Abstract
Purpose
Xylitol is used in the food and pharmaceutical industries as a sweetener, and its consumption rate has remarkably increased over the years. Currently, xylitol is produced through a chemical reduction method. However, considering the global climate change issue, the development of eco-friendly, renewable, and sustainable substrates for the production of high-value platform chemicals such as xylitol is the need of the hour. Hence, the present study aimed to use a microbial process for xylitol production.
Methods
Escherichia coli M15 platform was used to overexpress the D-xylose reductase (XR) gene from a mesophilic yeast, Candida tropicalis GRA1. The 37-KDa CtXR sequence exhibited a highly conserved active site structure, where a tetrad of residues (Tyr51, Lys80, His113, and Asp46) was located at the base of the substrate-binding pocket. To mitigate the rate-limiting step of cofactor supply in the bioconversion of xylose to xylitol, overexpression of XR genes coupled with auxiliary substrates was used toward in vivo cofactor regeneration.
Results
In the presence of xylose as the only carbon source, the M15 CtXRΔ produced 2.1 g L−1 of xylitol. The xylitol titer showed a steady-state increase with the addition of auxiliary substrates glucose and glycerol to 3.4 and 6.4 g L−1, respectively. Further, M15 CtXRΔ as a whole-cell biocatalyst in alkali pretreated detoxified corncob hydrolysate produced xylitol titer of 3.7 g L−1.
Conclusion
Therefore, we successfully produced xylitol from corncob hydrolysates using the engineered E. coli M15 as whole-cell biocatalysts. Further, this process can be up scaled for the synthesis of eco-friendly high-value green chemicals.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Statement of Novelty
Xylitol is produced by recombinant whole-cell biocatalysts that carry xylose reductase (XR) using corn cob as a substrate. XR is the key enzyme that converts the xylose component of corn cob into xylitol.
Introduction
The increasing concern about global climate change and the energy crisis has led to the development of sustainable and clean sources for the production of high-value platform chemicals. Xylitol is a crystalline pentose sugar alcohol widely substituted for six-carbon sugars as it has almost the same sweetness as glucose and sucrose. Xylitol metabolism is insulin-independent, and has low energy value (2.5 cal g−1) compared with sucrose (4 cal g−1) [1]. Therefore, both the food and pharmaceutical industries use xylitol as an alternate sweetener [2]. The consumption rate of xylitol has increased from 6 million tons to 190.9 million tons from 1978 to 2017 and is likely to increase to 266.5 million tons by 2025 [3, 4].
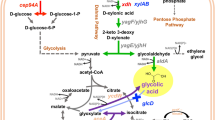
While searching for renewable substrates for xylitol production, lignocellulosic biomass (LCB) has become an eco-friendly, safe alternative for fossil fuels as it is more abundant and a non-food-oriented resource. LCB is composed of key macromolecules: cellulose (30–50%), hemicellulose (25–30%), and lignin (15–25%), where hemicellulose is a complex heteropolymer that includes xylose, arabinose, mannose, and galactose [5]. Xylose represents the second most abundant sugar in nature, accounting for 18–30% of LCB hydrolysate sugars [6]. Currently, chemical reduction of D-xylose is employed for xylitol production from xylan-rich LC sources such as birch and beech wood. The hydrogenation process involves a metal catalyst, preferably nickel (Ni), temperature ranging 353–413 K, and high pressure up to 5 MPa with a xylitol yield of 50–60% [7]. However, the nickel catalyzed hydrogenation process has certain demerits, such as intensive purification steps, high energy demand, wastewater pollution, and expensiveness [7, 8]. In contrast, the biotechnological process of xylitol production is environmentally safe, and a higher yield can be obtained with more specificity. The two important biotechnological approaches for xylitol production developed in recent years are the microbial and enzymatic approaches. For the enzymatic approach, bioconversion of d-xylose to xylitol is achieved by purified d-xylose reductase (XR) from engineered yeasts or bacteria. Although the enzymatic approach is more efficient, it requires pure d-xylose, which is expensive [9], whereas the microbial process involves xylitol production through a fermentative pathway by bacteria and yeast strains. Albeit of the bacterial species investigated, Corynebacterium sp., Enterobacter liquefaciens, or Mycobacterium smegmatis [8, 10], several yeast strains are more efficient xylitol producers [7, 11]. For instance, natural xylitol producers are non-conventional yeasts, Candida sp., Kluyveromyces sp., and Meyerozyma guilliermondii [10, 12,13,14]. Among these microorganisms, Penicillium crustosum consumed 76% of d-xylose with higher mycelial production [15]. The obtained xylitol from these strains ranged from 0.14 to 0.52 g L−1, and substrate consumption varied from 15 to 79%. Metabolically engineered S. cerevisiae, Pichia pastoris, C. tropicalis, and E. coli have been considered as alternatives for enhanced xylitol recovery [16,17,18]. Considering both biotechnological approaches, microbial xylitol production from lignocellulosic is more feasible because the starting material xylose need not be in a purified form [7, 8, 14, 19]. However, the feedstocks have to be pretreated depending on the microbes to be used.
Xylitol production from LCB is greatly limited by the selection and engineering of microbial strains resulting in the low xylose conversion efficiency and product specificity. Very few microbes possess the native xylose metabolic pathway [20], and the xylose metabolism is inhibited by glucose because of a regulatory phenomenon termed carbon catabolite repression (CCR) [21]. CCR routes to diauxic growth, impacting xylose metabolism and other LCB-derived secondary sugars. This phenomenon impedes the xylose conversion efficiency during the fermentation process. Further, the transmembrane transport rate of xylose also terminates the metabolic fluxes. Hence, for improving cell growth and xylose bioconversion efficiency, an increased xylose transport metabolism rate is important [22, 23]. Microorganisms metabolize xylose into xylitol in two ways. Yeast and fungi follow a two-step process, whereas bacteria use a single step to convert xylose to xylulose [24]. Wild-type S. cerevisiae and other yeast strains lack efficient and specific xylose transporters and show poor cell growth rates with xylose as a sole carbon source [25]. Several researchers have attempted to bioengineer yeast, and E. coli strains to enhance xylitol production. Among the bioengineered strains, E. coli is more preferred as a cell factory for xylitol production. Even though the presence of CCR preferentially metabolizes glucose over xylose, E. coli, with its native transporters and xylose metabolism pathways, can utilize mixed sugars derived from LCB [21, 26]. XylFGH, an ABC transporter, and XylE, a major facilitator superfamily protein (D-xylose/proton symporter), are the native xylose transporters. Besides, AraE, an arabinose symporter, also plays a role in transporting xylose across membranes in some specific situations [27]. XylE exhibits a relatively low affinity for xylose and has a Km value of 63 to 169 µM [26, 28]. XylE transports xylose by proton-motive force rather than a phosphotransferase or other direct energy drives. XylF is a periplasmic xylose-binding protein, XylG is an ATP-binding protein, and XylH is a membrane component of the ABC transporter [29, 30]. Among the transporter systems, XylFGH is the dominant xylose transporter in E. coli [27].
The natural xylose metabolic pathway enzymes in E. coli and yeast are xylose isomerase (XI) and XR- xylitol dehydrogenase (XDH) complexes. Xylose metabolic pathways have been constructed successfully in S. cerevisiae, E. coli, Corynebacterium glutamicum, and Zymomonas mobilis to improve xylose utilization [31,32,33,34,35].
Therefore, the reconstruction of the xylose metabolic pathway in microorganisms that do not use xylose naturally can derepress glucose inhibition. Another strategy to improve the xylose utilization efficiency is the introduction of heterologous xylose metabolic genes in microbes that assimilate xylose naturally [33].
The replacement of native cyclic AMP receptor protein (CRP) E. coli with a cyclic AMP independent mutant CRP facilitated a higher xylose uptake from the mixture of glucose and xylose in which glucose served as a growth substrate [36]. In the overexpression studies, nicotinamide adenine dinucleotide phosphate (NADPH)-dependent CtXR and xylitol encoding gene XDH increased the xylitol production by three folds compared with the wild strain with a yield of about 0.837 g g−1 within 22 h [36, 37]. A coupled system using XR from Rhizopus oryzae and glucose dehydrogenase (GDH) from Exiguobacterium sibiricum constructed in E. coli converted 150 g L−1 of xylose into xylitol, and the productivity was about 21.2 g L−1 h−1 [38]. Therefore, recombinant E. coli co-expressing XR-XDH enzymes will be an efficient candidate for xylitol production using xylose.
Furthermore, the cofactor-specificity of XR (preferentially with NADPH) and XDH (strictly with nicotinamide adenine dinucleotide (NAD+) resulted in a limited supply of NAD+ for XDH during the xylitol accumulation [39]. Therefore, changing the coenzyme specificity and effective recycling of cofactors between XR and XDH improves the xylose fermentation efficiency. The quadruple mutant harboring four mutations such as R276F, S271G, K270S, N272P, and indicated a 13-fold improved preference for NAD+ compared with NADP+ and an 86% decrease in unfavorable xylitol accumulation compared with the control strain carrying the wild-type XDH [40, 41]. Despite these results, the biotechnological production of xylitol from LCB is still unsuccessful due to many disadvantages, such as a longer fermentation period and co-products, which complicate the downstream process [42].
In recent years, the hemicellulose containing corncob biomass hydrolysate and its vital metabolites were explored for the production of high-value industrial commodities. Corncob hemicelluloses contain 33–35% arabinose, 3–6% galactose, 6–16% glucuronic acid, and 48–53% xylose. However, the presence of inhibitors such as lignin, furfural, and hydroxymethylfurfural interfere with xylitol metabolism. Consequently, detoxification of the corncob hydrolysate before the fermentation step is necessary, which makes the process costlier [43]. This study presents a single-step xylitol production strategy from corncob hydrolysate using a recombinant E. coli strain expressing the XR gene from a yeast C. tropicalis GRA1. In our previous studies, the gene encoding for XR from a mesophilic yeast strain C. tropicalis GRA1 was cloned into a pGEMT vector and maintained in E. coli DH5α cells. It exhibited the closest similarity with the XR C. tropicalis [44]. The metabolically engineered E. coli M15 whole cells were used to convert D-xylose in the corncob to xylitol, and the cells were recycled under shake flask fermentation. Xylitol produced was further purified and crystallized. The biotransformation efficiency of 70% was achieved within two hours. We also observed the internal redox balance where the cells could recycle NADPH generated through the glycerol catabolism. The addition of glycerol as a co-substrate enhanced the xylitol productivity. Consequently, in this study, we aimed to manipulate the xylitol metabolism in E. coli and evaluate the feasibility of the xylitol production from the corncob.
Materials and Methods
Strains, Media, and Culture Conditions
All enzymes, primers, and molecular markers were bought from BIO-RAD (India) and Biolabs (New England). Molecular reagents were purchased from Sigma-Aldrich (St. Louis, MO) and Qiagen (Valencia, CA). The corncob biomass was obtained from Central Farm, Tamil Nadu Agricultural University (Tamil Nadu, India). The strains and plasmids used in this study are presented in Table S1. Mesophilic C. tropicalis GRA1, previously isolated from grapes by our team, was used as the wild variant for genetic manipulation [44]. The expression host E. coli M15 used in the study was provided by the Indian Institute of Science Education and Research, Kolkata, India. The E. coli cells (XL1 and M15 strains) were cultured in Luria–Bertani (LB) medium (5 g L−1 yeast extract, 10 g L−1 peptone, and 10 g L−1 NaCl) supplemented with 100 µg mL−1 ampicillin and 25 µg mL−1 kanamycin at 37 °C. The xylitol production efficiency of the engineered strain was evaluated in a fermentation medium containing xylose (100 mM).
Cloning and Characterization of XR of C. tropicalis GRA 1
The primers used in the study are listed in Table S2. The overnight-grown E. coli DH5α cells harboring CtXR in pGEM®-T vector (pGEMTCtXR) knocked out [44] were cultured in 50 mL LB broth containing ampicillin and kept in a shaker incubator at 200 rpm and 37 °C. The open reading frame of Xr (1000 bp size) was amplified from the plasmid using CtXR BamH1 F and CtXR Kpn1 R primers following an initial denaturation at 95 °C for 1 min, 35 denaturation cycles at 94 °C for 1 min, annealing at 55 °C for 1 min, primer extension at 72 °C for 1 min, and final extension at 72 °C for 10 min. The PCR amplicons were further sequenced using the M13 primer walk (M/s Scigenome Pvt Ltd., India). The homologous regions of CtXR were analyzed using the basic local alignment search tool (BLAST) (https://www.blast.ncbi.nlm.nih.gov).
Genetic Modifications of pQE30Xa and Transformation of CtXR in E. coli
For knocking out the putative CtXR, pGEMTCtXR was amplified by PCR using the primers (CtXR 2F and CtXR 2R) and cloned upstream and downstream of the Amp-selection cassette of pQE30Xa to generate pQE30XaCtXR under the control of a constitutive T5-lac promoter. The knockout cassette was transformed into E. coli XL1 to generate CtXR::Tet. The transformants were screened on LB plates with Tet 25 µg mL−1 and Amp 100 µg mL−1 (LB + Tet + Amp), resulting in the CtXRΔ strain. The successful homologous recombination was verified by PCR (primers CtXR 2F and CtXR 2R) and restriction digestion with Bam HI and KpnI enzymes. The presence of the CtXR in pQE30Xa was further confirmed by PCR amplification of the upstream T5 promoter region using T5 specific primers. Sequencing of amplicon confirmed successful cloning of the CtXR into pQE30Xa introduced in the coding sequence with no mutation. The identity of the CtXR sequence was established by performing a similarity search against the GenBank database in NCBI. To overexpress putative CtXR, pQE30Xa—CtXR-T5-lac was transformed into the E. coli M15 cells. The clones positive for CtXR (CtXRΔ) were further used for protein induction. Additionally, the pQE30Xa-Amp-T5 cassette transformed into M15 without an insert served as a control.
Competent Cell Preparation
The method of Chang et al. [45] was adapted for competent cell preparation. The E. coli XL and M15 cells were inoculated in 50 mL LB broth separately and grown at 37 °C to obtain an OD600 of 0.6. The whole E. coli culture (M15 and XL separately) was kept on ice for 10 min, and the harvested cells were re-suspended in 10 mL of ice-cold 0.1 M MgCl2. After washing the pellet with 0.1 M CaCl2, the cells were incubated on ice for 1 h followed by resuspension in 0.5 mL of ice-cold CaCl2 containing 15% glycerol. CaCl2 treated E. coli M15, and XL cells were stored in aliquots at − 80 °C.
Transformation
The purified CtXR digested with BamH1 and Kpn1 restriction enzymes was ligated into the pQE30Xa vector at a vector: insert ratio of 1:3. Ten microliters of ligation mixture were gently mixed with 100 µL of the competent E. coli XL1 cells and incubated on ice for 30 min followed by a heat shock at 42 °C for 90 s in a water bath. The tubes were immediately cooled on ice for 2–3 min and, after adding 800 µL of freshly-prepared pre-warmed (37 °C) LB broth, incubated at 37 °C for 1 h. Approximately 50 µL of cell suspension was spread on the surface of LB agar plates containing ampicillin (100 µg mL−1) and tetracycline (20 µg mL−1), and incubated at 37 °C overnight. To transform pQE30Xa –CtXR-T5-lac into the E. coli M15 cells, the cocktail comprised of 2 µL recombinant pQE30XaCtXR plasmid and 100 µL of E. coli M15 competent cells and bacterial transformation were performed as mentioned for the E. coli XL1 strain. After 1 h, 100 µL of the bacterial culture was plated on selection plates and incubated overnight at 37 °C for growth. Plasmid construction is schematically represented in Fig S1.
Overexpression and Purification of Recombinant XR Protein
Three transformants generated from the expression plasmid were selected for the protein expression study. The E. coli M15 pQE30-CtXR cells (CtXRΔ) were induced to produce CtXR by the addition of 1 mM isopropyl-ß-d-thiogalactopyranoside (IPTG) to obtain a final OD600 of 0.6. The culture was kept at 28 °C for 6 h for the induction of XR protein with orbital shaking at 220 rpm. The M15 CtXRΔ cells harboring pQE30-CtXR without IPTG induction served as a control.
Preparation of Crude Cell Lysate
The cells from both uninduced and induced cultures of E. coli M15 CtXRΔ were harvested by centrifugation at 6000 rpm for 5 min and suspended in phosphate-buffered saline (pH 8.0). The cell pellets were sonicated using a probe ultrasonicator for 2 min (30 s sonication and 10 s “gap” cycle) an amplitude of 40%. After the sonication, the cell suspension was centrifuged at 10,000 rpm for 20 min, chilled at 4 °C, and the supernatants were loaded onto 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels (Bio-Rad®) for analysis of the XR expression. The gels were stained in a 0.2% Coomassie Brilliant Blue R250 solution with continuous shaking for 2 h followed by destaining in methanol:water:acetic acid (2:7:1, v/v/v) until clear bands were observed [46].
Purification of the XR Protein
CtXR extracted from the induced E. coli M15 cells was purified using the Ni–NTA column (Qiagen, Germany) following the manufacturer’s instructions. The different fractions collected were analyzed and confirmed in SDS-PAGE gels. Protein concentrations in cell extracts were quantified by Bradford assay (Bio-Rad) with bovine serum albumin as a standard [47].
XR protein Characterization Using Matrix-Assisted Laser Desorption/Ionization-Time of Flight (MALDI-TOF)
The purified XR protein resolved in SDS-PAGE gels was excised using a sterile scalpel and suspended in sterile water. Mass spectrometry detection of the trypsin digested protein with the AutoFlex MALDI-TOF system (Bruker Dalton, Germany) was performed with a cyano-4-hydroxycinnamic acid matrix (CHCA; Sigma), where 30 mg of the matrix dissolved in 200 µL acetonitrile and 100 µL of 0.3% trifluoroacetic acid (TFA). The matrix mixture was vortexed for 5 min and then centrifuged at maximum speed (16,000×g) for 2 min followed by the addition of 100 µL of the supernatant to 300 µL of isopropanol and vortexed for 10 s. Approximately 30 µL of this matrix mixture was spotted to the center of the MALDI plate and quickly smeared throughout the grid using the pipette tip. The “precoat” was allowed to air dry at 37 °C for 3 min. Similarly, the second round of 5 µL of the matrix supernatant was added to 5 µL of each sample in a 0.2-mL tube and mixed thoroughly. Two microliters of the sample mixture were loaded onto the precoated grid spots on the MALDI plate and kept to air dry for 3 min. MALDI-TOF analysis was performed by the Proteomics Facility at the Indian Institute of Sciences, Bangalore, India.
MALDI-TOF Analysis and Database Search
The identification and differentiation of XR proteins were done by peptide mass fingerprinting. Search parameters of 300 ppm mass error tolerance for parent ions and missed cleavage of trypsin digestion were used. Oxidation was chosen as a possible modification. The top MASCOT scoring hit for each MALDI spot was used to determine the peptide type. The peptide coverage, score obtained, and unique peptide matches were also considered for peptide characterization. The m/z values and the peak file obtained from MALDI-TOF results were analyzed in the MASCOT server (https://www.matrixscience.com).
Bioinformatics Analysis
The XR gene sequences were translated into their respective amino acids, and then aligned using the multiple sequence alignment tools in BioEdit version 7.2.5. The protein sequences of various XR were retrieved from NCBI (https://www.ncbi.nlm.nih.gov) or RCSB (https://www.rcsb.org) data repositories. The retrieved protein sequences were subjected to homology modeling using the SWISS-MODEL [48], an automated protein structure homology modeling server. The Computed Atlas of Surface Topography of proteins (CASTp) 3.0 was used to predict the active sites in the protein structure [49]. This online server was used to identify and measure voids in 3D protein structures. The modeled protein 3D structure of XR was submitted to the server, and the binding site residues were predicted.
XR Activity
XR activity was determined using the protocol described by Smiley and Bolen [50] and Veras et al. [51]. E. coli pQE30-CtXR (M15 CtXRΔ) cells were harvested from an exponentially growing culture and washed with phosphate buffer. The cells were lysed in a probe sonicator, and a cell-free supernatant was prepared. The total protein in the cell-free lysate was determined by the Bradford assay [47]. The XR reaction mixture in a 96-well microtiter plate consisted of the following components: 20 µL phosphate buffer (pH 7.4), 20 µL mercaptoethanol (0.1 M), 20 µL enzyme solution, 20 µL NADPH (3.4 mM), 20 µL d-xylose (0.5 M), and 100 µL distilled water. The changes in the optical density at 340 nm (OD340) at an interval of 10 s were detected in a microplate reader up to 10 min, which corresponds to a cofactor conversion. XR activity (U mg−1 protein) was calculated using 6.22 mL (µmol cm−1) as the molar absorption coefficient. One enzyme unit (1 U) was defined as 1 µmol of cofactor oxidation or reduction min−1.
Optimization of Assay Conditions
Different assay parameters such as pH, temperature, and carbon sources were optimized for XR activity. To determine the optimal pH for XR activity, the pH of the medium was maintained using different buffers in the range of 3–10 (citrate buffer pH 3, 4, and 5; phosphate buffer pH 6, 7, and 8; and carbonate buffer pH 9 and 10). Similarly, temperature between 20 and 45 °C was used to determine the optimal temperature for XR activity. The optimal substrate was determined using different 5-C sugars (xylose, arabinose, and rhamnose) and 6-C sugars (glucose, galactose, mannose, and mannitol).
Kinetic Parameters
Michaelis–Menten kinetics and Lineweaver–Burk plots for a single-substrate reaction were determined for the XR synthesized by M15 CtXRΔ. The Michaelis–Menten equation is as follows:
where v is the reaction velocity, Vmax is the maximum velocity, Km is the Michaelis constant, and [S] is the substrate concentration.
The apparent kinetic parameters (Vmax and Km) of XR were determined at different levels of substrate up to 80 mM of xylose. Data were analyzed using the Graph Pad Prism software (version 5.0). The Km and Vmax values were compared with the Lineweaver–Burk plot using reaction rates (1/v) and substrate concentrations (1/S). The significance and best fit of the plot were obtained using their mean R2 values.
Biotransformation of d-Xylose to Xylitol Using Recombinant E. coli M15
Overnight-grown E. coli M15 CtXRΔ culture was inoculated at 2% in a 50 mL minimal medium of pH 7 (3.5 g KH2PO4, 5.0 g K2HPO4, 3.5 g (NH4)H2PO4, 15 mg CaCl2·2H2O, 0.25 g MgSO4·7H2O, 1 mL trace elements, and 1000 mL demineralized water) containing 25 µg mL−1 kanamycin and 100 µg mL−1 ampicillin and incubated at 37 °C. After attaining a 0.6 optical density at 600 nm (OD600), IPTG was added at a final concentration of 1 mM. d-xylose, glucose, and glycerol were added separately in the following combinations: d-xylose (100 mM); d-xylose (100 mM) with glucose (50 mM); and d-xylose (100 mM) with glycerol (15 mL L−1). The inoculated culture was incubated at 30 °C for 24 h. The samples were withdrawn every 1 h and centrifuged to collect the cell-free supernatant followed by filtration through a 0.22 µm filter. The supernatant was used for further purification. The cell-free supernatants were analyzed for xylitol production and d-xylose consumption by a high-performance liquid chromatography (HPLC) system.
Xylitol Production from Corncob Hydrolysate Using Recombinant E. coli
Pretreatment of Corncob Biomass
Approximately 100 g of pulverized corncob was treated with 60 mL of 2% 1 M NaOH, incubated in a water bath at 70 °C for 5 h and then allowed to cool at room temperature. The treated alkaline mixture of corncob was centrifuged at 14,000 rpm for 20 min, and the supernatant was adjusted to pH 5 using 6 N HCl. For the precipitation of sugars in the supernatant, three-volume of cold ethanol was added and centrifuged at 3000 rpm. The obtained pellet was treated with 1.5% diluted H2SO4 and kept in a water bath at 75 °C for 30 min. The solution was neutralized with 1 M NaOH and detoxified using 2% activated charcoal. The detoxified hydrolysate was used as a substrate for xylitol production (Kumar et al., 2015). Actively growing E. coli M15 CtXRΔ cells were inoculated in the corncob hydrolysate containing minimal medium with an initial reaction volume of 5 mL and incubated at 30 °C for 24 h. The cells were reused in multiple rounds of reaction and scaled up by ten folds (50 mL final volume).
To remove the impurities and discoloration of the fermented broth, 4 g 100 mL−1 activated charcoal was added to the fermentation broth and kept for 1 h at 60 °C. After incubation, the fermentation broth was filtered through Whatman filter paper grade no. 1.
Xylitol Crystallization
To attain saturation, the purified fermentation broth was concentrated in a vacuum evaporator (Eppendorf concentrator Plus™) at 60 °C to attain saturation. The concentrated solution was mixed with four volumes of ethanol, stirred, and incubated at − 40 °C at 48 h. After incubation, the solution was centrifuged at 4000 rpm for 10 min. The final pellet was dried in a vacuum drier, and the crystallized sugar was weighed. The crystals containing xylitol were dissolved in water, and their purity was analyzed by an HPLC system. The xylose to xylitol bioconversion percentage was calculated using the molar concentration of xylose present at the beginning of the reaction and the molar concentration of xylitol produced at the end of the reaction.
Xylitol Quantification
Xylitol quantification was carried out in an HPLC using an Agilent system equipped with Agilent Technologies 1260 infinity evaporative light scattering detection (ELSD) (Palo Alto, USA). All samples and standards were filtered through 0.25 µm (Millipore) filters. The separation was done in a Shimadzu-NH2 column (250 mm × 4.6 mm, i.e., 5 µm particle size). The mobile phase consisted of acetonitrile and water (80:20 v/v) in an isocratic elution with a flow rate of 1.5 mL min−1. The column temperature was maintained at 20 °C. The detection was carried out by the ELSD detector. The evaporative temperature and nebulizing temperature were set at 95 °C and 80 °C, respectively, and the N2 gas flow rate was set at 1.60 standard liters per minute.
Statistical Analysis
All experiments were performed in triplicate. Statistical analyses were performed using the GraphPad Software Prism v8.0.2 (San Diego, California, USA). Data were reported as means ± standard deviation. *p < 0.05, **p < 0.005 and ***p < 0.005.
Results
To identify the putative XR genes in the mesophilic C. tropicalis GRA1 isolated from grapes, investigations on the xylose to xylulose reversible pathways of Candida sp., Saccharomyces sp., and Hansenula sp. were carried out on the Kyoto encyclopedia of genes and genomes platform. BLAST analysis revealed 98.76% sequence similarity with the previously described XR of C. tropicalis (NCBI MF143598.1). The activity of NADP-dependent XR (EC 1.1.1.307) in the wild C. tropicalis was 355.15 U. In our previous studies CtXR gene was knocked out and cloned into pGEMT vector and maintained in E. coli DH5α cells [44]. Before sub-cloning, the CtXR gene was authenticated by sequencing and similarity analysis (Fig. 1).
Cloning and Overexpression of CtXR in a Prokaryotic System
PCR amplification of CtXR from C. tropicalis GRA1 with primers CtXR 2F and CtXR 2R exhibited a 1000 bp fragment, later cloned in the expression vector pQE30Xa-T5-Amp. Successful cloning of the resulting pQE30Xa-CtXR expression plasmid was confirmed by releasing insert using restriction digestion BamH1 and Kpn1 and performing sequencing analysis. Restriction digestion of pQE30Xa–CtXR plasmid isolated from the positive transformants with BamH1 and Kpn1 restriction sites released the CtXR insert of size 1000 bp. The PCR amplification of pQE30–CtXR plasmid with the T5 upstream promoter region yielded a PCR amplicon with an additional 189 bp and CtXR (Fig. 2).
The pQE30Xa–CtXR expression cassette was transformed in M15 E. coli. The transformants that grew successfully on the selection plates containing X-gal were selected based on beta-galactosidase alpha-complementation and further confirmed using CtXR specific colony PCR analyses. To characterize CtXR, E. coli pQE30–CtXR cells were induced to produce CtXR protein by adding 1 mM IPTG (0.6 OD600).
CtXR Characterization by MALDI-TOF
To characterize CtXR protein, SDS-PAGE and enzyme activity assays with crude cell lysates of E. coli M15 strains overexpressing CtXR were performed. SDS-PAGE of the cell lysates showed the presence of 37 kDa CtXR in the induced E. coli M15 cells harboring pQE30-CtXR, which was absent in the uninduced cells (Fig S2). In addition, pQE30-CtXR harboring E. coli M15 contained 5.32 mg mL−1 protein. MALDI-TOF results reported peptide coverage, m/z value, and area coverage of peptides (Fig. 3 and Table S3). The m/z and peak files analyzed in the MASCOT server (https://www.matrixscience.com) reported peptide match, score, and other functional properties (Fig. S3). CtXR of C. tropicalis XYL1 showed a higher score value of 101 and 23 peptide matches. The results provide evidence that CtXR encoded the predicted XR enzymes.
Bioinformatics Analysis of XR
The three-dimensional structure of XR was developed by the SWISS-MODEL server. The binding site of XR was predicted by the CASTp server by submitting the three-dimensional structure of XR to their protein data bank file format. The active site residues were determined from the predicted binding site (Fig. 4). The XR consisted of the following 18 amino acids in its binding site: TRP, LEU, SER, GLN, VAL, MET, LYS, THR, GLY, PRO, ILE, ASN, GLU, TYR, CYS, PHE, ALA, and ASP.
XR Activity of CtXR E. coli
The XR activity was analyzed using the purified recombinant CtXR. The assay conditions were optimized with pH ranging from 3.0 to 9.0 (Fig. 5). The maximum XR activity was observed at pH 7 (244.24 U mg−1), followed by pH 6 (188.52 U mg−1), and a minimum XR activity was observed at pH 3 (52.76 U mg−1). Similarly, the lysates showed maximum XR activity (270.15 U mg−1) at 35 °C followed by 40 °C (183.77 U mg−1), which declined after that. However, CtXR activity was considerably reduced at psychrophilic temperature, which was 44.46 U mg−1 at 20 °C. When xylose and NADPH were provided as the substrates, lysates of E. coli M15 CtXRΔ showed a 195% increase in NADPH to NADP+ conversion. The relative activity with galactose as a substrate showed a 462.8% increase, followed by mannitol 295.5% and xylose 206%. Lysates of E. coli M15 CtXRΔ exhibited a decreased conversion when mannose and NADPH were provided. This provides clear evidence that CtXR encoded the predicted, which converted the expected substrates and cofactors (xylose and NADPH for CtXR) (Fig. 5).
Optimization of CtXR protein a CtXR activity was measured between the temperature range of 20–45 °C, and the optimal temperature was found to be 35 °C. b pH optima of CtXR protein was measured from pH 3 to 9. Optimum pH for CtXR activity was found to be pH 7. Activity of CtXR protein was determined using 0.5 M substrate and 3.4 mM of NADPH. c Substrate specificity was carried out using 0.5 M substrate at pH 7, temperature 35 °C and 3.4 mM of NADPH. Arabinose was the preferred substrate registering maximum activity followed by xylose and rhamnose. d Relative activity was measured for all the substrates
Kinetic Analysis
XR was assayed with the substrate D-xylose in limiting concentrations and NADPH in non-limiting concentrations. The Michaelis–Menten plot (Fig. 6) showed the best fit values of Km and Vmax in the range of 193 to 221 mM and 7.6 to 11.9 µmol min−1, respectively at a 95% confidence interval (Table 1). The optimized Vmax and Km values for M15 CtXRΔ are 207.4 mM D-xylose and 9.77 µmol min−1, respectively. The Lineweaver–Burk plot considers the rate of reaction where the Vmax and Km values are 58.44 mM and 27.39 µmol min−1, respectively. Both the models reported the best fit with an R2 value of 0.96 and 0.92, respectively (Fig. 6).
Xylitol Production by Genetically Engineered M15 Strain
Xylitol production by M15 CtXRΔ whole cells was analyzed in a shake flask fermentation using different glucose, D-xylose, and glycerol combinations. Overexpression of CtXR synthesized XR protein in the cytoplasm upon IPTG induction. Both the wild strain and uninduced recombinant did not produce xylitol. Xylitol production was detected periodically by HPLC. A minimum quantity of xylitol (1.5 g L−1) at 12 h of fermentation with very low xylose was observed. When the fermentation duration was increased up to 20 h, the xylose utilization rate also gradually increased. However, the conversion rate of xylitol was relatively low (2.1 g L−1). The second combination containing glucose as an NADPH generating source (xylose + glucose) ensued a two-fold increase in xylitol accumulation after 24 h of fermentation, yielding a xylitol titer of 3.4 g L−1. E. coli M15 CtXRΔ cells continued to grow utilizing xylose when glucose was depleted in the fermentation medium. Therefore, the initial uptake of xylose was lower than that of glucose. Glucose was completely utilized after 24 h of fermentation, and unutilized xylose was left in the fermentation medium. Xylose, along with glycerol as a co-substrate, significantly increased xylose consumption, which resulted in a higher yield of xylitol. After 24 h of fermentation, production of xylitol increased by three folds yielding 6.4 g L−1 with the specific productivity of 0.67 g L−1 xylose and the volumetric productivity of 0.28 g L−1 h−1 by consuming all the xylose and glycerol present in the fermentation medium (Table 2). The authors have previously studied the process curve for xylitol production from different carbon sources using engineered M15 cells [31].
Enhanced Xylitol Production from Corncob Hydrolysate Using M15 pQE30CtXR Whole Cells
To better assess the potential of M15 CtXRΔ for xylitol production using lignocellulosic biomass on a laboratory scale, a mini bioreactor with optimized conditions was used for fermentation. Proximate compositional analysis of corncob reported the total cellulose, hemicellulose, and lignin contents as 35.86%, 31.1%, and 19.87%, respectively. Corncob hydrolysate with glycerol as a co-substrate was used as a fermentation medium for CtXRΔ cells. The xylitol production rate increased after 12 h, and a yield of 3.7 g L−1 hydrolysate was obtained after 24 h of fermentation with specific productivity of 0.57 g L−1 xylose. The conversation rate of xylose to xylitol in the corncob hydrolysate was 57.8%. Finally, the xylitol pellets obtained after purification were dried, and the yield after the crystallization process was 3.2 g L−1 with a recovery of 70.27% (Fig. 7).
Discussion
Corncob hydrolysate is an abundant feedstock and biomass byproduct in India, which can be optimized to overproduce xylitol using engineered whole cells of E. coli M15 using a suitable co-substrate. The natural metabolic pathway of xylose isomerization to xylitol and conversion to xylulose 5-phosphate is well known in E. coli and other bacteria. The conversion of xylose to xylitol by XR is the first step of the pentose phosphate pathway-dependent xylose metabolism in yeast and fungi. The two-step pathway in yeast and fungi is mediated by the enzyme XR and XDH accompanied by cofactor consumption and regeneration [52]. Yeast shows a slow growth rate because of the higher affinity of hexose transporters to glucose than that with xylose [25, 53]. Metabolic reconstruction of xylose metabolism in E. coli is one of the important strategies because of the fast growth, easy genetic manipulation, and low cost of E. coli. More importantly, E. coli has native xylose transporters, and the CRP-cAMP complex positively regulates the catabolic operons for secondary sugars derived from lignocellulosic biomass under glucose-limited conditions [53,54,55]. Even so, the presence of CCR makes E. coli preferentially metabolize glucose over xylose [21]. The XR of a mesophilic yeast strain, C. tropicalis GRA1 was overexpressed in E. coli M15 cells to improve xylitol productivity.
In this study, we analyzed the XR nucleotide sequence and provided data that reported the function of XR in xylose metabolism by MALDI-TOF. The XR gene was cloned, and the heterologously expressed XR encoded for a putative XR protein. Overexpression of XR in M15 CtXRΔ enabled higher xylitol production in the minimal medium supplemented with xylose. Moreover, lysates of XR overexpressed in E. coli strain M15 CtXRΔ exhibited the higher enzymatic activity with xylose as a substrate and NADPH as a cofactor. The CtXR yield synthesized by the recombinant M15 CtXRΔ was about 5.32 mg mL−1 and the protein size was 37 kDa. CtXR contained a highly conserved active sites region with a tetrad of residues (Tyr51, Lys80, His113, and Asp46) situated at the substrate-binding pocket base. A side-stereochemistry of transferred hydride from NADH and a positional conserved catalytic tetrad residue of aldo–keto reductase (AKR) (Tyr51, His113, Lys80, and Asp46 in XR from C. tenuis CBS 4435) were in good agreement with the classification of XR in the AKR superfamily. Starting from a common tyrosine, yeast XRs contained two conserved sequence motifs corresponding to the catalytic signatures of the single-domain reductases/epimerases/dehydrogenases and AKRs. AKR2B5 (XR from C. tenuis) is a well-characterized member of the AKR superfamily. Members of the AKR superfamily and single-domain reductase/epimerase/dehydrogenase superfamily do not show detectable similarities at their primary structure level and three-dimensional scaffold [9]. MALDI-TOF results reported a higher score of 101 with 23 peptide matches with C. tropicalis 5842 NADPH-dependent XR (EC = 1.1.1.307) [56].
M15 CtXRΔ lysates reported more relative enzymatic activity with xylose as a substrate and NADPH as a cofactor than other sugars. XR preferentially uses NADPH as a cofactor, and a conserved Isopentenyl Phosphate Kinases (IPKs) amino acid motif determines its specificity [57,58,59]. CtXRΔ lysates exhibited a decreased conversion when mannose and NADPH were provided. These results reported that different metabolic patterns due to different carbohydrates such as glucose or mannose and not xylose strongly regulate the sugar metabolism genes through CCR. The specific XR activity was maximum at pH 7 and 35 °C. Thus, CtXR encoded the predicted XR, which converted the expected substrates and cofactors (xylose and NADPH for CtXR). Previous reports suggest that NADPH-linked XR activity linearly increased when the temperature increased from 20 to 38 °C and when the pH increased from 3 to 9 [60,61,62]. Further, the affinity of M15 CtXRΔ is greater for 6-C sugars than for 5-C sugars. Such variations in the substrate specificity of CtXR can be due to minor modifications in the surface active sites (aldo–keto residues). The structure of XR also decides the substrate affinity.
The NADPH/NADP+ cofactor plays a major role in xylitol production by oxidizing NADPH to NAPD+ in recombinant organisms. An enzyme system (such as NADPH) that oxidizes a co-substrate is normally used in whole-cell biocatalytic processes involving oxidoreductases to regenerate NADPH. Two different cofactor regeneration systems were used with xylose (100 mM) as a control to study the xylitol production kinetics. The xylose catabolic reaction by M15CtXRΔ whole cells diverts the flux through xylose isomerization and pentose phosphate pathway so that the overexpression of CtXR substantially increases the intracellular NADPH availability. When xylose was the sole carbon source, the xylitol yield was 0.21 g. The Vmax and Km values for D-xylose are 207.4 mM and 9.77, respectively. The enzyme exhibited a greater cofactor specificity for NADPH and preferred xylose as a carbon source among all the investigated 5-C sugars [62]. While augmenting glucose, it increased the cell biomass yield compared with xylose as the sole C source, with the simultaneous increase in xylitol yield. Inhibiting xylose by glucose significantly reduces the xylitol production rates in C. mogi [63]. Interestingly, M15CtXRΔ whole cells in the synthetic medium augmented with the combination of glucose and xylose yielded more xylitol compared with only xylose. At low glucose concentrations, the negative effect of glucose had not affected the xylitol productivity and NADP + accumulation. As shown in Fig. 7, M15CtXRΔ cells utilized glucose more preferentially compared with xylose. A similar phenomenon was previously reported in Kluyveromyces marxianus [64], S. cerevisiae [65], and C. tropicalis [18]. The present study also reported that the recombinant CtXR exhibited an oxidoreductase coupling system in the presence of NADPH for the conversion of xylose to xylitol. Previous studies have reported the cytosolic NADPH/NADP + ratios in the range of 15–60, which drives the synthesis of nucleic acid and fatty acids [66, 67]. Glycerol as an auxiliary substrate along with xylose significantly improved the xylitol yield by three folds. Previous reports suggest that glycerol as a substrate can increase NADPH supply [68]. Co-expression of P. stipitis glycerol dehydrogenase genes in C. tropicalis BSXDH-3 facilitated the abundant supply of NADPH cofactor and increased xylitol productivity by ∼30% [69]. The glycerol addition is hypothesized to increase xylose to xylitol conversion by NADPH regeneration and diverting the entire XR pathway toward the xylitol synthesis. The cells must recycle NADPH generated through glycerol catabolism for the efficient utilization of xylose in the presence of glycerol.
Hemicellulosic hydrolysates from different lignocellulosic biomasses such as corncob, sugarcane bagasse, wheat straw, rapeseed straw, brewer’s spent grain, cashew apple, and many other substrates are the most common renewable substrates for xylitol production [14, 70, 71]. The pretreatment and costly steps of detoxification are the major bottlenecks for biorefineries that produce xylitol. Other drawback includes the low tolerance of yeast toward inhibitory compounds such as acetic acid, furfural, and hydroxymethylfurfural [71, 72]. In this study, the production of xylitol from alkali pretreated corncob hydrolysate yielded 0.57 g xylitol for 1 g xylose and 0.15 g L−1 h−1 volumetric productivity. After crystallization and purification, we achieved a total recovery of 70%. The xylitol yield and productivity from corncob hydrolysate by different yeast strains are given in Table 3. Cofactor regeneration is a rate-limiting step in the valorization of biomass hydrolysate to xylitol. Here, E. coli strain M15 reconstructed by heterologous recombination with XR from C. tropicalis GRA1 lead to the carbon flux toward the pentose phosphate pathway/transhydrogenase activity on NADPH generation.
Previous studies have reported the significance of direct NADPH supply by NADP+-utilizing enzymes for driving the heterologous NADPH-dependent reactions (Table 3). Our study also attempted strain modifications to improve the coupling between glucose catabolism (oxidation) and xylose reduction. Our results also indicated that co-utilization of cellobiose and xylose in the hydrolysate could improve xylitol production through increased xylose uptake and efficient cofactor regeneration [73]. As promising results were obtained from this two-step bioconversion of corncob to xylitol, upscaling the production in optimized bioreactors will pave the way for xylitol production from lignocellulosic biomass on an industrial scale.
Conclusion
Overexpression of the endogenous XR from C. tropicalis GRA1 in the robust E. coli M15 with inherent xylose transporters resolved the limitations of xylose bioconversion to xylitol in yeast. Genetic modifications coupled with glucose catabolism followed by xylose reduction and cofactor regeneration increased the xylitol productivity. The auxiliary substrate glycerol in the fermentation medium diverted the carbon flux toward pentose phosphate pathway/transhydrogenase activity on NADPH generation. Under the optimized conditions, the metabolically engineered E. coli M15 biocatalyzed the production of xylitol in shake flask fermentation. In addition, we also obtained a remarkable yield of xylitol from alkali pretreated and detoxified corncob. The step-wise engineering strategy and xylitol production process from corncob hydrolysate can be further up scaled for the synthesis of ecofriendly and high-value green chemicals.
Data Availability
All data generated or analyzed during this study are included in this article and its supplementary information files.
References
Chattopadhyay, S., Raychaudhuri, U., Chakraborty, R.: Artificial sweeteners—a review. J. Food Sci. Technol. 51, 611–621 (2014)
Lugani, Y., Sooch, B.S.: Xylitol, an emerging prebiotic: a review. Int J. Appl. Pharm. Biol. Res. 2, 67–73 (2017)
Arcaño, Y.D., García, O.D.V., Mandelli, D., Carvalho, W.A., Pontes, L.A.M.: Xylitol: a review on the progress and challenges of its production by chemical route. Catal. Today 344, 2–14 (2020)
Ravella SR, Gallagher J, Fish S, Prakasham RS: Overview on commercial production of xylitol, economic analysis and market trends. In: d-Xylitol, pp. 291–306. Springer, New York (2012)
de Paula, R.G., Antoniêto, A.C.C., Ribeiro, L.F.C., Srivastava, N., O’Donovan, A., Mishra, P.K., Gupta, V.K., Silva, R.N.: Engineered microbial host selection for value-added bioproducts from lignocellulose. Biotechnol. Adv. 37, 107347 (2019)
Van Dyk, J.S., Pletschke, B.I.: A review of lignocellulose bioconversion using enzymatic hydrolysis and synergistic cooperation between enzymes—factors affecting enzymes, conversion and synergy. Biotechnol. Adv. 30, 1458–1480 (2012)
Granström, T.B., Izumori, K., Leisola, M.: A rare sugar xylitol. Part II: biotechnological production and future applications of xylitol. Appl. Microbiol. Biotechnol. 74, 273–276 (2007)
Rafiqul, I.S.M., Sakinah, A.M.M.: Processes for the production of xylitol—a review. Food Rev. Intl. 29, 127–156 (2013)
Nidetzky, B., Klimacek, M., Mayr, P.: Transient-state and steady-state kinetic studies of the mechanism of NADH-dependent aldehyde reduction catalyzed by xylose reductase from the yeast Candida tenuis. Biochemistry 40, 10371–10381 (2001)
Winkelhausen, E., Kuzmanova, S.: Microbial conversion of D-xylose to xylitol. J. Ferment. Bioeng. 86, 1–14 (1998)
Jeffries, T.W., Jin, Y.S.: Metabolic engineering for improved fermentation of pentoses by yeasts. Appl. Microbiol. Biotechnol. 63, 495–509 (2004)
Atzmüller, D., Ullmann, N., Zwirzitz, A.: Identification of genes involved in xylose metabolism of Meyerozyma guilliermondii and their genetic engineering for increased xylitol production. AMB Express 10, 1–11 (2020)
Chen, X., Jiang, Z.-H., Chen, S., Qin, W.: Microbial and bioconversion production of D-xylitol and its detection and application. Int. J. Biol. Sci. 6, 834 (2010)
Rao, L.V., Goli, J.K., Gentela, J., Koti, S.: Bioconversion of lignocellulosic biomass to xylitol: an overview. Biores. Technol. 213, 299–310 (2016)
Sampaio, F.C., Silveira, W.B.D., Chaves-Alves, V.M., Passos, F.M.L., Coelho, J.L.C.: Screening of filamentous fungi for production of xylitol from D-xylose. Braz. J. Microbiol. 34, 325–328 (2003)
Akinterinwa, O., Cirino, P.C.: Heterologous expression of D-xylulokinase from Pichia stipitis enables high levels of xylitol production by engineered Escherichia coli growing on xylose. Metab. Eng. 11, 48–55 (2009)
Louie, T.M., Louie, K., DenHartog, S., Gopishetty, S., Subramanian, M., Arnold, M., Das, S.: Production of bio-xylitol from d-xylose by an engineered Pichia pastoris expressing a recombinant xylose reductase did not require any auxiliary substrate as electron donor. Microb. Cell Fact. 20, 1–13 (2021)
Zhang, L., Chen, Z., Wang, J., Shen, W., Li, Q., Chen, X.: Stepwise metabolic engineering of Candida tropicalis for efficient xylitol production from xylose mother liquor. Microb. Cell Fact. 20, 1–12 (2021)
de Albuquerque, T.L., da Silva Jr, I.J., de Macedo, G.R., Rocha, M.V.P.: Biotechnological production of xylitol from lignocellulosic wastes: a review. Process Biochem. 49, 1779–1789 (2014)
Buijs, N.A., Siewers, V., Nielsen, J.: Advanced biofuel production by the yeast Saccharomyces cerevisiae. Curr. Opin. Chem. Biol. 17, 480–488 (2013)
Park, J.M., Vinuselvi, P., Lee, S.K.: The mechanism of sugar-mediated catabolite repression of the propionate catabolic genes in Escherichia coli. Gene 504, 116–121 (2012)
Parachin, N.S., Bergdahl, B., van Niel, E.W.J., Gorwa-Grauslund, M.F.: Kinetic modelling reveals current limitations in the production of ethanol from xylose by recombinant Saccharomyces cerevisiae. Metab. Eng. 13, 508–517 (2011)
Young, E.M., Tong, A., Bui, H., Spofford, C., Alper, H.S.: Rewiring yeast sugar transporter preference through modifying a conserved protein motif. Proc. Natl. Acad. Sci. 111, 131–136 (2014)
Karhumaa, K., Fromanger, R., Hahn-Hägerdal, B., Gorwa-Grauslund, M.-F.: High activity of xylose reductase and xylitol dehydrogenase improves xylose fermentation by recombinant Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 73, 1039–1046 (2007)
Du, J., Li, S., Zhao, H.: Discovery and characterization of novel d-xylose-specific transporters from Neurospora crassa and Pichia stipitis. Mol. BioSyst. 6, 2150–2156 (2010)
Khankal, R., Chin, J.W., Cirino, P.C.: Role of xylose transporters in xylitol production from engineered Escherichia coli. J. Biotechnol. 134, 246–252 (2008)
Hasona, A., Kim, Y., Healy, F.G., Ingram, L.O., Shanmugam, K.T.: Pyruvate formate lyase and acetate kinase are essential for anaerobic growth of Escherichia coli on xylose. J. Bacteriol. 186, 7593–7600 (2004)
Sumiya, M., Davis, E.O., Packman, L.C., McDonald, T.P., Henderson, P.J.: Molecular genetics of a receptor protein for D-xylose, encoded by the gene xylF, in Escherichia coli. Recept. Chann. 3, 117–128 (1995)
Ahlem, C., Huisman, W., Neslund, G., Dahms, A.S.: Purification and properties of a periplasmic D-xylose-binding protein from Escherichia coli K-12. J. Biol. Chem. 257, 2926–2931 (1982)
Sofia, H.J., Burland, V., Daniels, D.L., Plunkett Iii, G., Blattner, F.R.: Analysis of the Escherichia coli genome. V. DNA sequence of the region from 76.0 to 81.5 minutes. Nucleic Acids Res. 22, 2576–2586 (1994)
Ariyan, M., Uthandi, S.: Xylitol production by xylose reductase over producing recombinant Escherichia coli M15. Madras Agric. J. 106, 205–209 (2019)
Kawaguchi, H., Vertès, A.A., Okino, S., Inui, M., Yukawa, H.: Engineering of a xylose metabolic pathway in Corynebacterium glutamicum. Appl. Environ. Microbiol. 72, 3418–3428 (2006)
Wang, Y., Shi, W.-L., Liu, X.-Y., Shen, Y., Bao, X.-M., Bai, F.-W., Qu, Y.-B.: Establishment of a xylose metabolic pathway in an industrial strain of Saccharomyces cerevisiae. Biotech. Lett. 26, 885–890 (2004)
Young, E., Lee, S.-M., Alper, H.: Optimizing pentose utilization in yeast: the need for novel tools and approaches. Biotechnol. Biofuels 3, 1–12 (2010)
Zhang, M., Eddy, C., Deanda, K., Finkelstein, M., Picataggio, S.: Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science 267, 240–243 (1995)
Cirino, P.C., Chin, J.W., Ingram, L.O.: Engineering Escherichia coli for xylitol production from glucose-xylose mixtures. Biotechnol. Bioeng. 95, 1167–1176 (2006)
Qi, X., Zhang, H., Magocha, T.A., An, Y., Yun, J., Yang, M., Xue, Y., Liang, S., Sun, W., Cao, Z.: Improved xylitol production by expressing a novel D-arabitol dehydrogenase from isolated Gluconobacter sp. JX-05 and co-biotransformation of whole cells. Bioresour. Technol. 235, 50–58 (2017)
Jin, L.-Q., Yang, B., Xu, W., Chen, X.-X., Jia, D.-X., Liu, Z.-Q., Zheng, Y.-G.: Immobilization of recombinant Escherichia coli whole cells harboring xylose reductase and glucose dehydrogenase for xylitol production from xylose mother liquor. Biores. Technol. 285, 121344 (2019)
Ostergaard, S., Olsson, L., Nielsen, J.: Metabolic engineering of Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 64, 34–50 (2000)
Watanabe, S., Kodaki, T., Makino, K.: Complete reversal of coenzyme specificity of xylitol dehydrogenase and increase of thermostability by the introduction of structural zinc. J. Biol. Chem. 280, 10340–10349 (2005)
Watanabe, S., Saleh, A.A., Pack, S.P., Annaluru, N., Kodaki, T., Makino, K.: Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein engineered NADP+-dependent xylitol dehydrogenase. J. Biotechnol. 130, 316–319 (2007)
Yan, N.: Structural advances for the major facilitator superfamily (MFS) transporters. Trends Biochem. Sci. 38, 151–159 (2013)
Mussatto, S.I., Santos, J.C.: Roberto IeC: effect of pH and activated charcoal adsorption on hemicellulosic hydrolysate detoxification for xylitol production. J. Chem. Technol. Biotechnol. 79, 590–596 (2004)
Valliammai G: Xylan derived products by fermentation. Master Thesis 2017.
Chang, A.Y., Chau, V.W., Landas, J.A., Pang, Y.: Preparation of calcium competent Escherichia coli and heat-shock transformation. JEMI Methods 1, 22–25 (2017)
He F: Laemmli-sds-page. Bio-protocol 2011:e80–e80.
He F: Bradford protein assay. Bio-protocol 2011:e45–e45.
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., Heer, F.T., de Beer, T.A.P., Rempfer, C., Bordoli, L.: SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018)
Tian, K., Zhao, X., Zhang, Y., Yau, S.: Comparing protein structures and inferring functions with a novel three-dimensional Yau-Hausdorff method. J. Biomol. Struct. Dyn. 37, 4151–4160 (2019)
Smiley, K.L., Bolen, P.L.: Demonstration of D-xylose reductase and D-xylitol dehydrogenase in Pachysolen tannophilus. Biotechnol. Lett. 4, 607–610 (1982)
Veras, H.C.T., Parachin, N.S., Almeida, J.R.M.: Comparative assessment of fermentative capacity of different xylose-consuming yeasts. Microb. Cell Fact. 16, 1–8 (2017)
Rajakumari, S., Grillitsch, K., Daum, G.: Synthesis and turnover of non-polar lipids in yeast. Prog. Lipid Res. 47, 157–171 (2008)
Saloheimo, A., Rauta, J., Stasyk, V., Sibirny, A.A., Penttilä, M., Ruohonen, L.: Xylose transport studies with xylose-utilizing Saccharomyces cerevisiae strains expressing heterologous and homologous permeases. Appl. Microbiol. Biotechnol. 74, 1041–1052 (2007)
Görke, B., Stülke, J.: Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6, 613–624 (2008)
Kim, J.-H., Block, D.E., Mills, D.A.: Simultaneous consumption of pentose and hexose sugars: an optimal microbial phenotype for efficient fermentation of lignocellulosic biomass. Appl. Microbiol. Biotechnol. 88, 1077–1085 (2010)
Yokoyama, S.-I., Suzuki, T., Kawai, K., Horitsu, H., Takamizawa, K.: Purification, characterization and structure analysis of NADPH-dependent D-xylose reductases from Candida tropicalis. J. Ferment. Bioeng. 79, 217–223 (1995)
Kostrzynska, M., Sopher, C.R., Lee, H.: Mutational analysis of the role of the conserved lysine-270 in the Pichia stipitis xylose reductase. FEMS Microbiol. Lett. 159, 107–112 (1998)
Petschacher, B., Leitgeb, S., Kavanagh, K.L., Wilson, D.K., Nidetzky, B.: The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and X-ray crystallography. Biochem. J. 385, 75–83 (2005)
Silva, S.S., Vitolo, M., Pessoa, A., Jr., Felipe, M.G.A.: Xylose reductase and xylitol dehydrogenase activities of D-xylose-xylitol-fermenting Candida guilliermondii. J. Basic Microbiol. 36, 187–191 (1996)
Suzuki, T., Yokoyama, S.-I., Kinoshita, Y., Yamada, H., Hatsu, M., Takamizawa, K., Kawai, K.: Expression of xyrA gene encoding for D-xylose reductase of Candida tropicalis and production of xylitol in Escherichia coli. J. Biosci. Bioeng. 87, 280–284 (1999)
Zhang, J., Zhang, B., Wang, D., Gao, X., Hong, J.: Improving xylitol production at elevated temperature with engineered Kluyveromyces marxianus through over-expressing transporters. Biores. Technol. 175, 642–645 (2015)
Zhang, X., Shen, Y., Shi, W., Bao, X.: Ethanolic cofermentation with glucose and xylose by the recombinant industrial strain Saccharomyces cerevisiae NAN-127 and the effect of furfural on xylitol production. Biores. Technol. 101, 7093–7099 (2010)
Tochampa, W., Sirisansaneeyakul, S., Vanichsriratana, W., Srinophakun, P., Bakker, H.H.C., Chisti, Y.: A model of xylitol production by the yeast Candida mogii. Bioprocess Biosyst. Eng. 28, 175–183 (2005)
Khankal, R., Luziatelli, F., Chin, J.W., Frei, C.S., Cirino, P.C.: Comparison between Escherichia coli K-12 strains W3110 and MG1655 and wild-type E. coli B as platforms for xylitol production. Biotechnol. Lett. 30, 1645–1653 (2008)
Gonzalez, R., Tao, H., Shanmugam, K.T., York, S.W., Ingram, L.O.: Global gene expression differences associated with changes in glycolytic flux and growth rate in Escherichia coli during the fermentation of glucose and xylose. Biotechnol. Prog. 18, 6–20 (2002)
Carmel-Harel, O., Storz, G.: Roles of the glutathione-and thioredoxin-dependent reduction systems in the Escherichia coli and Saccharomyces cerevisiae responses to oxidative stress. Ann. Rev. Microbiol. 54, 439–461 (2000)
Hedeskov, C., Capito, K., Thams, P.: Cytosolic ratios of free [NADPH]/[NADP+] and [NADH]/[NAD+] in mouse pancreatic islets, and nutrient-induced insulin secretion. Biochem. J. 241, 161–167 (1987)
Arruda, P.V., Felipe, M.G.: Role of glycerol addition on xylose-to-xylitol bioconversion by Candida guilliermondii. Curr Microbiol 58, 274–278 (2009)
Ahmad, I., Shim, W.Y., Kim, J.-H.: Enhancement of xylitol production in glycerol kinase disrupted Candida tropicalis by co-expression of three genes involved in glycerol metabolic pathway. Bioprocess Biosyst. Eng. 36, 1279–1284 (2013)
Kumar, S., Dheeran, P., Singh, S.P., Mishra, I.M., Adhikari, D.K.: Bioprocessing of bagasse hydrolysate for ethanol and xylitol production using thermotolerant yeast. Bioprocess Biosyst. Eng. 38, 39–47 (2015)
López-Linares, J.C., Romero, I., Cara, C., Castro, E., Mussatto, S.I.: Xylitol production by Debaryomyces hansenii and Candida guilliermondii from rapeseed straw hemicellulosic hydrolysate. Biores. Technol. 247, 736–743 (2018)
Pereira, I., Madeira, A., Prista, C., Loureiro-Dias, M.C., Leandro, M.J.: Characterization of new polyol/H+ symporters in Debaryomyces hansenii. PLoS ONE 9, e88180 (2014)
Dasgupta, D., Bandhu, S., Adhikari, D.K., Ghosh, D.: Challenges and prospects of xylitol production with whole cell bio-catalysis: a review. Microbiol. Res. 197, 9–21 (2017)
Cheng, H., Wang, B., Lv, J., Jiang, M., Lin, S., Deng, Z.: Xylitol production from xylose mother liquor: a novel strategy that combines the use of recombinant Bacillus subtilis and Candida maltosa. Microb. Cell Fact. 10, 1–12 (2011)
Abd Rahman, N.H., Jahim, J.M., Munaim, M.S.A., Rahman, R.A., Fuzi, S.F.Z., Illias, R.M.: Immobilization of recombinant Escherichia coli on multi-walled carbon nanotubes for xylitol production. Enzyme Microb. Technol. 135, 109495 (2020)
Yuan, X., Tu, S., Lin, J., Yang, L., Shen, H., Wu, M.: Combination of the CRP mutation and ptsG deletion in Escherichia coli to efficiently synthesize xylitol from corncob hydrolysates. Appl. Microbiol. Biotechnol. 104, 2039–2050 (2020)
Reshamwala, S.M.S., Lali, A.M.: Exploiting the NADPH pool for xylitol production using recombinant Saccharomyces cerevisiae. Biotechnol. Prog. 36, e2972 (2020)
da Silva, E.G., Borges, A.S., Maione, N.R., Castiglioni, G.L., Suarez, C.A.G., Montano, I.D.C.: Fermentation of hemicellulose liquor from Brewer’s spent grain using Scheffersomyces stipitis and Pachysolen tannophilus for production of 2G ethanol and xylitol. Biofuels Bioprod. Biorefin. 14, 127–137 (2020)
Kim, S.-Y., Oh, D.-K., Kim, J.-H.: Evaluation of xylitol production from corn cob hemicellulose hydrolysate by Candida parapsilosis. Biotech. Lett. 21, 891–895 (1999)
Li, M., Meng, X., Diao, E., Du, F.: Xylitol production by Candida tropicalis from corn cob hemicellulose hydrolysate in a two-stage fed-batch fermentation process. J. Chem. Technol. Biotechnol. 87, 387–392 (2012)
Ping, Y., Ling, H.-Z., Song, G., Ge, J.-P.: Xylitol production from non-detoxified corncob hemicellulose acid hydrolysate by Candida tropicalis. Biochem. Eng. J. 75, 86–91 (2013)
Mardawati E, Andoyo R, Syukra KA, Kresnowati M, Bindar Y: Production of xylitol from corn cob hydrolysate through acid and enzymatic hydrolysis by yeast. 2018. IOP Publishing.
Prabhu, A.A., Ledesma-Amaro, R., Lin, C.S.K., Coulon, F., Thakur, V.K., Kumar, V.: Bioproduction of succinic acid from xylose by engineered Yarrowia lipolytica without pH control. Biotechnol. Biofuels 13, 1–15 (2020)
Prabhu, A.A., Thomas, D.J., Ledesma-Amaro, R., Leeke, G.A., Medina, A., Verheecke-Vaessen, C., Coulon, F., Agrawal, D., Kumar, V.: Biovalorisation of crude glycerol and xylose into xylitol by oleaginous yeast Yarrowia lipolytica. Microb. Cell Fact. 19, 1–18 (2020)
Baptista, S.L., Costa, C.E., Cunha, J.T., Soares, P.O., Domingues, L.: Metabolic engineering of Saccharomyces cerevisiae for the production of top value chemicals from biorefinery carbohydrates. Biotechnol. Adv. 47, 107697 (2021)
Yuan, X., Wang, J., Lin, J., Yang, L., Wu, M.: Efficient production of xylitol by the integration of multiple copies of xylose reductase gene and the deletion of Embden–Meyerhof–Parnas pathway-associated genes to enhance NADPH regeneration in Escherichia coli. J. Ind. Microbiol. Biotechnol. 46, 1061–1069 (2019)
Shah, S.S.M., Luthfi, A.A.I., Low, K.O., Harun, S., Manaf, S.F.A., Illias, R.M., Jahim, J.M.: Preparation of kenaf stem hemicellulosic hydrolysate and its fermentability in microbial production of xylitol by Escherichia coli BL21. Sci. Rep. 9, 1–13 (2019)
Camargo, D., Sene, L., Variz, D.I.L.S.: de Almeida Felipe MdG: Xylitol bioproduction in hemicellulosic hydrolysate obtained from sorghum forage biomass. Appl. Biochem. Biotechnol. 175, 3628–3642 (2015)
Acknowledgements
The authors acknowledge the Indian Institute of Science Education and Research, (IISER) Kolkata, India, for providing us with the expression host E. coli M15.
Funding
This work was carried out with financial assistance from University Core Project (B27NV-CP016) and previous financial assistance to SU through Indo Russia joint collaboration supported by DBT, GoI, New Delhi (No.DBT/IC2/Indo-Russia/2014–16/04) and DBT-BIOCARe (Sanction No. BT/PR18134/BIC/101/795/2016).
Author information
Authors and Affiliations
Contributions
Sivakumar Uthandi: Conceptualized the research, Funding acquisition, Designed the experiments, and corrected the manuscript. Manikandan Ariyan: Performed the experiments, drafted the manuscript, editing, and software. Sugitha Thankappan: Redrafted the manuscript, Assisting with experimental data analyses, Editing, and software. Priyadarshini Ramachandran: Assisted in the experimentation.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ariyan, M., Thankappan, S., Ramachandran, P. et al. Xylitol Production from Corncob Hydrolysate by an Engineered Escherichia coli M15 as Whole-Cell Biocatalysts. Waste Biomass Valor 14, 3195–3210 (2023). https://doi.org/10.1007/s12649-022-01860-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12649-022-01860-4