Abstract
Three molecular markers, including start codon targeted (SCoT) polymorphism, directed amplification of minisatellite-region DNA polymerase chain reaction (DAMD-PCR), and inter simple sequence repeat (ISSR) markers, were compared in terms of their informativeness and efficiency for analysis of genetic relationships among 38 accessions of eight annual Cicer species. The results were as follows: (1) the highest level of detected polymorphism was observed for all three marker types; (2) the rate of diversity for the three marker techniques was approximately equal, and the correlation coefficients of similarity were statistically significant for all three marker systems; (3) the three molecular markers showed relatively similar phylogenetic grouping for examined species. Diversity analysis showed that Cicer reticulatum is the closest wild species to the cultivated chickpea, and this finding supports the hypothesis that C. reticulatum is the most probable progenitor of the cultivated species. C. bijugum, C. judaicum, and C. pinnatifidum were clustered together, and in other clusters C. yamashitae and C. cuneatum were grouped close together. To our knowledge, this is the first detailed comparison of performance among two targeted DNA region molecular markers (SCoT and DAMD-PCR) and the ISSR technique on a set of samples of Cicer. The results provide guidance for future efficient use of these molecular methods in genetic analysis of Cicer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The phylogeny of the genus Cicer has attracted considerable attention because it harbors the third most important grain legume crop worldwide—the chickpea. Chickpea (C. arietinum L.) is the only cultivated species belonging to the Cicer genus, which is classified in Fabaceae, tribe Cicereae Alef (van der Maesen 1987). The genus Cicer contains 42 wild species including nine annuals and 33 perennials with chromosome number 2n = 2x = 16 in almost all the species (Labdi et al. 1996). Chickpea is an important self-pollinated grain legume crop, grown mainly in West Asia, North Africa, and the Indian Subcontinent, where it is a basic component of the human diet. Vavilov (1926) was the first to identify areas with similar physiographic characteristics with maximum variability for the major cultivated species. He recognized the Near Eastern, Central Asian, Indian, and Mediterranean regions as the probable centers of origin for chickpea. C. arietinum is an economically important crop in Iran, India, the Middle East, North Africa, and Ethiopia, and is the third most important pulse crop in the world next to Phaseolus vulgaris and Pisum sativum. However, the average annual yield worldwide (0.78 ton/ha) is considered to be somewhat lower than its potential yield (Singh et al. 1994; Sudupak et al. 2002). Therefore, many chickpea breeding programs are focused on improving the genetic potential both to increase yield and to provide protection against abiotic and biotic stresses. It has been recognized that interspecific hybridization will increase the variation and can be useful for plant breeding purposes in a recalcitrant crop such as chickpea (Singh et al. 1994; Van Rheenen et al. 1993). This is further exemplified by the utilization of two related wild species, C. reticulatum and C. echinospermum, in plant breeding programs (Singh and Ocampo 1993, 1997; Singh et al. 1994). Wild relatives of chickpea were reported to bear traits conferring resistance to a number of biotic and abiotic stresses and have potential to improve yield (Singh et al. 1994; Muehlbauer et al. 1994; Singh and Ocampo 1997; Akem et al. 2000). Investigation of genetic variation and relationships among accessions collected from diverse geographic regions where the closest wild relatives exist and were presumed to be first domesticated is critical for genetic improvement of the chickpea. Most of the interspecific relationship studies in Cicer have been carried out using plant morphology (Robertson et al. 1997), karyotype (Ocampo et al. 1992; Tayyar et al. 1994), crossability data (Ladizinsky and Adler 1976; Pundir and Vander Maesen 1983; Ahmad et al. 1987), restriction fragment length polymorphism (RFLP) (Patil et al. 1995), seed storage protein analyses (Vairinhos and Murray 1983; Ahmad and Slinkard 1992), allozyme markers (Kazan and Muehlbauer 1991), amplified fragment length polymorphism (AFLP) markers (Nguyen et al. 2004), inter simple sequence repeat (ISSR) markers (Sudupak 2004), and more recently random amplified polymorphic DNA (RAPD) markers (Talebi et al. 2009). In addition, allelic variation at a microsatellite locus [(TAA)n] (Udupa et al. 1999) and also in Ty1-copia-like retrotransposon sequence (Sant et al. 2000) have been utilized to study diversity in Cicer. In recent years, many new alternative and promising marker techniques have emerged. These techniques include start codon targeted (SCoT) polymorphism (Collard and Mackill 2009) and directed amplification of minisatellite-region DNA polymerase chain reaction (DAMD-PCR) to direct the amplification of tandemly repeated region of a genome (Karaca et al. 2002; Kang et al. 2002). However, to the best of our knowledge, there has been no application of SCoT and DAMD-PCR in Cicer. Coupled with the rapid growth of genomics research, there has been a trend away from random DNA markers towards gene-targeted markers (Andersen and Lubberstedt 2003; Gupta and Rustgi 2004). Genome sequence data offers enormous potential for the development of new markers in diverse plant species (Collard and Mackill 2009). The present study was carried out to test the utility of the SCoT and DAMD-PCR markers in comparison with ISSR markers to determine intra- and interspecies genetic diversity and relationships among accessions representing eight annual Cicer species.
Materials and methods
Plant materials
Thirty-eight accessions, representing eight annual Cicer species, originating from the main Cicer centers of the world diversity, were obtained from the Australian Temperate Field Crops Collection (ATFCC) at the Victorian Institute for Dryland Agriculture (VIDA), Horsham, Australia and International Center for Agricultural Research in the Dry Areas (ICARDA), Alepo, Syria (Table 1).
Genomic DNA extraction
DNA was extracted from 2 g of young leaves collected from 10-day-old seedling plants of each accession using the cetyltrimethylammonium bromide (CTAB) method according to Lassner et al. (1989) with the modification described by Torres et al. (1993). The DNA final concentration was determined by agarose-gel electrophoresis using a known concentration of uncut λ DNA as standard.
ISSR and DAMD-PCR
For ISSR analysis, a set of 15 primers representing di-, tri-, tetra-, and pentamer repeats (UBC set #9) was procured from the Biotechnology Laboratory, University of British Columbia, Canada. Following the optimization of polymerase chain reaction (PCR) conditions and prescreening of the first 15 primers on a sample set that included each species, 10 primers providing clear and informative amplicon profiles across species were selected to survey ISSR variation in the accessions listed in Table 2. PCR reactions were composed as described previously by Rajesh et al. (2002). DAMD-PCR markers [originally derived from the repeat elements of weed rice (Kang et al. 2002)] were used in this study. Of 10 primers screened, 6 with GC content of 50–60 % were selected to generate the DNA fingerprint profiles of all the accessions at 48 °C annealing temperature. PCR reactions were performed in a volume of 20 μl in Eppendorf thermocyclers. PCR products were separated on 1.5 % agarose gels, stained with ethidium bromide, and scored for presence or absence of bands.
SCoT PCR
Start codon targeted primers were designed from consensus sequences derived from the studies by Collard and Mackill (2009). For primer design, the ATG codon (+1, +2, and +3), G at position +4, and A, C, and C at positions +7, +8, and +9, respectively, were fixed (Table 2). All primers were 18-mer and ranged in GC content between 50 and 72 % (Table 2). Primers were checked for dimers and hairpin loops. Using the FAST PCR program (Kalendar 2007). PCR was optimized for 10 primers as described previously by Collard and Mackill (2009). PCR reactions were performed in volume of 20 μl in Eppendorf thermocyclers. PCR products were separated on 1.2 % agarose gels, stained with ethidium bromide, and scored for presence or absence of bands.
Data analysis
DNA bands obtained with all the markers were scored visually for presence (1) or absence (0) of bands for all 38 accessions. DARwin version 5.0 was used for calculating pairwise genetic distances and for constructing the dissimilarity matrix (Perrier et al. 2003). The dissimilarity matrix thus obtained was subjected to cluster analysis using the unweighted neighbor-joining analyses (UNJ) (Gascuel 1997), followed by bootstrap analysis with 1,000 permutations to obtain a dendrogram for all 38 accessions (Perrier et al. 2003). Mantel statistic was used to compare the similarity matrices as well as the dendrograms produced by the ISSR, URP, and SCoT techniques. All these procedures were performed by appropriate routines in NTSYSpc version 2.0 (Rohlf 1997). Polymorphic information content (PIC) values were calculated for each ISSR primers according to the formula: \( {\text{PIC}} = 1 - \sum {(P_{ij} )}^{2} , \) where P ij is the frequency of the ith pattern revealed by the jth primer summed across all patterns revealed by the primers (Botstein et al. 1980). Partitioning of variation within and among the groups by each marker system was achieved by analysis of molecular variance (AMOVA) using Arlequin version 2000 software as described by Excoffier et al. (1992). The Mantel test of significance (Mantel 1967) was also used to compare each pair of similarity matrices produced.
Results
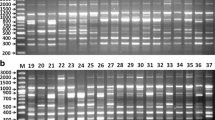
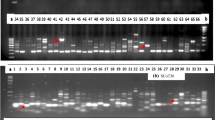
DNA fingerprint database has been reported using the three different PCR-based molecular marker (ISSR, DAMD-PCR, and SCoT) systems for 38 Cicer accessions belonged to eight annual species. Our results indicated that primers which were obtained from the different regions of genomic DNA successfully amplified accession template DNAs (Fig. 1). All three molecular markers used in this study were able to distinguish and identify each of 38 accessions to referred species. Salient features of the fingerprint database obtained using the different markers are given below.
ISSR analysis
A total of 81 bands were detected using seven ISSR markers, of which 78 were polymorphic (Table 3). ISSR DNA bands varied between 15 (UBC828) and 6 (UBC874), with an average of 11.5 ± 0.9 per primer. Percent polymorphism ranged from 85.7 % to as high as 100 % with average polymorphism of 96.75 % across all accessions. ISSR markers showed relatively high level of polymorphism in the examined germplasm; the PIC value for ISSRs was calculated in the range of 0.29–0.49 with an average of 0.4 across the genotypes assayed (Table 3). A significant correlation (r = 0.938; P < 0.01) was observed between the total number of bands and the number of polymorphic bands. As seen in Table 3, dinucleotides (AG)8, (TG)8, (TC)8, and (CA)8 with single-nucleotide anchor T/A at the 3′ end were found to be the most polymorphic compared with all others.
DAMD-PCR analysis
A PCR-based approach involving the DAMD with seven minisatellite core sequences as primers was used to analyze diversity in 38 annual Cicer accessions. Analysis indicated that the PCR profile and the optimized chemical concentrations resulted in reproducible and reliable DNA amplification (Fig. 1). In the accessions of Cicer species the number of amplified DAMD-PCR products varied from 11 to 15 fragments depending on the primers used. A total of 92 bands were scored, all of which were polymorphic (Table 3). PIC values ranged from 0.23 to 0.60, with an average value of 0.39 per locus. Based on the independent replications of DAMD-PCR, we observed that reproducible DNA markers were amplified and also noted that all DAMD-PCR primers used in this study produced RAPD-like results, but the numbers of bands were sharp and clear. The relatively high PCR stringencies in DAMD-PCR application effectively limited the PCR artifacts which commonly occur in RAPDs (Karaca et al. 2002; Ince et al. 2009).
SCoT analysis
A total of 112 bands were detected among 38 accessions belonging to eight annual Cicer species using nine SCoT markers, of which 109 were polymorphic (Table 3). The number of bands ranged from 17 (SCoT13) to 7 (SCoT1) with an average of 12.4 per primer. The overall size of amplified products ranged from 220 to 2,250 bp. Percent polymorphism ranged from 86.6 % to as high as 100 % with average polymorphism of 97 % across all accessions. PIC values ranged from 0.39 to 0.78, with an average value of 51.7 per primer. Evaluation using a representative set of Cicer species has indicated that SCoT primers generate a DNA fingerprint similar to those generated by RAPD markers, but the bands were sharp, clear, and 100 % polymorphic (Fig. 1).
Correlation between the similarity values measured using the three marker systems
The values of the Mantel test showed a positive correlation between the three marker types. The correlation coefficient (r) was 0.629 between ISSR and DAMD-PCR, 0.650 between ISSR and SCoT (significant, P > 0.01), and 0.50 (significant, P > 0.05) between DAMD-PCR and SCoT (Table 4). The AMOVA showed apparent differences in partitioning of variation within and between species accomplished by the three different marker types (Table 5). All three marker types showed greater variance between species than within species. SCoT and ISSR markers showed relatively greater variance between the species, while DAMD-PCR markers showed greater variance among species compared with other marker types (Table 5).
Interrelationships among species
All three marker types showed high similarity in dendrogram topologies. The dendrogram constructed by DARwin (version 5.0) software using an unweighted neighbor-joining method. Cluster analysis using ISSR data grouped genotypes into five distinct clusters (Fig. 2). Group I included C. arietinum, C. reticulatum, and C. yamashitae, group II included C. echinospermum, and group III included C. pinnatifidum. C. bijugum and C. cuneatum were grouped in cluster IV. Group V had C. judacium and C. cuneatum. Generally, all accessions were positioned into species clusters. The dendrogram constructed by SCoT markers demonstrated that four main groupings existed in these accessions (Fig. 3). Group I [the primary and secondary crossability groups documented by Ladizinsky and Adler (1976)] clustered C. arietinum and C. reticulatum close together, and C. yamashitae and C. pinnatifidum also grouped in this cluster. Group II and group III included accessions belonging to C. echinospermum and C. judcium, respectively. In the fourth cluster C. bijugum and C. cuneatum were grouped close together. In cluster analysis based on DAMD-PCR markers, all accessions that belonged to C. arietinum and C. echinospermum grouped in cluster I, with C. judacium, C. bijugum, and C. cuneatum in the second cluster (Fig. 4). Also C. yamashitae and C. pinnatifidum were grouped in one cluster very close together (Fig. 4).
Discussion
For genetic analysis, it is of utmost importance to know which type of markers and how many of them truly represent variation in the entire genome and should be used to derive reliable estimates of diversity. There have been a number of efforts to transfer agro-economically important genes from wild species into cultivated species through conventional breeding practices; For example, interspecific hybridizations were performed to introgress perennial germplasm into cultivated alfalfa (McCoy and Echt 1993) and a nematode resistance gene in peanut (Garcia et al. 1996). However, similar attempts were not successful to develop hybrids between C. arietinum and any wild species of Cicer except C. reticulatum and C. echinospermum. A few efforts using plant tissue culture techniques such as embryo rescue to develop hybrids between C. arietinum and C. pinnatifidum were reported (Anonymous 1995). High hybrid vigor was obtained in F1 and F2 generations, when C. arietinum was crossed with C. reticulatum and C. echinospermum (Singh and Ocampo 1997), confirming the potential of increasing the chickpea yield using interspecific hybridization. However, knowledge of genetic relationships among various wild species is necessary for successful and efficient exploitation of genetic diversity present in the wild species, and such information is very poorly available in the genus Cicer, especially using molecular markers. In this study, we compared the marker datasets produced using three different marker systems, namely ISSR, DAMD-PCR, and SCoT, to define genetic relationships within a set of 38 genotypes representing eight annual Cicer species, and to determine whether these marker systems can be effectively used and/or complement identification of Cicer species. This technique was more informative than previously used biochemical and molecular methods to study variation and genetic relationships in Cicer such as isozymes, storage proteins (Ahmad et al. 1992; Labdi et al. 1996), and RAPD markers (Sudupak et al. 2002; Talebi et al. 2009). All three molecular markers detected abundant polymorphisms at both inter- and intraspecies level, implying that all of them could be applied to germplasm identification and genetic diversity assessment in the genus Cicer. Results of the three types of markers in this study corroborated the first and second crossability groups of Ladizinsky and Adler (1976), and grouping in the first crossability group revealed that C. reticulatum is the closest wild species to C. arietinum while C. echinospermum is relatively distant from both, consistent with allozyme- (Ahmad et al. 1992), RAPD- (Talebi et al. 2009; Sudupak et al. 2002; Iruela et al. 2002), and AFLP-based groupings (Nguyen et al. 2004). Groupings of the accessions of the second crossability group species indicated that C. pinnatifidum is closer to C. judaicum, and C. bijugum was the next closer species to this group, consistent with results obtained with different markers (Sudupak et al. 2002; Rajesh et al. 2002; Nguyen et al. 2004). It is interesting to note that all three (ISSR, DAMD-PCR, and SCoT) datasets showed high levels of correlation (Table 4). This is not surprising as these markers are known to target different genomic fractions involving repeat and/or unique sequences, which may have been differentially evolved or preserved in due course of natural or human selection. The levels of genetic variation detected by these three types of markers within and between Cicer species were relatively higher than the amount computed from RAPD and AFLP markers (Sudupak et al. 2002; Nguyen et al. 2004). This can be attributed to the hypervariable nature of these markers, which are expected to reveal higher levels of variation. Consistent with our previous analyses of RAPDs, C. pinnatifidum was the most polymorphic species followed by C. reticulatum, while C. arietinum had the least amount of RAPD variation. Results obtained from these three marker systems also confirm that chickpea has a narrow genetic base. Previous studies have already shown that studies of DNA polymorphisms across multiple regions of the genome enable discrimination between closely related cultivars or accessions (Saini et al. 2004; Aruna et al. 2008). Although a larger number of species and accessions are needed to identify species-specific DAMD-PCR and SCoT, and genetic relationships, our studies indicated that accessions represented their corresponding species in Cicer. Our results demonstrate that high genetic diversity exists between the investigated accessions belonging to eight annual species. As the examined species are diploid and have relatively similar pedigree, this range of diversity is interesting. Estimates of genetic diversity were nearly the same using SCoT, ISSR, and DAMD-PCR markers. Although the level of diversity for the three marker techniques was approximately equal, we anticipate that the source of detected diversity is different, as each technique targets different regions of the genome. For precise phylogeny and evolutionary analysis of Cicer, studies with more perennial and annual species and with a large number of accessions of each species are required. However, the scarcity of seeds of Cicer species and their poor growing ability at various locations are the major limitations to carrying out this exercise. In conclusion, in this study we provide additional data for identification of accessions and species in Cicer in addition to previous analyses of morphological characteristics and RAPD, AFLP, and isozyme studies. We have demonstrated that DAMD-PCR and SCoT markers can be used as reliable techniques for detecting levels of DNA polymorphism and genetic relationship in Cicer.
References
Ahmad F, Slinkard AE (1992) Genetic relationships in the genus Cicer L. as revealed by polyacrylamide gel electrophoresis of seed storage proteins. Theor Appl Genet 84:688–692
Ahmad F, Slinkard AE, Scoler GJ (1987) The cytogenetic relationship between Cicer judaicum Boiss and Cicer chroassannicum (Bge). M Pop Genome 29:883–886
Ahmad F, Gaur PM, Slinkard AE (1992) Isozyme polymorphism and phylogenetic interpretations in the genus Cicer L. Theor Appl Genet 83:620–627
Akem C, Caccarelli S, Erkine W, Lenne J (2000) Using genetic diversity for disease resistance in agricultural production. Outlook Agric 29:25–30
Andersen JR, Lubberstedt T (2003) Functional markers in plants. Trends Plant Sci 8:554–560
Anonymous (1995) Kabuli chickpea improvement. Annu rep germplasm program: legumes. ICARDA, Aleppo, pp 59–132
Aruna R, Rao DM, Sirvaramakrishnan S, Reddy LJ, Bramel P, Upadhaya H (2008) Efficiency of three DNA markers in revealing genetic variation among wild Cajanus species. Plant Genet Resour 7(2):113–121
Botstein B, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Ann J Hum Genet 32:314–331
Collard BCY, Mackill DJ (2009) Start Codon Targeted (SCoT) polymorphism: a simple novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol Biol 27:86–93
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: applications to human mitochondrial DNA restriction data. Genetics 131:479–491
Garcia GM, Stalker HT, Shroeder E, Kochert G (1996) Identification of RAPD, SCAR and RFLP markers tightly linked to nematode resistance genes introgressed from Arachis cardenasii into Arachis hypogaea. Genome 39:836–845
Gascuel O (1997) Concerning the NJ algorithm and its unweighted version, UNJ. In: Mirkin B, McMorris FR, Roberts FS, Rzhetsky A (eds) Mathematical hierarchies and biology. DIMACS series in discrete mathematics and theoretical computer science. Providence, RI. Amer Math Soc 149–170
Gupta PK, Rustgi S (2004) Molecular markers from the transcribed/expressed region of the genome in higher plants. Funct Integr Geonomics 4:139–162
Iruela M, Rubio J, Cubero JI, Gil J, Milan T (2002) Phylogenetic analysis in the genus Cicer and cultivated chickpea using RAPD and ISSR markers. Theor Appl Genet 104:643–651
Kalendar R (2007) FastPCR: a PCR primer design and repeat sequence searching software with additional tools for the manipulation and analysis of DNA and protein. Available at www.biocenter
Kang HW, Park DS, Go SJ, Eun MY (2002) Fingerprinting of diverse genomes using PCR with universal rice primers generated from repetitive sequence of Korean weedy rice. Mol Cells 13:281–287
Karaca M, Saha S, Zipf A, Jenkins JN, Lang DJ (2002) Genetic diversity among forage bermudagrass (Cynodon spp.): evidence from chloroplast and nuclear DNA fingerprinting. Crop Sci 42:2118–2127
Kazan K, Muehlbauer FJ (1991) Allozyme variation and phylogeny in annual species of Cicer (Leguminosae). Plant Syst Evol 175:11–21
Labdi M, Robertson LD, Singh KB, Charrier A (1996) A genetic diversity and phylogenetic relationships among the annual Cicer species as revealed by isozyme polymorphism. Euphytica 88:181–188
Ladizinsky and Adler (1976) Genetic relationships among the annual species of Cicer L. Theor Appl Genet 48:197–203
Lassner MW, Peterson P, Yoder JI (1989) Simultaneous amplification of multiple DNA fragments by polymerase chain reaction in the analysis of transgenic plants and their progeny. Plant Mol Biol Rep 7:116–128
Mantel NA (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
McCoy TJ, Echt CS (1993) Potential of trispecies bridge crosses and random amplified polymorphic DNA markers for introgression of Medicago daghestanica and Medicago pironae germplasm into alfalfa (Medicago sativa). Genome 36:594–601
Muehlbauer FJ, Kaiser WJ, Simon CJ (1994) Potential for wild species in cool season food legume breeding. Euphytica 73:109–114
Nguyen TT, Taylor PWJ, Redden RJ, Ford R (2004) Genetic diversity estimates in Cicer using AFLP analysis. Plant Breed 123:173–179
Ocampo B, Venora G, Errico A, Singh KB, Saccardo F (1992) Karyotype analysis in the genus Cicer. J Genet Breed 46:229–240
Patil P, Vrinten PL, Scoles GJ, Slinkard AE (1995) Variation in the ribosomal RNA units of the genera Lens and Cicer. Euphytica 83:33–42
Perrier X, Flori A, Bonnot F (2003) Data analysis methods. In: Hamon P, Seguin M, Perrier X, Glaszmann JC (eds) Genetic diversity of cultivated tropical plants. Science, Enfield, pp 43–76
Pundir RPS, Vander Maesen LJG (1983) Interspecific hybridization in Cicer. Int Chickpea Newsl 8:4–5
Rajesh PN, Sant VJ, Gupta VS, Muehlbauer FJ, Ranjekar PK (2002) Genetic relationships among annual and perennial wild species of Cicer using inter simple sequence repeat (ISSR) polymorphism. Euphytica 129:15–23
Robertson LD, Ocampo B, Singh KB (1997) Morphological variation in wild annual Cicer species in comparison to the cultigens. Euphytica 95:309–319
Rohlf FJ (1997) NTSYS-Pc. Numerical taxonomy and multivariate analysis system version 2.1. Exeter Software, Setauket, p 7
Saini N, Jain N, Jain RK (2004) Assessment of genetic diversity within and among Basmati and non-Basmati rice varieties using AFLP, ISSR and SSR markers. Euphytica 140:133–146
Sant VJ, Sainani MN, Sami-subbu R, Ranjekar PK, Gupta VS (2000) Ty1-copia retrotransposon-like elements in chickpea genome: their identification, distribution and use for diversity analysis. Gene 257:157–166
Singh KB, Ocampo B (1993) Interspecific hybridization in annual Cicer species. J Genet Breed 47:199–204
Singh KB, Ocampo B (1997) Exploitation of wild Cicer species for yield improvement in chickpea. Theor Appl Genet 95:418–423
Singh KB, Malhotra RS, Halila MH, Knights EJ, Verma MM (1994) Current status and future strategy in breeding chickpea for resistance to biotic and abiotic stresses. Euphytica 73:137–149
Sudupak MA (2004) Inter and intra-species Inter Simple Sequence Repeat (ISSR) variations in the genus Cicer. Euphytica 135:229–238
Sudupak MA, Akkaya MS, Kence A (2002) Analysis of genetic relationships among perennial and annual Cicer species growing in turkey using RAPD markers. Theor Appl Genet 105:1220–1228
Talebi R, Jelodar NB, Mardi M, Fayaz F, Furman BJ, Bagheri NA (2009) Phylogenetic diversity relationship among annual Cicer species using random amplified polymorphic DNA. Gener Appl Plant Physiol 35:3–12
Tayyar RI, Lukaszewiski AJ, Waines JG (1994) Chromosome banding patterns in the annual species of Cicer. Genome 37:656–663
Torres AM, Weeden NF, Martin A (1993) Linkage among isozyme, RFLP and RAPD markers in Vicia faba. Theor Appl Genet 85:935–945
Udupa SM, Robertson LD, Weigand F, Baum M, Kahl G (1999) Allelic variation at (TAA) microsatellite loci in a world collection of chickpea (Cicer arietinum L) germplasm. Mol Gen Genet 261:354–363
Vairinhos F, Murray DR (1983) The seed proteins of chickpea comparative studies of Cicer arietinum, C. reticulatum and C.echinospermum (Leguminosae). Plant Syst Evol 142:15–22
van der Maesen LJG (1987) Origin, history and taxonomy of chickpea. In: Saxena MC, Singh KB (eds) The chickpea. CAB Int Publ, UK, pp 11–34
van Rheenen HA, Pundir PRS, Miranda JH (1993) How to accelerate the genetic improvement of a recalcitrant crop species such as chickpea. Curr Sci 65:414–417
Vavilov NI (1926) The origin of the cultivation of ‘primary’ crops, in particular cultivated hemp. In: Studies on the origin of cultivated plants, Institute of applied botany and plant breeding, Leningrad, USSR, pp 221–233
Acknowledgments
We would like to thank Islamic Azad University, Sanandaj Branch for support of this study. Also, we wish to present our special thanks to the International Center for Agricultural Research in the Dry Areas (ICARDA), Aleppo, Syria and the Australian Temperate Field Crops Collection (ATFCC) at the Victorian Institute for Dryland Agriculture (VIDA), Horsham, Australia for kindly supplying seeds of Cicer species.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Amirmoradi, B., Talebi, R. & Karami, E. Comparison of genetic variation and differentiation among annual Cicer species using start codon targeted (SCoT) polymorphism, DAMD-PCR, and ISSR markers. Plant Syst Evol 298, 1679–1688 (2012). https://doi.org/10.1007/s00606-012-0669-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-012-0669-6