Abstract
Although most patients with papillary thyroid cancer (PTC) are curable, there are still a few patients showing poor outcomes and increased risk of secondary cancers after therapies. In this study, we aimed to investigate the correlation between miR-4500 and PTC and to explore its molecular functions. A total of 50 patients were included, and sonography and histological examinations were used for diagnosis. Quantitative real-time polymerase chain reaction (qRT-PCR) was applied for detection of mRNA levels while Western blotting was used for measuring protein expression. Cell proliferation was tested using CCK-8 and colony formation assays. Caspase-3 activity and nucleosomal fragmentation assays were employed to test cell apoptosis. Cell invasive ability was measured using transwell assay. MiR-4500 target was identified using luciferase assay and RNA pull-down assay. MiR-4500 expression was significantly decreased in five PTC cell lines compared with Nthy-ori 3-1 cells and in PTC tissues compared with adjacent normal thyroid tissues, respectively. Decreased expression of miR-4500 showed lower survival rate, higher cancer stage, and lymphatic metastasis. Therefore, our results implied that miR-4500 could serve as a potential biomarker for PTC prognosis. Overexpression of miR-4500 repressed colony formation, proliferation, and invasiveness of PTC cells whereas increased cell apoptosis. We identified that PLXNC1 was a direct target of miR-4500. PLXNC1 knockdown showed similar effects on cell viability, colony formation, and cell apoptosis as overexpression of miR-4500 in PTC cells. In conclusion, miR-4500 inhibits the malignant transformation of PTC cells by directly targeting and repressing PLXNC1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Thyroid cancer (TC) is the most common endocrine cancer. Although TC is less likely to cause death, the incidence of TC remains high and has been increasing over the last several decades, which may be caused by advanced screening and detection methods. There are four main TC subtypes, including papillary, follicular, medullary, and metamorphic TC [1]. However, among all the incidences of these TC types, the most prevailing one is papillary thyroid carcinoma (PTC). The prognosis of PTC is commonly good. However, there are still a few patients showing poor differentiation of tumor cells and metastasis, and are confronted with recurrence and death [1, 2]. Therefore, it becomes urgent to explore the molecular mechanism of PTC malignant transformation, which could provide theoretical basis for the development of effective diagnosis and therapeutic targets.
MicroRNAs (miRNAs, miRs) are a group of evolutionarily conserved endogenous non-coding RNAs. These RNAs commonly are 18–25 bp in length and bind to the 3′-untranslated region (UTR) of its target genes [3], eliciting mRNA degradation or blocking of translation. MiRNAs play plenty of roles in different cellular or biological activities such as cell proliferation, angiogenesis, and metastasis [4]. In cancers, miRNA expression is frequently observed to be aberrant, leading to a possible utilization of miRNA as prognostic markers or potential therapeutic targets [4, 5]. In fact, there are numerous miRNAs that have been documented to be related to PTC tumors. For example, the expression levels of miR-221, miR-222, and miR-146 were significantly higher in PTC tumors than those of normal thyroid [6].
In previous studies, miR-4500 was reported to be repressed in liver cancer, colorectal cancer, and non-small cell lung cancer [7,8,9,10]. Patients with low miR-4500 level showed higher tumor stage and declined survival rate after surgery, implying an association between miR-4500 expression and prognosis. However, no research has focused on miR-4500 involvement and function in PTC. In this study, we aimed to investigate the roles of miR-4500 in PTC and its underlying mechanism.
Methods
Cell Culture
Five PTC cell lines (HTH83, TPC-1, K1, NIM-1, and B-CPAP) and a benign human thyroid follicular cell line, Nthy-ori 3-1, were purchased from American Type Culture Collection (ATCC, Manassas, VA). K1 cells were cultured in Dulbecco’s modified Eagle medium (DMEM Gibco, BRL, NY, USA): Ham′s F12: MCDB 105 (2:1:1), supplemented with 2 mM glutamine and 10% fetal bovine serum (FBS, Gibco-BRL) as suggested. Nthy-ori 3-1 cells were culture in RPMI 1640 with 2 mM glutamine and 10% FBS. For other cells, DMEM (Gibco) containing 10% FBS was used. All cells were incubated at 37 °C and 5% CO2 in a humidified atmosphere.
Patients
A total of 50 PTC patients who underwent thyroid sonography and thyroidectomy surgery were included in this study. All enrolled patients signed the informed consents. The study was approved by the Ethics Committee of Provincial Hospital Affiliated to Shandong University, Shandong University. Patient characteristics were obtained from records.
Ultrasound
All the PTC patients were diagnosed by both cytological and histological examinations. Patients were diagnosed with sonography to detect thyroid nodule and lymphatic metastasis. The procedure was performed using Philips IU22 (Philips, Best, the Netherlands) and a color Doppler ultrasound system, Mindray Resona 7S (Shenzhen Mindray Bio-Medical Electronics Co., LTD, Shenzhen, China). Two experienced pathologists appraised the pathological stage, grade, and nodal metastasis status. All PTC patients included in this study were conventional subtype. The criteria for lymphatic infiltration were according to previously reported studies [11,12,13].
Quantitative Real-Time Polymerase Chain Reaction
TRIzol reagent (Invitrogen, CA, USA) was used to isolate total RNA according to the manufacturer’s instructions. QuantMir RT Kit (System Biosciences, CA, USA) was employed for reverse transcription using 1 μg of RNA. For detection of miR-4500, SYBR Green (Takara, Dalian, China) was applied for amplification of target miRNA. The results were calculated using 2−ΔΔCt method. The primers used in this study are listed below. MiR-4500 forward primer: 5′-TGAGGTAGTAGTTTCTTGCGCC-3′, and reverse primer: 5′-CTCTACAGCTATATTGCCAGCCAC-3′. U6 and GAPDH were used as controls. U6 forward primer: 5′-ATTGGAACGATACAGAGAAGATT-3′, reverse primer: 5′-GGAACGCTTCACGAATTTG-3′. PLXNC1 forward primer: 5′-AACTGTTCCCTTCCTTGACTAC-3′, R: 5′-TCGTTGGCGTCTCTGTTATG-3′. GAPDH forward primer: 5′-GGCTGAGAACGGGAAGCTTGTCAT-3′; R: 5′-CAGCCTTCTCCATGGTGGTGAAGA-3′.
Cell Viability and Colony Formation
Cell viability was tested using a CCK-8 kit (Beyotime, Shanghai, China) following the manufacturer’s instruction. Briefly, cells were seeded at a density of 5 × 103 cells/well. After 24 h, CCK-8 was added and the values were detected at 450 nm using the Epoch microplate spectrophotometer (Bio-Tek, VT, USA). Cell viability was tested each 24 h for 5 days. For colony formation, 6-well plate was used and cells were plated at a density of 1500 cells/well. After 2 weeks of culture, colonies were fixed with methanol and stained by crystal violet. Only colony containing > 50 cells was included for statistical analysis. Each group contained three parallel wells.
Transwell Invasion Assay
For testing cell invasive ability, Matrigel (Corning Incorporated, NY, USA) was used to pre-coat the transwell chambers (8-mm pore size). Then, the chamber was filled with 2.5 × 104 cells in cell medium without FBS and placed into a 24-well plate filled with cell medium containing 10% FBS. The invaded cells in the plate were fixed by methanol and stained with crystal violet. The invaded cell numbers were counted.
Cell Apoptosis
Cell apoptosis was analyzed by testing nucleosomal fragmentation using Cell Death Detection ELISA PLUS (Roche Applied Science, IN, USA). The absorbance was detected and normalized to calculate the nucleosomal enrichment factor. Caspase-3/CPP32 Colorimetric Assay Kit (BioVision, CA, USA) was employed to measure caspase-3 activity. The assay was conducted according to the manufacturer’s instructions and a previous study [14].
Luciferase Assay
The luciferase assay used luciferase reporter plasmids including PLXNC1 3′UTR-wildtype (WT), PLXNC1 3′UTR-mt, psi-CHECK2. PLXNC1 3′UTR-WT plasmid was constructed using the primers shown below. Forward primer: 5′-CTACTCGAGGCACTCTGGGGCC-3′ and reverse primer: 5′-CTAGCGGCCGCCACAGTTACATCAAAAT-3′. The PLXNC1 3′UTR-mt was constructed using forward primer: 5′-TATAATATTTCTAAGATCCGTGA-3′, and reverse primer: 5′-TCACGGATCTTAGAAATATTATA-3′. Plasmids were co-transfected in cells as indicated in the results. MiR-4500 mimics were transfected for miR-4500 overexpression, which was compared with its control mimics. All groups of cells were co-transfected with a Renilla luciferase gene for transfection control. After 24 h of transfection, cells were collected and Dual-Luciferase® Reporter Assay System (Promega, WI, USA) was used for the detection of luciferase activity according to the manufacturer’s instructions.
Pull-down Assay of miR-4500
The direct binding of miR-4500 to PLXNC1 was further assessed using RNA pull-down assay. Briefly, cells were harvested by cell lysis buffer (0.05% NP-40, 25 mM Tris-HCl, pH 7.5, 2.5 mM EDTA, 70 mM KCl). The lysis buffer was added with diluted protease inhibitor cocktail (Sigma-Aldrich, MO, USA) and RNase Inhibitor (80 U/mL, Life Technologies, CA, USA). Cell lysate was incubated with biotinylated double-stranded RNA (8 nmol) of miR-4500 at 4 °C for 30 min with shaking at 8 rpm. Then, the tubes were continued to be shaken at 30 °C for 1 h at 30 rpm. To pull down the biotinylated RNA, Streptavidin Mutein Matrix (Roche Applied Science, Mannheim, Germany) was added, and the combined RNAs were collected by centrifugation at 5000×g for 30 s and washed 5 times at 4 °C with cell lysis buffer. Then, elution buffer (5 mM biotin, 0.5% NP-40, 20 mM Tris-HCl, pH 7.4, 400 mM KCl) containing 80 U/mL RNase inhibitor was used for elution of the complex. The PLXNC1 gene was detected using quantitative real-time polymerase chain reaction (qRT-PCR), and the results were normalized to GAPDH (as a negative control).
Western Blotting
Protein expression was detected by Western blotting. Total protein extracts were obtained using cell lysis buffer as mentioned above and were separated by 10% SDS-PAGE. Then, the proteins on the gel were transferred to polyvinylidene difluoride membranes (PVDF) and the membranes were blocked using 5% non-fat milk (in TBST, Tris-buffered saline, 0.1% Tween 20) for 1 h at 37 °C. Primary antibodies were diluted in TBST as 1:1000. Membranes were incubated with the diluted primary antibodies at 37 °C for 1 h. After three times washing with TBST, the membranes were next incubated with anti-mouse secondary antibody (Santa Cruz Biotechnology, CA, USA) at 37 °C for 1 h. The signals were developed using a chemiluminescent HRP substrate (EMD Millipore, MA, USA). The primary antibodies used in this study were anti-PLXNC1 (AF3887, R&D Systems Inc., MN, USA) and anti-actin (A-5060, Sigma, Germany).
Statistical Analysis
For data analysis, SPSS 20.0 Software (SPSS Inc., Chicago, IL) was used. The comparisons between groups were determined by Student’s t test or ANOVA analysis followed by a Bonferroni post hoc test. Kaplan-Meier overall survival curve was used for analyzing the survival difference between miR-4500 low and high expressing patients. All data were shown as mean ± standard deviation (SD). A P value less than 0.05 was considered as statistical significant.
Results
Downregulation of miR-4500 Is Correlated with Poor Prognosis of Patients with PTC
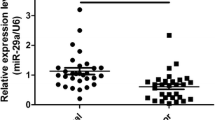
To investigate the association between miR-4500 and PTC, we firstly detected miR-4500 levels in five different PTC cell lines and a benign human thyroid follicular cell line, Nthy-ori 3-1. It was found that miR-4500 expression level was significantly lower in PTC cell lines than Nthy-ori 3-1 cells (Fig. 1a). We next analyzed the miR-4500 expression levels in the 50 PTC patients using their PTC and adjacent normal tissues. It was observed that miR-4500 level was dramatically downregulated in PTC tissues compared with the adjacent normal tissues (Fig. 1b). We found that miR-4500 level was positively correlated to metastasis as remarkably lower level of miR-4500 was observed in patients with metastasis compared with those without metastasis (Fig. 1c). To reveal whether miR-4500 was associated with prognosis, overall survival rates were compared between patients with high or low expression of miR-4500 (Fig. 1d). Significantly reduced survival rate was shown in patients with low level of miR-4500.
Downregulation of miR-4500 predicts poor prognosis for patients with PTC. a qRT-PCR assay was performed to determine miR-4500 expression in five PTC cell lines (HTH83, TPC-1, K1, NIM-1, and B-CPAP) and a benign human thyroid follicular cell line Nthy-ori 3-1. U6 was used as the endogenous controls. b qRT-PCR analysis of miR-4500 expression in papillary thyroid cancer (PTC) and the adjacent non-cancerous (normal) tissues (n = 50). U6 was used as the endogenous control. c qRT-PCR showed expression level of miR-4500 in metastasis and non-metastasis PTC tissues. d Kaplan-Meier overall survival curve of 50 PTC patients divided by miR-4500 expression levels. *P < 0.05; **P < 0.01
In addition, we analyzed the correlation between miR-4500 expression and clinicopathologic characteristics and found that miR-4500 level was significantly correlated with tumor stage, lymph node metastasis, extrathyroidal extension, and multifocality (Table 1). There results suggested that miR-4500 could be a potential biomarker for PTC prognosis. Furthermore, in BRAF (V600E)-mutated patients, more than 60% showed low miR-4500 expression (Table 1). Another prognostic genetic marker, Telomerase reverse transcriptase (TERT) promoter mutation was also found to be prevalent in patients with low miR-4500 expression.
MiR-4500 Suppresses Cell Proliferation and Invasive Ability
To reveal the roles of miR-4500 in PTC, miR-4500 overexpressing cell lines were established (Fig. 2a). Cell viability and colony formation assays were performed. Overexpression of miR-4500 markedly repressed cell viability of both K1 and TPC-1 cells and reduced their abilities of colony formation (Fig. 2b–d). To assess cell apoptosis activity, caspase-3 activity was measured and nucleosomal fragmentation assay was performed (Fig. 2 e and f). We found that miR-4500 overexpression significantly promoted cell apoptosis. Using transwell assay, miR-4500 overexpression was observed to reduce cell invasive ability (Fig. 2g), further demonstrating miR-4500 as a tumor suppressor.
MiR-4500 suppresses proliferation and invasive ability of PTC cells. a The transfection efficiency of miR-4500 in K1 and TPC-1 cells was confirmed by qRT-PCR assay. b–d CCK-8 and colony formation assays showed the growth of K1 and TPC-1 cells transfected with miR-4500 mimics (miR-4500) or the negative control (miR-NC). e, f The caspase-3 activity and enrichment factor were analyzed to indicate the apoptosis of K1 and TPC-1 cells transfected with miR-4500 mimics (miR-4500) or the negative control (miR-NC). g Transwell assay was performed to detect the invasive capacity of K1 and TPC-1 cells transfected with miR-4500 mimics (miR-4500) or the negative control (miR-NC). The data represent the mean ± SD from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001, (two-way ANOVA for b and c, student’s t test for others)
MiR-4500 Directly Targets PLXNC1
Next, to reveal the molecular mechanism of miR-4500, it is important to identify its target genes. We therefore employed Targetscan (http://www.targetscan.org/vert_71/) (Fig. 3a) to predict the potential miR-4500 target genes, and found that PLXNC1 was a potential one target. To confirm this prediction, we constructed two reporter plasmids containing wild-type (WT) PLXNC1 3′UTR or the mutated PLXNC1 3′UTR of the binding site. As shown in Fig. 3 b and c, miR-4500 overexpression was able to inhibit WT PLXNC1 reporter activity but not the mutant one. To further confirm the direct binding of miR-4500 to PLXNC1 mRNA, we employed pull-down assay with biotin-labeled RNA and found that PLXNC1 was detected in the RNA pull-down complex (Fig. 3d). Taken together, PLXNC1 was indeed a bona fide target of miR-4500. Then, we tested PLXNC1 mRNA and protein levels in miR-4500 overexpressing cells and found that PLXNC1 levels were significantly repressed (Fig. 3 e and f). In contrast, miR-4500 inhibitor led to a significant increase of PLXNC1 expression. These results indicated that PLXNC1 was a direct target of miR-4500.
MiR-4500 directly targets PLXNC1 in PTC cells. a The prediction for miR-4500 binding sites on the 3′UTR of PLXNC1 and schematic of luciferase reporter vector constructs wild-type (PLXNC1 3′UTR-wt) PLXNC1 3′UTR and the miR-4500-binding-site mutated (PLXNC1 3′UTR-mt) one. b, c Luciferase reporter assay of K1 and TPC-1 cells co-transfected with constructed luciferase reporter vectors (PLXNC1 3′UTR-wt, PLXNC1 3′UTR-mt, psi-CHECK2) and miR-4500 or negative control mimics (miR-NC). d Detection of PLXNC1 mRNAs in biotinylated miRNA/target mRNA complex by real-time RT-PCR. The relative level of PLXNC1 mRNA in the complex pulled down by using biotinylated miR-4500 (Bio-miR-4500) was compared with that of the complex pulled down by using the biotinylated control random RNA (Bio-miR-NC). e, f The relative expression levels of PLXNC1 in K1 and TPC-1 cells transfected with miR-4500 mimics and miR-4500 inhibitors or their respective negative controls were detected by qRT-PCR and Western blot. The data represent the mean ± SD from three independent experiments. *P < 0.05; **P < 0.01; ns, not significant, (Student’s t test)
Knockdown of PLXNC1 Inhibits Proliferation and Invasion of PTC Cells
To explore the role of PLXNC1 in PTC, we used si-RNA to repress PLXNC1 expression (Fig. 4 a and b). Cell viability and colony formation were tested in PLXNC1 WT and knockdown cells. As expected, PLXNC1 knockdown showed the same trends of impacts on cell viability and colony formation ability as miR-4500 overexpression (Fig. 4c–e). We further tested caspase-3 activity and nucleosomal fragmentation. Knockdown of PLXNC1 was observed to promote cell apoptosis (Fig. 4 f and g). Transwell assay also evidenced that PLXNC1 knockdown cells exhibited inhibited invasive behavior compared with WT cells (Fig. 4h). Therefore, PLXNC1 could serve as a target proto-oncogene of miR-4500.
Knockdown of PLXNC1 inhibits proliferation and invasion of PTC cells. a, b mRNA and protein levels of PLXNC1 were determined by qRT-PCR and Western blot in K1 and TPC-1 cells transfected with PLXNC1 siRNAs (si-PLXNC1-1 and si-PLXNC1-2) or negative control (si-NC). c–e CCK-8 and colony formation assays showed the growth of K1 and TPC-1 cells transfected with PLXNC1 siRNAs (si-PLXNC1-1 and si-PLXNC1-2) or negative control (si-NC). f, g The caspase-3 activity and enrichment factor were analyzed to indicate the apoptosis of K1 and TPC-1 cells transfected with PLXNC1 siRNAs (si-PLXNC1-1 and si-PLXNC1-2) or negative control (si-NC). h Transwell assay was performed to detect the invasive capacity of K1 and TPC-1 cells transfected with PLXNC1 siRNAs (si-PLXNC1-1 and si-PLXNC1-2) or negative control (si-NC). The data represent the mean ± SD from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001, (two-way ANOVA for c and d, Student’s t test for others)
Overexpression of PLXNC1 Impairs miR-4500-Induced Inhibition of PTC Malignancy
To further confirm that PLXNC1 could mediate miR-4500 functions, we transfected cells with PLXNC1 in miR-4500 overexpression or WT cells (Fig. 5 a and b). It was observed that, although miR-4500 overexpression reduced cell viability, this effect was abolished when PLXNC1 was also overexpressed (Fig. 5c). The number of colonies was significantly increased in PLXNC1 and miR-4500 overexpressing cells compared with miR-4500 overexpressing cells alone (Fig. 5d), indicating the inhibitory effect of miR-4500 on colony formation could be blocked by PLXNC1 overexpression. MiR-4500-induced cell apoptosis was also significantly inhibited by PLXNC1 overexpression (Fig. 5 e and f). Cell invasion was repressed in miR-4500 overexpressing cells compared with control. However, PLXNC1 overexpression showed significantly promoted invasive ability compared with miR-4500 overexpressing cells alone (Fig. 5g). Taken together, these results suggested that miR-4500-induced inhibitory effects on PTC malignancy were dependent on PLXNC1 expression.
Overexpression of PLXNC1 impairs miR-4500-induced inhibition of malignancy of PTC cells. K1 cells were co-transfected with miR-NC + empty vector, miR-4500 mimics + empty vector, or miR-4500 mimics + PLXNC1-expressing plasmid. a, b The expression levels of PLXNC1 in these co-transfected K1 cells were detected by qRT-PCR and Western blot. c, d Cell viability of these co-transfected K1 cells was measured by CCK-8 and colony formation assays. e, f Cell apoptosis of these co-transfected K1 cells was determined by caspase-3 activity and nucleosomal fragmentation assays. g Cell invasion of these co-transfected K1 cells was assessed by transwell assay. The data represent the mean ± SD from three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001, (two-way ANOVA for c, Student’s t test for others)
Discussion
The incidence rate of TC, especially in females, has been increasing for several decades in numerous countries [15,16,17]. Although this cancer is commonly not lethal, its high recurrence and persistence rates may lead to increased morbidity and mortality of patients [18]. About 5% of patients with TC died due to aggressive progression. Among different types of TCs, a large portion of the increased incidence is the papillary type [19]. PTC, derived from follicular thyroid cells, is commonly curable but tends to show lymphatic metastasis. Surgical thyroidectomy and adjuvant radioiodine therapy are commonly used to treat PTC patients. However, the risk of radioiodine to cause secondary malignancies [20] limits the choices for patients. In particular, for patients with aggressive and radioiodine-insensitive TCs, few therapies are available. To identify a biomarker for patients with TCs is urgent, which could be utilized to show the prognosis of patients and also determine whether patients need therapies such as the high risky radioiodine. However, in this study, we focused on investigating a molecular marker of PTC and its underlying mechanism.
MiRNAs are suggested to serve as potential biomarkers in a set of cancer types. In several previous studies, miRNA expression profiles were investigated in TC, evidencing the key roles of miRNA expression to maintain tumorigenesis [21]. For example, in 2005, He H. et al. demonstrated that miR-221, miR-222, and miR-146 showed remarkably increased expression in PTC tumors compared with normal thyroids [6]. Since the first publication on the functions of miRNA expression in TC, numerous studies later endeavored to investigate the contributions of miRNAs to PTC further revealed the critical roles of miRNAs in different TC developments, including PTC [22, 23]. In addition to aforementioned miRNAs, miR-181b was also reported to be related to PTC aggressiveness [24,25,26].
Here, our results showed that miR-4500 expression was downregulated in PTC and negatively associated with metastasis. Patients with low level of miR-4500 showed a decline trend in overall survival rate after surgery, suggesting miR-4500 could be used as a biomarker for prognosis of PTC patients. However, it was difficult to confirm whether these patients died only due to PTC progression, because some of them passed away in a different hospital and from which we were not able to retrieve their records. To date, researches on miR-4500 are rare but its repressed expression was observed in both non-small cell lung cancer and colorectal cancer [9, 10], which are consistent with our results. Additionally, their studies also showed that miR-4500 level was negatively associated with tumor stage and survival rate, giving further support for miR-4500 as a biomarker of cancer. In 2003, BRAFV600E was reported to be the most frequent mutation in PTC [27, 28]. It is mainly resulted from a point mutation T1799A, which exchanges valine to glutamate and therefore consistently activates the MAPK signaling pathway [29]. Previous studies demonstrated that BRAFV600E was associated with higher risks of aggressive and recurrent PTC [30, 31]. Here, we found that more than 60% of patients with low miR-4500 expression had BRAFV600E mutations. TERT promoter mutation is also frequently observed in PTC, leading to increasing expression of TERT. Its mutation was reported to be associated with more aggressive tumor and poor outcomes of PTC patients [32]. In this study, we found that more that 70% patients with TERT promoter mutation showed low miR-4500 level. These results, on the other hand, suggest that low level of miR-4500 contributes to aggressive progression of PTC. However, it needs further study to investigate whether BRAFV600E activates MAPK signaling to regulate miR-4500 expression, and how TERT promoter mutation contributes to miR-4500 expression.
Downregulation of miR-4500 was demonstrated to inhibit cell proliferation, invasion, and migration in colorectal cancer. Consistently, our results showed similar inhibitory functions of miR-4500 on cell proliferation and invasive ability of PTC cells. Distinguished from their findings documenting that HMGA2 was regulated by miR-4500, we have demonstrated that PLXNC1 is a direct target of miR-4500. Mutation of PLXNC1 on its predicted miR-4500 binding site led to the ablation of miR-4500 effects on its transcriptional and translational levels. It is possible that PLXNC1 also mediates miR-4500 function in other cancers including colorectal cancer and lung cancer, which needs to be further investigated.
PLXNC1, also named VESPR, is a binding receptor for virally encoded SEMA7A [33]. The function of PLXNC1 was firstly described as a factor mediating neuronal cell adhesion in the optic tectum [34]. Interestingly, Comeau et al. later showed that PLXNC1 participated in A39R-induced cytokine production of monocytes, and affected the adhesion and migration of numerous immune cells including dendritic cells and neutrophils [35, 36]. Based on the roles of PLXNC1 in inflammation, it is possible that it is also involved in tumorigenesis. A previous study observed that overexpression of PLXNC1 led to significantly reduced cell viability and increased cell apoptosis in melanoma, where its mRNA level was regulated by miR-181a [37], suggesting that PLXNC1 could be a tumor suppressor in melanoma. These results are contrary to our findings, which may be caused by its differential functions in various cancers, which this needs to be further investigated. PLXNC1 was found to be highly expressed in TC [38]. A previous study comparing the gene expression profiles revealed that PLXNC1 expression was increased about 2-fold in PTC tissues in comparison with their paired normal tissues [39]. This finding is consistent with a recent study analyzing two data sets containing 16 PTC and paired normal samples [40]. In the current study, we repressed PLXNC1 using siRNA and found that cell viability, colony formation, and metastasis were all reduced compared with control K1 and TPC-1 cells. On the contrary, cell apoptosis was increased in PLXNC1 knockdown cells compared with control cells. These findings further extend the functions of PLXNC1 from cell adhesion to cell proliferation and apoptosis. However, deep insights and further investigation in to its molecular mechanism are still necessary.
In conclusion, we found that miR-4500 expression was repressed in PTC tissues. PTC with higher tumor stages and lymphatic metastasis showed lower levels of miR-4500. These evidences provide support of miR-4500 as a potential prognosis marker for PTC. Furthermore, investigation on its molecular mechanism has revealed that miR-4500 plays critical roles in cell proliferation and apoptosis. By predicting miR-4500 targets, we have identified that PLXNC1 as a target of miR-4500. This prediction was verified by luciferase and RNA pull-down assays. Knockdown PLXNC1 also exhibits inhibitory effects on cell viability, colony formation, and cell apoptosis, similar as overexpression of miR-4500. However, overexpression of PLXNC1 abrogates miR-4500-induced inhibition of malignancy of PTC cells.
References
Carling T, Udelsman R (2014) Thyroid cancer. Annu Rev Med 65:125–137
Cabanillas ME, McFadden DG, Durante C (2016) Thyroid cancer. Lancet 388:2783–2795
Croce CM, Calin GA (2005) miRNAs, cancer, and stem cell division. Cell 122:6–7
Croce CM (2009) Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10:704–714
Pereira DM, Rodrigues PM, Borralho PM, Rodrigues CM (2013) Delivering the promise of miRNA cancer therapeutics. Drug Discov Today 18:282–289
He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, Kloos RT, Croce CM, de la Chapelle A (2005) The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A 102:19075–19080
Lin Q, Zheng H, Xu J, Zhang F, Pan H (2019) LncRNA SNHG16 aggravates tumorigenesis and development of hepatocellular carcinoma by sponging miR-4500 and targeting STAT3. J Cell Biochem. https://doi.org/10.1002/jcb.28440
Kim K, Shin EA, Jung JH, Park JE, Kim DS, Shim BS, Kim SH (2018) Ursolic acid induces apoptosis in colorectal cancer cells partially via upregulation of MicroRNA-4500 and inhibition of JAK2/STAT3 phosphorylation. Int J Mol Sci 20:E114
Yu FY, Tu Y, Deng Y, Guo C, Ning J, Zhu Y, Lv X, Ye H (2016) MiR-4500 is epigenetically downregulated in colorectal cancer and functions as a novel tumor suppressor by regulating HMGA2. Cancer Biol Ther 17:1149–1157
Zhang L, Qian J, Qiang Y, Huang H, Wang C, Li D, Xu B (2014) Down-regulation of miR-4500 promoted non-small cell lung cancer growth. Cell Physiol Biochem 34:1166–1174
Han RL, Wang J, Zhang FJ, Zhao N, Gao BL (2018) Ultrasound risk assessment combined with molecular markers of Galectin-3, c-MET, HBME-1 and CK19 for diagnosis of malignant and benign thyroid nodules. Pathol Oncol Res. https://doi.org/10.1007/s12253-018-0485-6
Sun B, Zhong Z, Wang F, Xu J, Xu F, Kong W, Ling Z, Shu N, Li Y, Wu T, Zhang M, Zhu L, Liu X, Liu L (2018) Atorvastatin impaired glucose metabolism in C2C12 cells partly via inhibiting cholesterol-dependent glucose transporter 4 translocation. Biochem Pharmacol 150:108–119
Snozek CL, Chambers EP, Reading CC, Sebo TJ, Sistrunk JW, Singh RJ, Grebe SK (2007) Serum thyroglobulin, high-resolution ultrasound, and lymph node thyroglobulin in diagnosis of differentiated thyroid carcinoma nodal metastases. J Clin Endocrinol Metab 92:4278–4281
Zhao H, Tang H, Huang Q, Qiu B, Liu X, Fan D, Gong L, Guo H, Chen C, Lei S, Yang L, Lu J, Bao G (2016) MiR-101 targets USP22 to inhibit the tumorigenesis of papillary thyroid carcinoma. Am J Cancer Res 6:2575–2586
Kahn C, Simonella L, Sywak M, Boyages S, Ung O, O’Connell D (2012) Pathways to the diagnosis of thyroid cancer in New South Wales: a population-based cross-sectional study. Cancer Causes Control 23:35–44
Kilfoy BA, Zheng T, Holford TR, Han X, Ward MH, Sjodin A, Zhang Y, Bai Y, Zhu C, Guo GL (2009) International patterns and trends in thyroid cancer incidence, 1973–2002. Cancer Causes Control 20:525–531
Morris LG, Sikora AG, Tosteson TD, Davies L (2013) The increasing incidence of thyroid cancer: the influence of access to care. Thyroid 23:885–891
Xing M, Alzahrani AS, Carson KA, Shong YK, Kim TY, Viola D, Elisei R, Bendlová B, Yip L, Mian C (2015) Association between BRAF V600E mutation and recurrence of papillary thyroid cancer. J Clin Oncol 33:42–50
Nikiforov YE (2011) Molecular diagnostics of thyroid tumors. Arch Pathol Lab Med 135:569–577
Yoshinaga S, Mabuchi K, Sigurdson AJ, Doody MM, Ron E (2004) Cancer risks among radiologists and radiologic technologists: review of epidemiologic studies. Radiology 233:313–321
Fuziwara CS, Kimura ET (2014) MicroRNA deregulation in anaplastic thyroid cancer biology. Int J Endocrinol 2014:743450
Chou C-K, Chen R-F, Chou F-F, Chang H-W, Chen Y-J, Lee Y-F, Yang KD, Cheng J-T, Huang C-C, Liu R-T (2010) miR-146b is highly expressed in adult papillary thyroid carcinomas with high risk features including extrathyroidal invasion and the BRAFV600E mutation. Thyroid 20:489–494
Rosignolo F, Memeo L, Monzani F, Colarossi C, Pecce V, Verrienti A, Durante C, Grani G, Lamartina L, Forte S, Martinetti D, Giuffrida D, Russo D, Basolo F, Filetti S, Sponziello M (2017) MicroRNA-based molecular classification of papillary thyroid carcinoma. Int J Oncol 50:1767–1777
Zhou Y-L, Liu C, Dai X-X, Zhang X-H, Wang O-C (2012) Overexpression of miR-221 is associated with aggressive clinicopathologic characteristics and the BRAF mutation in papillary thyroid carcinomas. Med Oncol 29:3360–3366
Wang Z, Zhang H, He L, Dong W, Li J, Shan Z, Teng W (2013) Association between the expression of four upregulated miRNAs and extrathyroidal invasion in papillary thyroid carcinoma. OncoTargets Ther 6:281
Stokowy T, Gawel D, Wojtas B (2016) Differences in miRNA and mRNA profile of papillary thyroid cancer variants. Int J Endocrinol 2016:1–10
Cohen Y, Xing M, Mambo E, Guo Z, Wu G, Trink B, Beller U, Westra WH, Ladenson PW, Sidransky D (2003) BRAF mutation in papillary thyroid carcinoma. J Natl Cancer Inst 95:625–627
Xu X, Quiros RM, Gattuso P, Ain KB, Prinz RA (2003) High prevalence of BRAF gene mutation in papillary thyroid carcinomas and thyroid tumor cell lines. Cancer Res 63:4561–4567
Czarniecka A, Oczko-Wojciechowska M, Barczynski M (2016) BRAF V600E mutation in prognostication of papillary thyroid cancer (PTC) recurrence. Gland Surg 5:495–505
Jo YS, Li S, Song JH, Kwon KH, Lee JC, Rha SY, Lee HJ, Sul JY, Kweon GR, Ro HK, Kim JM, Shong M (2006) Influence of the BRAF V600E mutation on expression of vascular endothelial growth factor in papillary thyroid cancer. J Clin Endocrinol Metab 91:3667–3670
Kebebew E, Weng J, Bauer J, Ranvier G, Clark OH, Duh QY, Shibru D, Bastian B, Griffin A (2007) The prevalence and prognostic value of BRAF mutation in thyroid cancer. Ann Surg 246:466–470 discussion 470-461
Alzahrani AS, Alsaadi R, Murugan AK, Sadiq BB (2016) TERT promoter mutations in thyroid cancer. Horm Cancer 7:165–177
Chen H, Chédotal A, He Z, Goodman CS, Tessier-Lavigne M (1997) Neuropilin-2, a novel member of the neuropilin family, is a high affinity receptor for the semaphorins Sema E and Sema IV but not Sema III. Neuron 19:547–559
Ohta K, Mizutani A, Kawakami A, Murakami Y, Kasuya Y, Takagi S, Tanaka H, Fujisawa H (1995) Plexin: a novel neuronal cell surface molecule that mediates cell adhesion via a homophilic binding mechanism in the presence of calcium ions. Neuron 14:1189–1199
Rosenberger P, Schwab JM, Mirakaj V, Masekowsky E, Mager A, Morote-Garcia JC, Unertl K, Eltzschig HK (2009) Hypoxia-inducible factor–dependent induction of netrin-1 dampens inflammation caused by hypoxia. Nat Immunol 10:195–202
Walzer T, Galibert L, De Smedt T (2005) Dendritic cell function in mice lacking Plexin C1. Int Immunol 17:943–950
Ghaffar M, Khodahemmati S, Li J, Shahzad M, Wang M, Wang Y, Li C, Chen S, Zeng Y (2018) Long non-coding RNA LINC01234 regulates proliferation, invasion and apoptosis in esophageal cancer cells. J Cancer 9:4242–4249
Chen Y, Soong J, Mohanty S, Xu L, Scott G (2013) The neural guidance receptor Plexin C1 delays melanoma progression. Oncogene 32:4941–4949
Kim HS, Kim DH, Kim JY, Jeoung NH, Lee IK, Bong JG, Jung ED (2010) Microarray analysis of papillary thyroid cancers in Korean. Korean J Intern Med 25:399–407
Qu T, Li YP, Li XH, Chen Y (2016) Identification of potential biomarkers and drugs for papillary thyroid cancer based on gene expression profile analysis. Mol Med Rep 14:5041–5048
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, R., Teng, X., Zhu, H. et al. MiR-4500 Regulates PLXNC1 and Inhibits Papillary Thyroid Cancer Progression. HORM CANC 10, 150–160 (2019). https://doi.org/10.1007/s12672-019-00366-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12672-019-00366-1