic material, while correlating with traits as well (Mulpuri et al. 2013). SCoT markers have been successfully employed in revealing plant diversity and population structures among plant species, e.g. safflower (Talebi et al. 2018), Cactaceae (Abouseadaa et al. 2020), grape (Guo et al. 2012), Sugarcane (Que et al. 2014), wild relatives of wheat (Ghobadi et al. 2021; Pour-Aboughadareh et al. 2017, 2018, 2022; Khodaee et al. 2021), Cicer (Amirmoradi et al. 2012), peanut (Xiong et al. 2011), Damask Rose (Asadi and Shooshtari 2021), and durum-wheat (Etminan et al. 2016; Heidari et al. 2017).
Universal rice primer (URP) technique is another type of the DNA markers, designed based on repetitive sequences of the rice-species genetic materials (Kang et al. 2002). The URP primers are basically random, but their length and higher annealing temperature cause them to be more reproducible than many other PCR-based markers, e.g. ISSR and RAPD (Mehrparvar et al. 2012). The URP marker system is simple, powerful, and reliable in a wide range of genomes, especially in evaluating plant variations (Kang et al. 2002; Jana et al. 2005; Mehrparvar et al. 2012; Amirmoradi et al. 2012; Kandan et al. 2014; Kumar et al. 2018; Salahlou et al. 2019). Several different markers rely on plant variations at a genetic level, representing differences among flax populations. Molecular evaluation has been successfully undertaken on flax germplasm using different DNA marker-assisted approaches, e.g. ISSR (Uysal et al. 2010; Kumari et al. 2018; El Sayed et al. 2018; Mhiret and Heslop-Harrison 2018), AFLP (Everaert et al. 2001), SSR (Soto-Cerda et al. 2013; Yadav et al. 2018), IRAP (Mhiret and Heslop-Harrison 2018), and RAPD (Fu et al. 2003; Kumari et al. 2018).
The current research aimed at evaluating plant variations among 93 flax genotypes belonging to five Linum usitatissimum subspecies based on SCoT and URP markers.
Materials and methods
DNA extractions and plant materials
Ninety-three flax genotypes of four populations (35 accessions of usitatissimum subspecies, 27 accessions of elongatum subspecies, 12 accessions of mediterranian subspecies, and 19 accessions of an unknown subspecies) were considered for evaluation of plant variation via SCoT as well as URP molecular-based markers. A detailed list of accessions with gene bank codes is provided in Table 1. The seeds of 93 accessions were grown under greenhouse conditions. DNA of genomes was obtained through the CTAB protocol (Doyle and Doyle 1987) from fresh leaf tissue of a single plant per accession. DNA was extracted in high-quality form and checked through 0.8% (w/v) agarose-gel electrophoresing.
SCoT and URP assays
Based on sequences, suggested by Kang et al. (2002), 10 URP-primers of sequence-based materials were considered. Also, the SCoT primers were selected based on the primer developed by Collard and Mackill (2009) and Luo et al. (2010). In previous study on the genetic variation in Damask rose (Mostafavi et al. 2021), the primers of the two markers were used. Based on the results of the previous studies, the primers had a good ability to reveal genetic variation among the plants, therefore, they were used in the present study.
PCR reactions were carried out with a 20 μL solution of 10 μL master-mix PCR, 2 μL primer, 6 μL ddH2O (double distilled water), and 2μL DNA template (approximately 50 ng of template DNA). Amplifications of PCR were carried out in a Biorad thermal Cycler (Model: T-100) by denaturing (94 °C) for 5 min, and then 45 times denaturing (94 °C) within a timeframe of 45 s, primer-annealing (48–56 °C) (which varied depending on SCoT and URP primers) for 45 s, and extension at 72 °C within 90 s, leading to an ultimate extension-step (72 °C, 10 min), and then remaining at 10 °C. The products of PCR were separated on 1.5% agarose-based gel, marked with Safestaine-II, and digitally photographed using a gel documentation device.
Analyses of data
Amplified fragments were described as 1 or 0, if bands were present or absent, respectively, across the 93 genotypes. To compare the efficiency of markers, polymorphism information content (PIC) was measured according to Anderson et al. (1993). The marker index (MI) was designated based on a method proposed by Powell et al. (1996). Measurement of genetic distances relied on Jaccard’s dissimilarity coefficient matrix and cluster analysis was based on the neighbor joining (NJ) algorithm, using DARwin software ver. 6.0.13 (Perrier and Jacquemoud-Collet 2006). These were applied for principal-coordinate analysis (PCoA) in visualizing genetic correlations between 93 flax accessions. A fan dendrogram was constructed with MEGA software ver. 5.1 (Tamura et al. 2011) by combined (SCoT + URP) data. Molecular variation was assessed by GenAlEx v6.5 (Peakall and Smouse 2006) (AMOVA) to calculate genetic variations, observed alleles (Na), number of effective alleles (Ne), Nei’s gene diversity (He), Shannon’s information index (I), and polymorphic loci percentage. Population structures were analyzed via STRUCTURE (Pritchard et al. 2000). Clusters were counted through ten independently-running modes. Structural analyses were determined in an estimation of subpopulations (ΔK) through STRUCTURE HARVESTER (Earl and vonHoldt 2012).
Results
Markers diversity pattern
Twelve SCoT primers were tested, among which only 10 primers generated distinct, scorable band patterns. Ten SCoT primers generated 109 distinct banding patterns with a polymorphic appearance. Maximal and minimal counts of the polymorphic banding appeared through SCoT-5 (14 bands) as well as SCoT-10 (8 bands). SCoT primers had PIC values that occurred between 0.22 (SCoT-2) and 0.43 (SCoT-7 and SCoT-8) while averaging 0.35 for each primer. Moreover, the mean values of MI for each SCoT primer were 3.8 and ranged from 2.4 to 5.5 for primers SCoT-2 and SCoT-8, respectively (Table 2). Ten URP primers generated a total of 93 bands of polymorphic appearance. Amplified fragments were counted as 7 (by URP-1, URP-7, and URP-10) to 16 (by URP-4), averaging 9.3 bands for each primer. The minimal and maximal values of PIC occurred in URP-5 (0.29) and URP-7 (0.43), averaging 0.34 for each primer. MI values for these primers spanned between 2.2 (by URP-10) to 5.4 (by URP-4). The MI average for all URP primers was 3.2 (Table 2). In summarizing the pattern of bands, each population was described differently through combined data (URP + SCoT) (Fig. 1).
Genetic distances and grouping of the accessions
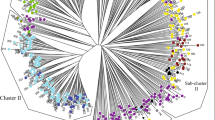
According to the dendrogram, from the cluster analysis of SCoT data (Fig. S1) and URP data (Fig. S2), the 93 flax accessions were classified into three and four clusters, respectively. Neither Scot nor URP marker data grouped the flax accessions exactly according to their sub-species. There were levels of admixture between the clusters. Based on the SCoT data, pairwise genetic distances between 93 flax genotypes had a range between 0.23 and 0.89, averaging at 0.65. Maximal genetic distance occurred among an accession of unknown origin (TN-97–95) and accession 1504, belonging to mediterranian population. Meanwhile, two accessions of the elongatum (LIN-939 and TN-382) showed minimal genetic distance. According to the URP data, pairwise genetic dissimilarity had a range between 0.12 and 0.91, averaging 0.58. Maximal genetic dissimilarity (0.91) happened among accession 2358 (of the usitatissimum population) and accession TN-97–10 (from an unknown population). Minimal genetic dissimilarity (0.12) occurred among two accessions from usitatissimum population (1172 and 1182). Moreover, for more reliable classification and a better explanation of genetic relationships, cluster analysis was performed using combined (SCoT + URP) data. A fan dendrogram resulted from cluster analysis on both markers (SCoT + URP) data, and classified all 93 accessions into four groups (Fig. 2). A clearer classification for accessions was made according to their genetic construct in a complimentary dendrogram, where the accessions of each population are shown in different colors (Fig. 3). The principal-coordinate-analysis made use of combined data in accordance with cluster analysis (Fig. 4).
Fan-dendrogram generated by neighbor joining based clustering of 93 Flax accessions using Jaccard’s similarity coefficients according to the combined (SCoT + URP) data. The accession codes, are presented in Table 1
Genetic diversity analysis
According to molecular variance (AMOVA), based on SCoT data, the variance between populations covered 11% of the total diversity, but intra-population diversity stood at 89%. Nonetheless, based on URP data, intra- and interspecies genetic diversity stood at 14% and 86%, respectively, which was almost similar to the results of SCoT data. Moreover, the AMOVA analysis according to the combined (SCoT + URP) data, revealed a major intra-population diversity (88%), whereas a mere 12% was attributed to inter-population diversity (Fig. 5). To compare the genetic diversity among populations, several attributes were considered for plant variation per population. Based on combined (SCoT + URP) data, effective alleles (Ne) were between 1.46 (for the unknown population) and 1.51 (for elongatum), averaging at 1.49. According to Shannon’s index (I), there was a variation between 0.40 and 0.44, with 0.42 as the average. Meanwhile, elongatum and unknown populations showed maximal and minimal (I) values, respectively. Nei’s genetic diversity (He) had an average (0.28) but differed between 0.26 (for the unknown population) and 0.30 (for elongatum). Besides, polymorphism loci (PPL) on average stood at 84%, whereas the highest and lowest values of PPL belonged to elongatum (90%) and mediterranian (75%) populations, respectively (Table 3).
Population structure and pattern of classification
In analyzing population structures, through SCoT data, maximal ΔK value occurred at K = 3, indicating that the 93 accessions should be categorized into 3 sub-populations (Fig. 6). Accordingly, membership by each sample had a threshold of ≥ 0.5 in each subpopulation. According to SCoT values, 22, 31, and 25 accessions formed the first, second, and third subpopulations, respectively, and fifteen accessions (16%) were not included in any of the subpopulations. Population structures were analyzed through URP values, showing a maximal peak of K = 4 and the occurrence of four subpopulations (Fig. 7). An optimal subpopulation count was revealed by maximal ΔK values via a structure harvester. Thresholds of membership per sample in each subpopulation were described as ≥ 0.5, along with the analysis of SCoT data. Bar plotting the models were clustered upon 28 genotypes (30.1%) and attributed to sub-population 1. Subpopulation 2 comprised 14 accessions (15.1%), including elongatum subspecies. Subpopulation 3 had 15 accessions (16.1%). Twenty-seven accessions (29%) were clustered into the fourth subpopulation. Nine accessions (9.7%) were not included in any of the subpopulations.
Discussion
The knowledge of genetic variation in flax and analysis of plant variation as intra- and inter-populations of flax is essential for sampling genetic resources when adopting suitable programs for the development of unprecedented varieties and for continuing germplasm conservation. In the case of linseed, different studies have been reported on evaluating plant variations and the relationships that exist between using different molecular markers and morphological characteristics (Fu et al. 2002; Cloutier et al. 2010; Uysal et al. 2010; Patial et al. 2019; Hoque et al. 2020; Ghobadi-Namin et al. 2021; Thakur et al. 2021). However, the current study failed to discover applications of gene-specific marker molecules for the evaluation of flax diversity. Here, twenty SCoT and URP primers were applied when analyzing plant variation to recognize different populations, comprising 93 flax accessions. SCoT marker as a gene-targeted marker is a reliable marker to assess genetic diversity in plant germplasms. These markers are diagnostic markers with many important advantages such as more reproducibility compared to arbitrary markers, low cost and convenient applications in lab settings, direct link with loci and alleles that are important, with acceptable polymorphism across plants (Collard and Mackill 2009; Talebi et al. 2018).
The number of polymorphic fragments generated by these 20 primers span between 7 and 16, averaging 10.1 bands for each primer (10.9 bands per SCoT primer and 9.3 bands per URP primer). This mean value in polymorphic fragments for each primer happened to be higher than those reported in other evaluations of plant variations within flax populations (Fu et al. 2002; Bhawna et al. 2017; Chandrawati et al. 2017). The higher number of polymorphic fragments could have resulted from largely different genotypes and suitable genome coverage of the markers used in this study. The average values of PIC for SCoT primers (0.35) and URP primers (0.34) were found to be higher than some other applications of SCoT and URP molecular markers to investigate the genetic diversity in different species including durum wheat (Heidari et al. 2017), Trigonella foenum-graecum (Daneshmand et al. 2017), Papaver bracteatum (Qaderi et al. 2019), and chickpea (Pakseresht et al. 2013). The PIC index depends on the frequency of alleles and reveals the discriminatory ability of the used primers to produce informative fragments. Regarding the maximum amount of PIC for dominant markers, (0.5), the average values of PIC in the present study confirmed the efficiency of SCoT and URP markers for further genetic diversity analyses in Flax and other plants.
In both SCoT and URP analysis, the investigated accessions were clustered relatively based on the sub-species with amounts of admixtures. The flax accessions based on SCoT and URP analyses were classified into three and four clusters, respectively. The URP marker data grouped the flax accessions exactly according to the four sub-species. This indicated that the URP marker was more useful to classify the flax accessions than the SCoT marker. In addition, cluster analysis using combined data revealed a clearer classification of accessions not only based on their sub-species but also approximately according to the place where they were collected. For example, as shown in Fig. 2, about 70% of the accessions collected from Iran were clustered together in a distinct group and this group was consisting of 18 accessions of which 17 accessions were collected from Iran. These results suggested significant associations between genetic and geographical distances. Also, these findings might be due to the presence of local adaptation. Similar results were reported in the study of Uysal et al. (2010) who reported significant associations not only between genetic and geographical distances but also between genetic distances and elevation differences in the assessment of genetic diversity of different species of flax using ISSR markers.
In this study, 93 Flax accessions showed a high level of genetic diversity (the average of dissimilarity calculated based on Jaccard’s coefficient were 0.65 and 0.58 using SCoT and URP markers data respectively). The wide range of genetic distances between accessions (0.12 to 0.91 and 0.23 to 0.89 based on URP and SCoT data, respectively) suggested diverse accessions of Flax. However, further complementary genetic analysis on these accessions is required to verify our findings. However, this result was not in agreement with Fu et al. (2002), so these authors in the assessment of genetic diversity in a set of Flax genotypes using RAPD markers, reported a low level of genetic diversity in evaluated cultivars and landraces of Flax. These contradictions could be due to different reasons such as the type of DNA marker system, genome coverage of selected primers, origins of genetic materials, and the number of investigated genotypes. The result of the genetic diversity analysis of Flax accessions collected from different regions of Turkey using ISSR markers (Uysal et al. 2010) was in agreement with our results. Besides, in the other study, the genetic variation in a set of 220 Flax (Linum usitatissimum L.) accessions was evaluated based on agro-morphological traits and revealed high genetic variability among Flax accessions (Saroha et al. 2022). It seems that the high level of genetic variation in this core collection was connected to the frequency of allelic variation which is affected by different climatic conditions (Nowak et al. 2020). The direct impacts of climate change on a decrease in diversity of those alleles controlling plant responses to climate changes have been reported in earlier studies (Jump and Penuelas 2005). From the genetic structure point of view, the high levels of genetic diversity provide ample opportunity to create new favorite varieties. Owing to the lack of genetic information on some crop plants like Flax, in comparison with other crops such as wheat, barley, and maize, the characterization and utilization of diverse germplasms of Flax is needed to enhance the genetic resources for breeding programs.
Furthermore, the results of the analysis of molecular variance (AMOVA) based on combined data, indicated 88% of the genetic variation resided within the populations and 12% was due to differences among the four populations, suggesting the presence of specific alleles within the populations (Shaygan et al. 2021). This finding represents that the studied populations consist of unique individuals and the accessions in each population have a wide genetic differentiation. In an assessment of genetic diversity in linseed (Linum usitatissimum L.) using SSR markers it was noticed that 79% of the total genetic variation was within the population and 21% was among the population (Chandrawati et al. 2017). These findings not only allow for the partitioning of the total molecular variation into within- and between populations components but also revealed good efficiency of URP and SCoT markers regarding genome coverage to detect the genetic diversity among and within populations. Higher distribution of genetic diversity within populations in comparison to among populations has been reported based on the AMOVA in different evaluations of genetic diversity using molecular markers (Feng et al. 2015, 2016; Chandrawati et al. 2017; Etminan et al. 2019; Ghobadi et al. 2021; Khodaee et al. 2021; Shaygan et al. 2021). The results obtained from the STRUCTURE analysis were in agreement with those obtained from the NJ clustering and PCoA. In a comparison of SCoT and URP markers, both molecular markers showed an acceptable efficiency to detect polymorphism and genetic relationships in Flax accessions, indicating these DNA markers can be used as reliable techniques for analyzing genetic diversity. SCoT primers showed good amplification and polymorphism with an acceptable average of PIC and MI values. This finding is in accordance with those of the previous studies that reported good efficiency of SCoT markers to study the genetic diversity in different plant species (Collard and Mackill 2009; Pakseresht et al. 2013; Etminan et al. 2016; Bhawna et al. 2017; Pour-Aboughadareh et al. 2018; Feng et al. 2018; Mishra et al. 2019; Gogoi et al. 2020; Zhao et al. 2020; Mostafavi et al. 2021). Although SCoT primers generated more polymorphic bands than URP primers, URP markers provided a better clustering pattern which was more similar to clustering based on combined data and in agreement with STRUCTURE analysis for evaluated samples. The efficiency of URP primers for fingerprinting and estimating genetic diversity in different species were reported in earlier studies (Jana et al. 2005; Amirmoradi et al. 2012; Salahlou et al. 2019).
Conclusion
It is believed that climate change has affected agricultural products and treats food security in the world. Renewing the crop varieties is one of the effective strategies to adapt to these changes. Providing diverse genetic resources is crucial to improve new varieties with desirable characteristics. Moreover, maintaining genetic variation is important for adaptation to various biotic and abiotic environmental stresses. The study of genetic diversity in plant germplasms is an essential requirement for plant breeding programs and the efficient maintenance of genetic reservoirs and gene pools. Flax is a multipurpose crop grown in many regions for several different uses. The present study was conducted to evaluate the genetic diversity of a set of 93 Flax accessions belonging to different subspecies. The results revealed a high level of genetic diversity in the collected Flax germplasm and clustering the accessions based on SCoT and URP markers, showed a significant association between genetic and geographical distances. Besides, different populations were consisting of diverse accessions with specific alleles, suggesting the potential of these genetic materials for future Flax genetic improvement programs. Our findings also confirmed that both URP and SCoT techniques are useful and reliable markers for the detection of genetic diversity and relationships in flax germplasm. Whereas molecular markers may or may not show a correlation with our desirable traits, the use of gene-targeting markers like SCoTs which are generated from the functional regions of the genomes would be more useful for the evaluation of genetic diversity and elucidation of population structure in breeding programs.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
References
Abouseadaa HH, Atia MAM, Younis IY, Issa MY, Ashour HA, Saleh I, Osman GH, Arif IA, Engy M (2020) Gene-targeted molecular phylogeny, phytochemical profiling, and antioxidant activity of nine species belonging to family Cactaceae Saudi. J Biol Sci 27:1649–1658
Amirmoradi B, Talebi R, Karami E (2012) Comparison of genetic variation and differentiation among annual Cicer species using start codon targeted (SCoT) polymorphism, DAMDPCR, and ISSR markers. Plant Syst Evol 298:1679–1688
Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME (1993) Optimizing parental selection for genetic linkage maps. Genome 36:181–186
Asadi A, Shooshtari L (2021) Assessment of somaclonal variation in micropropagation of Damask Rose (Rosa damascena Mill.) using molecular markers. MGJ 63:327–335
Bassett CM, Rodriguez-Leyva D, Pierce GN (2009) Experimental and clinical research findings on the cardiovascular benefits of consuming flaxseed. Appl Physiol Nutr Metab 34(5):965–974
Bhawna G, Abdin MZ, Arya L, Verma M (2017) Use of SCoT markers to assess the gene flow and population structure among two different populations of bottle gourd. Plant Gene 9:80–86
Chandrawati SN, Kumar R, Kumar S, Singh PK, Yadav VK, Ranade SA, Yadav HK (2017) Genetic diversity, population structure and association analysis in linseed (Linum usitatissimum L.). Physiol Mol Biol Plants 23:1–14
Cloutier S, Ragupathy R, Niu Z, Duguid S (2010) SSR- based linkage map of flax (Linum usitatissimum L.) and mapping of QTLs underlying fatty acid composition traits. Mol Breed 28:437–451
Collard BC, Mackill DJ (2009) Start codon targeted (SCoT) polymorphism: a simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol Biol Report 27(1):86
Daneshmand H, Etminan AR, Qaderi A (2017) Diversity evaluation of Trigonella foenum-graecum populations using DNA markers and phytochemical characteristics. JMP 63:16
de Lucena CM, de Lucena RFP, Costa GM, Carvalho TKN, da Silva Costa GG, da Nóbrega Alves RR, Pereira DD, da Silva Ribeiro JE, Alves CAB, Quirino ZGM, Nunes EN (2013) Use and knowledge of Cactaceae in Northeastern Brazil. J Ethnobiol Ehnomed 9(1):62
Diederichsen A, Richards K (2003) Cultivated flax and the genus Linum L.: taxonomy and germplasm conservation. In: Muir AD, Westcott ND (eds) Flax: the genus Linum. CRC Press, London, pp 22–54
Ding Q, Li J, Wang F, Zhang Y, Li H, Zhang J, Li, H, Zhang J, Gao J (2015) Characterization and development of EST-SSRs by deep transcriptome sequencing in Chinese cabbage (Brassica rapa L ssp pekinensis). Int J Genom 2015: 473028
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Earl DA, vonHoldt BM (2012) Structure Harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361
El Sayed AA, Ezzat SM, Mostafa SH, Zedan SZ, Abdel-Sattar E, El Tanbouly N (2018) Inter simple sequence repeat analysis of genetic diversity and relationship in four egyptian flaxseed genotypes. Pharm Res 10(2):166
Etminan A, Pour-Aboughadareh A, Mohammadi R, Ahmadi-Rad A, Noori A, Mahdavian Z, Moradi Z (2016) Applicability of start codon targeted (SCoT) and inter-simple sequence repeat (ISSR) markers for genetic diversity analysis in durum wheat genotypes. Biotechnol Biotechnol Equip 30(6):1075–1081
Etminan A, Pour-Aboughadareh A, Mehrabi AA, Shooshtari L, Ahmadi-Rad A, Moradkhani H (2019) Molecular characterization of the wild relatives of wheat using CAAT-box derived polymorphism. Plant Biosyst 153(3):398–405
Everaert I, De Riek J, De Loose M, Van Waes J, Van Bockstaele E (2001) Most similar variety grouping for distinctness evaluation of flax and linseed (Linum usitatissimum L.) varieties by means of AFLP and morphological data. Plant Var Seeds 14(2): 69–87
Feng S, Yang T, Liu Z, Chen LV, Hou N, Wang Y, Wei A (2015) Genetic diversity and relationships of wild and cultivated Zanthoxylum germplasms based on sequence-related amplified polymorphism (SRAP) markers. Genet Resour Crop Evol 62:1193–1204
Feng SG, He RF, Jiang MY, Lu JJ, Shen XX, Lu JJ, Liu JJ, Wang ZA, Wang HZ (2016) Genetic diversity and relationships of medicinal Chrysanthemum morifolium revealed by start codon targeted (SCoT) markers. Sci Hortic 201:118–123
Feng S, Zhu Y, Yu C, Jiao K, Jiang M, Lu J, Shen C, Ying Q, Wang H (2018) Development of species-specific SCAR markers, based on a SCoT analysis, to Authenticate Physalis (Solanaceae) species. Front Genet 9:192
Fu YB, Peterson G, Diederichsen A, Richards KW (2002) RAPD analysis of genetic relationships of seven flax species in the genus Linum L. Genet Resour Crop Evol 49:253–259
Fu YB, Rowland GG, Duguid SD, Richards KW (2003) RAPD analysis of 54 north American flax cultivars. Crop Sci 43(4):1510–1515
Ghobadi G, Etminan A, Mehrabi AM, Shooshtari L (2021) Molecular diversity analysis in hexaploid wheat (Triticum aestivum L.) and two Aegilops species (Aegilops crassa and Aegilops cylindrica) using CBDP and SCoT markers. J Genet Eng Biotechnol 19(1):56
Ghobadi-Namin L, Etminan A, Ghanavati F, Azizinezhad R, Abdollahi P (2021) Screening and selection of one hundred flax (Linum usitatissimum) accessions for yield production. J Nat Fibers. 19:7296–7304
Gogoi B, Wann SB, Saikia SP (2020) Comparative assessment of ISSR, RAPD, and SCoT markers for genetic diversity in Clerodendrum species of North East India. Mol Biol 47:7365–7377
Govindaraj M, Vetriventhan M, Srinivasan M (2015) Importance of genetic diversity assessment in crop plants and its recent advances: an overview of its analytical perspectives. Genet Res Int 2015:431487–431487
Goyal A, Sharma V, Upadhyay N, Gill S, Sihag M (2014) Flax and flaxseed oil: an ancient medicine & modern functional food. J Food Sci Technol 51(9):1633–1653
Guo DL, Zhang JY, Liu CH (2012) Genetic diversity in some grape varieties revealed by SCoT analyses. Mol Biol Rep 39:5307–5313
Heidari P, Etminan A, Azizinezhad R, Khosroshahli M (2017) Genomic variation studies in durum wheat (Triticum turgidum ssp. durum) using CBDP, SCoT and ISSR markers. Indian J Genet Plant. 77(3):379–386
Hoque A, Fiedler JD, Rahman M (2020) Genetic diversity analysis of a flax (Linum usitatissimum L.) global collection. BMC Genom 21:557
Jana TK, Singh NK, Koundal KR, Sharma TR (2005) Genetic differentiation of charcoal rot pathogen, Macrophomina phaseolina, into specific groups using URP-PCR. Can J Microbiol 51:159–164
Jump AS, Penuelas J (2005) Running to stand still: adaptation and the response of plants to rapid climate change. Ecol 8(9):1010–1020
Kandan A, Akhtar J, Singh B, Dixit D, Chand D, Roy A, Rajkumar S, Agarwal PC (2014) Molecular diversity of Bipolaris oryzae infecting Oryza sativa in India. Phytoparasitica 43(1):5–14
Kang HW, Park DS, Par YJ, You CH, Lee BM, Eun MY, Go SJ (2002) Fingerprinting of diverse genomes using PCR with universal rice primers generated from repetitive sequence of Korean weedy rice. Mol Cells 13:281–287
Khodaee L, Azizinezhad R, Etminan A, Khosroshahi M (2021) Assessment of genetic diversity among Iranian Aegilops triuncialis accessions using ISSR, SCoT, and CBDP markers. J Genet Eng Biotechnol 19(1):5
Kumar P, Akhtar J, Kandan A, Singh B, Kiran R, Nair K, Dubey S (2018) Efficacy of URP and ISSR markers to determine diversity of indigenous and exotic isolates of Curvularia lunata. Indian Phytopathol 71:235–242
Kumari S, Nirala PRB, Rani N, Prasad BD (2017) Selection criteria of linseed (Linum usitatissimum L.) genotypes for seed yield traits through correlation and path coefficient analysis. J Oilseeds Res. 34(3):171–174
Kumari A, Paul S, Sharma V (2018) Genetic diversity analysis using RAPD and ISSR markers revealed discrete genetic makeup in relation to fibre and oil content in Linum usitatissimum L. genotypes. Nucl 61(1):45–53
Kurt O, Bozkurt D (2006) Effect of temperature and photoperiod on seedling emergence of flax (Linum usitatissimum L.). J Agron 5:541–545
Luo C, He XH, Chen H, Ou SJ, Gao MP (2010) Analysis of diversity and relationships among mango cultivars using Start Codon Targeted (SCoT) markers. Biochem System Ecol 38(6):1176–1184
Mehrparvar M, Mohammadi Goltapeh E, Safaie N (2012) Evaluation of genetic diversity of Iranian Lecanicillium fungicola isolates using URP marker. J Crop Prot 1(3):229–238
Mhiret WN, Heslop-Harrison JS (2018) Biodiversity in Ethiopian linseed (Linum usitatissimum L.): molecular characterization of landraces and some wild species. Genet Resour Crop Evol 65(6):1603–1614
Mishra SK, Nag A, Naik A, Rath SC, Gupta K, Gupta AK, Panigrahi J (2019) Characterization of host response to bruchids (Callosobruchus chinensis and C. maculatus) in 39 genotypes belongs to 12 Cajanus spp. and assessment of molecular diversity inter se. J Stored Prod Res 81:76–90
Mohammadi SA, Prasanna BM (2003) Analysis of genetic diversity in crop plants-salient statistical tools and considerations. Crop Sci 43(4):1235–1248
Mostafavi A, Omidi M, Azizinezhad R, Etminan A, Naghdi BH (2021) Genetic diversity analysis in a mini core collection of Damask rose (Rosa damascena Mill.) germplasm from Iran using URP and SCoT markers. J Genet Eng Biotechnol 19:144
Mulpuri S, Muddanuru T, Francis G (2013) Start codon targeted (SCoT) polymorphism in toxic and non-toxic accessions of Jatropha curcas L. and development of a codominant SCAR marker. Plant Sci 207:117–127
Nowak JL, Okon S, Wieremczuk A (2020) Molecular diversity analysis of genotypes from four Aegilops species based on retrotransposon–microsatellite amplifed polymorphism (REMAP) markers. Cereal Res Commun 49(1):37–44
Pakseresht F, Talebi R, Karami E (2013) Comparative assessment of ISSR, DAMD and SCoT markers for evaluation of genetic diversity and conservation of landrace chickpea (Cicer arietinum L.) genotypes collected from north-west of Iran. Physiol Mol Biol Plants 19(4):563–574
Patial R, Paul S, Sharma D, Sood VK, Kumar N (2019) Morphological characterization and genetic diversity of linseed (Linum usitatissimum L.). J Oilseeds Res. 36(1):8–16
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6(1):288–295
Perrier X, Jacquemoud-Collet J (2006) DARwin software at available at: http://darwin.cirad.fr/
Pour-Aboughadareh A, Ahmadi J, Mehrabi AA, Etminan A, Moghaddam M (2017) Assessment of genetic diversity among Iranian Triticum germplasm using agro-morphological traits and start codon targeted (SCoT) markers. Cereal Res Commun 45(4):574–586
Pour-Aboughadareh A, Ahmadi J, Mehrabi AA, Etminan A, Moghaddam M (2018) Insight into the genetic variability analysis and relationships among some Aegilops and Triticum species, as genome progenitors of bread wheat, using SCoT markers. Plant Biosyst 152:694–703
Pour-Aboughadareh A, Poczai P, Etminan A, Jadidi O, Kianersi F, Shooshtari L (2022) An analysis of genetic variability and population structure in wheat germplasm using microsatellite and gene-based markers. Plants 11:1205
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Qaderi A, Omidi M, Pour-Aboughadareh A, Poczai P, Shaghani J, Mehrafarin A, Nohooji M, Etminan A (2019) Molecular diversity and phytochemical variability in the Iranian poppy (Papaver bracteatum Lindl.): a baseline for conservation and utilization in future breeding programmes. Ind Crops Prod 130:237–247
Que YX, Pan YB, Lu YH, Yang C, Yang Y, Huang N, Xu L (2014) Genetic analysis of diversity within a Chinese local sugarcane germplasm based on start codon targeted polymorphism. Bio Med Res Int 2014:1–10
Salahlou R, Safaie N, Shamsbakhsh M (2019) Using ISSR and URP-PCR markers in detecting genetic diversity among Macrophomina phaseolina isolates of sesame in Iran. J Crop Prot 8(3):293–309
Saroha A, Pal D, Kaur V, Kumar Bartwal A, Aravind J, Radhamani J, Rajkumar S, Kumar R, Sunil S, Gomashe SA, Wankhede DP (2022) Agro-morphological variability and genetic diversity in linseed (Linum usitatissimum L.) germplasm accessions with emphasis on flowering and maturity time. Genet Resour Crop Evol 69:315–333
Shaygan N, Etminan A, Majidi Hervan E, Azizinezhad R, Mohammadi R (2021) The study of genetic diversity in a minicore collection of durum wheat genotypes using agro-morphological traits and molecular markers. Cereal Res Commun 49(1):141–147
Soto-Cerda BJ, Diederichsen A, Ragupathy R, Cloutier S (2013) Genetic characterization of a core collection of flax (Linum usitatissimum L.) suitable for association mapping studies and evidence of divergent selection between fiber and linseed types. BMC Plant Biol 13(1):78
Talebi R, Nosrati S, Etminan A, Naji AM (2018) Genetic diversity and population structure analysis of landrace and improved safflower (Cartamus tinctorious L.) germplasm using arbitrary functional gene-based molecular markers. Biotechnol Biotechnol Equip 32(5):1183–1194
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thakur R, Paul S, Thakur G, Kumar A, Dogra R (2021) Assessment of genetic diversity among linseed (Linum usitatissimum L.) germplasm based on morphological traits. J Pharmacogn Phytochem 9(6):1785–1790
Uysal H, Fu YB, Kurt O, Peterson GW, Diederichsen A, Kusters P (2010) Genetic diversity of cultivated flax (Linum usitatissimum L.) and its wild progenitor pale flax (Linum bienne Mill.) as revealed by ISSR markers. Genet Resour Crop Evol 57:1109–1119
Vinu V, Singh N, Vasudev S, Kumar Yadava D, Kumar S, Naresh S, Ramachandra Bhat S, Vinod PK (2013) Assessment of genetic diversity in Brassica juncea (Brassicaceae) genotypes using phenotypic differences and SSR markers. Rev Biol Trop 61(4):1919–1934
Xiong FQ, Zhong RC, Han ZQ, Jiang J, He LQ, Zhuang WJ, Tang R (2011) Start codon targeted polymorphism for evaluation of functional genetic variation and relationships in cultivated peanut (Arachis hypogaea L.) genotypes. Mol Biol Rep 38:3487–3494
Yadav HK, Chandrawati D, Singh N, Kumar R, Kumar S, Ranade SA (2018) Agromorphological traits and microsatellite markers based genetic diversity in Indian genotypes of linseed (Linum usitatissimum L.). J Agric Sci Tech 19:707–718
Zhao X, Zou G, Zhao J, Hu L, Lan Y, He J (2020) Genetic relationships and diversity among populations of Paris polyphylla assessed using SCoT and SRAP markers. Physiol Mol Biol Plants 26:1281–1293
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
AE, LG and FG conceived the research idea and designed the experiments. LG and AE performed the experiments and analyzed the data. LG, RA and PA wrote the manuscript. AE and RA revised and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ghobadi-Namin, L., Etminan, A., Ghanavati, F. et al. Analysis of genetic diversity and population structure in a mini core collection of flax (Linum usitatissimum L.) based on URP and SCoT markers. J. Crop Sci. Biotechnol. 27, 43–55 (2024). https://doi.org/10.1007/s12892-023-00210-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12892-023-00210-7