Abstract
Objectives
To develop a xylose-nonutilizing Escherichia coli strain for ethanol production and xylose recovery.
Results
Xylose-nonutilizing E. coli CICIM B0013-2012 was successfully constructed from E. coli B0013-1030 (pta-ack, ldhA, pflB, xylH) by deletion of frdA, xylA and xylE. It exhibited robust growth on plates containing glucose, arabinose or galactose, but failed to grow on xylose. The ethanol synthesis pathway was then introduced into B0013-2012 to create an ethanologenic strain B0013-2012PA. In shaking flask fermentation, B0013-2012PA fermented glucose to ethanol with the yield of 48.4 g/100 g sugar while xylose remained in the broth. In a 7-l bioreactor, B0013-2012PA fermented glucose, galactose and arabinose in the simulated corncob hydrolysate to 53.4 g/l ethanol with the yield of 48.9 g/100 g sugars and left 69.6 g/l xylose in the broth, representing 98.6% of the total xylose in the simulated corncob hydrolysate.
Conclusions
By using newly constructed strain B0013-2012PA, we successfully developed an efficient bioprocess for ethanol production and xylose recovery from the simulated corncob hydrolysate.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The increasing worldwide demand for bioethanol has required the use of alternative sources of feedstocks to supplement or replace starch-based glucose because of the concern for competition with food resources. Development of bioprocesses based on non-food feedstocks such as lignocellulosic feedstocks will provide a sustainable route for commercial ethanol production (Banerjee et al. 2010).
Lignocellulosic biomass is an abundant, renewable source of carbohydrates for microbial conversion to chemicals or fuels and lignocellulosic-based bioethanol production represents the most scalable alternative route (Rubin 2008). The main components of lignocellulose are cellulose and hemicellulose, which is composed of various hexose and pentose sugars. Efficient conversion of all the monosaccharides, such as glucose and xylose, is a requisite for profitable production of bioethanol from lignocellulose biomass. However, no natural microorganisms are known to be capable of efficiently converting all monosaccharides of lignocellulose to ethanol. A great deal of effort has gone into the engineering of microbial strains that can consume xylose and glucose simultaneously for efficient ethanol production. For instance, enzymes of the xylose metabolic pathway were introduced to Saccharomyces cerevisiae (Ho et al. 1998; van Maris et al. 2007) or Zymomonas mobilis (Zhang et al. 1995), whereas enzymes of the ethanologenic pathway of Z. mobilis were introduced to strains with a complete xylose pathway to convert xylose to ethanol efficiently (Ingram et al. 1987; Ohta et al. 1991; Sun et al. 2004).
Xylose is the second most abundant carbohydrate in nature after glucose (Khankal et al. 2008). It is used for production of xylitol, a pentahydroxy sugar-alcohol and functional sweetener with various applications in food, dental and pharmaceutical industries due to its probiotic effects of reducing blood glucose, triglyceride, and cholesterol levels (Ur-Rehman et al. 2015). Commercial production of xylose usually involves isolation from the acidic/enzymatic hydrolysate of sugarcane bagasse or corncob and leaves other sugars including glucose, arabinose, galactose, and some minor monosaccharides as by-products or wastes. Thus, it is highly desirable to develop a new bioprocess with an engineered microbial strain capable of metabolizing all the monosaccharides but xylose in the lignocellulosic hydrolysate to produce industrial bioproducts, such as ethanol, and leaving xylose in the beer which can be used as a valuable source for xylitol production.
In the present study, an E. coli strain deficient in xylose transport and metabolism was constructed, followed by the introduction of an ethanol synthesis pathway. A fermentation process for converting all the monosaccharides except xylose in the simulated corncob hydrolysate to ethanol was developed. By using this novel strategy, efficient production of ethanol and complete recovery of xylose were achieved from the simulated corncob hydrolysate.
Materials and methods
Strains and plasmids
Strains and plasmids used in this study are listed in Table 1. Cultures were stored at − 70 °C in 15% glycerol in the Culture and Information Center of Industrial Microorganism of China Universities at Jiangnan University (CICIM-CU, http://CICIM-CU.jiangnan.edu.cn).
Escherichia coli B0013-1030 (Cao et al. 2010) was used as parent strain, in which xylH, encoding the high affinity xylose transporter (Hasona et al. 2004; Cirino et al. 2006), was inactivated by natural selection and identified recently (Sun et al. 2017). The frdA encoding fumarate reductase, xylA encoding xylose isomerase and xylE encoding the minor xylose transporter (Song and Park 1997) were further disrupted in B0013-1030 to create B0013-2012 according to the method described previously (Zhou et al. 2011, 2012). Primers used in this study are listed in Supplementary Table 1. Unless otherwise stated, standard molecular biology protocols (Sambrook and Russell 2001) were used for DNA manipulation. B0013-2012PA was developed by transforming pEtac-PA carrying Z. mobilis pdc and adhB for the ethanol pathway (Sun et al. 2004) into B0013-2012.
Media and growth conditions
Luria–Bertani medium (LB) (5 g/l yeast extract, 10 g/l tryptone, and 5 g/l NaCl) is used as regular cultivation medium. Agar (15 g/l) is added for solid medium. Modified M9 medium (Zhou et al. 2012) supplemented with 5 g/l glucose or xylose was used for strain selection. Modified M9 medium supplemented with 50 g/l xylose, 50 g/l glucose, or mixed sugars of 25 g/l xylose and 25 g/l glucose was used for shaking flask fermentation. Glucose and xylose solutions were sterilized separately by autoclaving at 115 °C for 20 min.
When it was necessary 100 μg/ml of ampicillin, 30 μg/ml of gentamycin or 50 μg/ml kanamycin were added into the media.
Fermentation experiments
Erlenmeyer flask fermentation was performed in 500-ml flasks containing 100 ml of medium at 37 °C in triplicate and all data points reported are the average of three experiments. To prepare inocula for Erlenmeyer flask fermentation, three colonies from a fresh LB plate was transferred into 50 ml of LB medium in 250-ml flasks and then cultivated at 37 °C and 200 rpm for 10–12 h to an OD600 of 2.0–2.5. Cells were harvested by centrifugation (4300×g, 10 min), washed and resuspended with M9 medium, and then inoculated into fermentation medium with final working volume of 100 ml and final cell density of 5 (OD600). After inoculation, the flasks were statically placed in incubator with no agitation at 37 °C for ethanol fermentation. The flasks were sealed with rubber stoppers drilled to allow the insertion of a needle through which carbon dioxide was vented and samples were taken.
Ethanol fermentation experiments in a 7-l bioreactor (Bioflo110; New Brunswick Scientific Co., Inc., Edison, NJ) were accomplished at 37 °C with an initial 3 l working volume. Cells were cultured and collected as mentioned above and inoculated into 3 l of modified M9 medium supplemented with 182 g (about 150 ml) of simulated corncob hydrolysate to an initial cell density of 0.1 (OD600). Fermentation process was carried out under aerobic conditions for about 11.5 h with agitation (200–1000 rpm) and sterile air continuously fed at a rate of 0.1–1 vvm. The fermentation process was then turned into anaerobic conditions for another 10.5 h by stopping air flow and reducing agitation speed to 100 rpm. During the anaerobic ethanol fermentation, three volumes of 150 ml of the simulated corncob hydrolysate were added into the bioreactor. The pH was measured on-line automatically by pH electrode (InPro 3253 l/SG/120, Mettler-Toledo, LLC) and maintained at 7.0 by automatically feeding concentrated NH4OH or 10% H2SO4 (v/v) during the fermentation phase. The simulated corncob hydrolysate used in this study was prepared based on 70% (w/w) concentrated xylose crystallization mother liquid (Futian Pharmaceutical Company Ltd., China) supplemented with glucose. The 1000 g of simulated corncob hydrolysate contained 440 g glucose, 330 g xylose, 190 g arabinose, and 40 g galactose.
Analytical methods
Sampling was performed periodically to measure the amounts of cell mass, glucose, xylose, arabinose, galactose and ethanol during fermentation. Glucose, xylose, arabinose, galactose and ethanol were determined by HPLC according to the method previously (Zhou et al. 2011) using a HPLC system equipped with Dionex p680 pump (Dionex Corporation, Sunnyvale, CA), a Shodex SH-1011 column (Shodex SH-1011 H610009; Showa Denko K.K., Kawasaki, Japan), and a refractive index detector. The samples were analysed at 60 °C and eluted at 1.0 ml/min with 0.01 M sulfuric acid. Cell density was monitored at 600 nm (1 cm light path) using a UNICO UV2000 spectrophotometer.
Results
Construction of xylose-nonutilizing ethanol producing strain E. coli B0013-2012PA
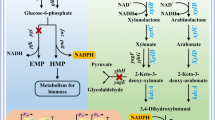
Metabolic pathway modification of xylose-nonutilizing E. coli B0013-2012PA for ethanol production is summarized in Fig. 1. E. coli B0013-1030 (pta-ack, ldhA, pflB, xylH) was used as parent strain, in which, pta-ack, ldhA, and pflB were deleted previously to block formation of acetate, lactate and formate (Cao et al. 2010) and xylH, encoding the membrane component of the high-affinity xylose transporter XylFGH, was inactivated by natural selection and recently identified (Sun et al. 2017). frdA was deleted and B0013-1031 (B0013-1030, ΔfrdA) was obtained. E. coli B0013-2010 was further developed as an intermediate strain by inactivating xylA in B0013-1031. E. coli B0013-2012 was obtained by disrupting xylE in B0013-2010. The ethanol synthesis pathway encoded by pEtac-PA was finally introduced into B0013-2012, resulting in ethanologenic E. coli B0013-2012PA (Fig. 1).
To examine the physiological properties of the engineered xylose-nonutilizing strain, E. coli B0013-2012 was cultivated on M9 plates supplemented with 5 g/l of various sugars (Fig. 2). Strains B0013-2012 and B0013-2010 displayed robust growth on plates containing glucose, galactose or arabinose, respectively, but failed to grow on xylose, while B0013 and B0013-1031 showed growth on all of the test plates.
Growth characteristics of E. coli B0013-2012 were further examined by cultivating in shaking flasks containing M9 medium with glucose or xylose. After cultivation, B0013-2012 exhausted glucose with cell density of 4.5 at OD600 but failed to grow on xylose (Fig. 3). The results further confirmed that E. coli B0013-2012 lost its xylose-utilizing ability.
The cell growth and substrates utilization of E. coli B0013-2012 and B0013-1031 on glucose or xylose. The strains were cultivated in 500-ml shaking flasks containing 100 ml of M9 medium with 5 g/l of glucose or 5 g/l of xylose at 37 °C and 200 rpm. Solid line: B0013-2012; dotted line: B0013-1031; solid circle: glucose; circle: cell density on glucose; solid square: xylose; square: cell density on xylose
E. coli B0013-2012PA fermented glucose and/or xylose to ethanol in Erlenmeyer flasks
To evaluate the fermentation performance of B0013-2012PA, experiments were conducted in 100 ml of medium with glucose and/or xylose in 500-ml Erlenmeyer flasks. Ethanol fermentation started with an initial cell density of 5.0 as described in Materials and methods section. The results are summarized in Fig. 4. After 18 h of fermentation, 24.2 g/l of ethanol was produced from glucose with the yield of 48.4 g/100 g glucose (94.7% of the theoretical yield) (Fig. 4a). Under the same condition, xylose was not metabolized and ethanol formation was not observed (Fig. 4b). With mixed 25 g/l of glucose and 25 g/l of xylose as substrates, B0013-2012PA produced 12.1 g/l of ethanol with xylose completely remained in the broth (Fig. 4c).
The substrates utilization and ethanol formation of E. coli B0013-2012PA during ethanol fermentation phase on glucose and/or xylose. M9 media with 50 g/l of glucose (a), 50 g/l of xylose (b) or 25 g/l of glucose and 25 g/l of xylose (c) were used for fermentation at 37 °C. Solid circle: glucose; solid square: xylose; triangle: ethanol; circle: cell mass
Ethanol fermentation by E. coli B0013-2012PA using simulated corncob hydrolysate as substrate
Ethanol fermentation performance of B0013-2012PA on the simulated corncob hydrolysate was further examined in a 7-l bioreactor with an initial 3 l working volume and 3 volumes of 150 ml of simulated corncob hydrolysate fed during the fermentation. The final volume of the completed fermentation was about 3.4 l. The results are summarized in Fig. 5 and Table 2. All glucose and galactose as well as almost all arabinose were metabolized and 53.4 g/l ethanol was produced with the yield of 48.9 g/100 g sugars after about 10.5 h of fermentation, while 69.6 g/l xylose remained in the broth, representing 98.6% of the total xylose in the simulated corncob hydrolysate added (Table 2, Fig. 5).
Ethanol fermentation by E. coli B0013-2012PA using the simulated corncob hydrolysate as substrate. Cells were pre-cultured and inoculated into 3 l of modified M9 medium supplemented with 150 ml of the simulated corncob hydrolysate. The process was conducted at 37 °C for 11.5 h under aerobic conditions for cell propagation. The ethanol fermentation was done at 37 °C for 10.5 h under anaerobic conditions. During ethanol fermentation, three volumes of 150 ml of the simulated corncob hydrolysate were fed into the bioreactor as indicated by dotted arrows
Discussion
Lignocellulosic ethanol production represents an important part of the world bio-economy. Lignocellulose comprises many different types of monosaccharides. The sugars in the corncob hydrolysate includes major sugars, glucose and xylose, and minor sugars, galactose and arabinose (Andersen et al. 2015). Previous studies were focused on efficient conversion of all the sugars in lignocellosic hydrolysate to desired products, such as ethanol (Ingram et al. 1987; Ohta et al. 1991; Zhang et al. 1995; Ho et al. 1998; van Maris et al. 2007; Rubin 2008). In this study we successfully developed a metabolically engineered E. coli strain which converted glucose, galactose and arabinose in the simulated corncob hydrolysate to ethanol but was incapable of metabolizing xylose, thus allowing its complete recovery from the fermentation beer as a valuable product with various applications.
The xylose non-utilizing strain E. coli B0013-2012 developed in this work carried triple mutations at xylH, xylE and xylA. Results on its physiological properties clearly indicated that it completely lost the ability to transport and metabolize xylose but its ability to utilize other sugars, such as glucose, galactose and arabinose for growth and for ethanol fermentation was not influenced and xylose remained unfermented in the fermentation broth.
As well-known, Saccharomyces cerevisiae, the conventional ethanol producing microorganism, cannot naturally ferment pentose sugars including xylose and arabinose (Jeffries 2006; Garcia Sanchez et al. 2010). The improved recovery of xylose was obtained by using S. cerevisiae to remove glucose in the corncob hydrolysates (Yoon et al. 2003). However, galactose is metabolized poorly by S. cerevisiae and C5 sugars, such as xylose and arabinose, are hardly metabolized and remained when glucose exists. To isolate xylose or its reduced product xylitol from the broth containing galactose and arabinose was still a challenge issue (Latif and Rajoka 2001; Kogje and Ghosalkar 2016).
Economic feasibility of ethanol production from biomass is always a challenge. Great efforts have been made, one of which is focus on engineering of microorganisms to efficiently utilize substrates and simplify fermentation process (Anasontzis and Christakopoulos 2014; Bátori et al. 2015; Puseenam et al. 2015; Treebupachatsakul et al. 2015). Here we report the construction of a xylose-nonutilizing E. coli strain which could convert all monosaccharides except xylose in the simulated corncob hydrolysate to ethanol. Ethanol could be then separated by distillation and recovery of xylose from the fermentation broth is simplified due to increased xylose purity. This strategy should contribute to lowering the costs of lignocellulosic bioconversion and increase the economic feasibility of bioethanol production, although the results obtained in this study were based on the simulated corncob hydrolysate and it may be more challenging when an actual corncob hydrolysate is used.
References
Anasontzis GE, Christakopoulos P (2014) Challenges in ethanol production with Fusarium oxysporum through consolidated bioprocessing. Bioengineered 5:393–395
Andersen RL, Jensen KM, Mikkelsen MJ (2015) Continuous ethanol fermentation of pretreated lignocellulosic biomasses, waste biomasses, molasses and syrup using the anaerobic, thermophilic bacterium Thermoanaerobacter italicus Pentocrobe 411. PLoS ONE 10:e0136060
Banerjee S, Mudliar S, Sen R, Giri B, Satpute D, Chakrabarti T, Pandey R (2010) Commercializing lignocellulosic bioethanol: technology bottlenecks and possible remedies. Biofuel Bioprod Bior 4:77–93
Bátori V, Ferreira JA, Taherzadeh MJ, Lennartsson PR (2015) Ethanol and protein from ethanol plant by-products using edible fungi Neurospora intermedia and Aspergillus oryzae. Biomed Res Int 2015:176371
Cao JL, Zhou L, Zhang L, Wang ZX, Shi GY (2010) Construction and fermentation of succinate-producing recombinant Escherichia coli. Chin J Appl Environ Biol 16:851–857
Cirino PC, Chin JW, Ingram LO (2006) Engineering Escherichia coli for xylitol production from glucose-xylose mixtures. Biotechnol Bioeng 95:1167–1176
Garcia Sanchez R, Karhumaa K, Fonseca C, Sànchez Nogué V, Almeida JR, Larsson CU, Bengtsson O, Bettiga M, Hahn-Hägerdal B, Gorwa-Grauslund MF (2010) Improved xylose and arabinose utilization by an industrial recombinant Saccharomyces cerevisiae strain using evolutionary engineering. Biotechnol Biofuels 3:13
Hasona A, Kim Y, Healy FG, Ingram LO, Shanmugam KT (2004) Pyruvate formate lyase and acetate kinase are essential for anaerobic growth of Escherichia coli on xylose. J Bacteriol 186:7593–7600
Ho NW, Chen Z, Brainard AP (1998) Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl Environ Microbiol 64:1852–1859
Ingram LO, Conway T, Clark DP, Sewell GW, Preston JF (1987) Genetic engineering of ethanol production in Escherichia coli. Appl Environ Microbiol 53:2420–2425
Jeffries TW (2006) Engineering yeasts for xylose metabolism. Curr Opin Biotechnol 17:320–326
Khankal R, Chin JW, Cirino PC (2008) Role of xylose transporters in xylitol production from engineered Escherichia coli. J Biotechnol 134:246–252
Kogje A, Ghosalkar A (2016) Xylitol production by Saccharomyces cerevisiae overexpressing different xylose reductases using non-detoxified hemicellulosic hydrolysate of corncob. 3. Biotech 6:127
Latif F, Rajoka MI (2001) Production of ethanol and xylitol from corn cobs by yeasts. Bioresour Technol 77:57–63
Ohta K, Beall DS, Mejia JP, Shanmugam KT, Ingram LO (1991) Metabolic engineering of Klebsiella oxytoca M5A1 for ethanol production from xylose and glucose. Appl Environ Microbiol 57:2810–2815
Puseenam A, Tanapongpipat S, Roongsawang N (2015) Co-expression of endoxylanase and endoglucanase in Scheffersomyces stipitis and its application in ethanol production. Appl Biochem Biotechnol 177:1690–1700
Rubin EM (2008) Genomics of cellulosic biofuels. Nature 454:841–845
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Song S, Park C (1997) Organization and regulation of the D-xylose operons in Escherichia coli K-12: XylR acts as a transcriptional activator. J Bacteriol 179:7025–7032
Sun JF, Xu M, Zhang F, Wang ZX (2004) Novel recombinant Escherichia coli producing ethanol from glucose and xylose. Acta Microbiol Sin 44:600–604
Sun JF, Tian KM, Shen W, Chen XZ, Wang ZX (2017) Genetic nature of xylose metabolism, differences between Escherichia coli strains. Chin J Food Ferment Ind 43:68–73
Treebupachatsakul T, Shioya K, Nakazawa H, Kawaguchi T, Morikawa Y, Shida Y, Ogasawara W, Okada H (2015) Utilization of recombinant Trichoderma reesei expressing Aspergillus aculeatus β-glucosidase I (JN11) for a more economical production of ethanol from lignocellulosic biomass. J Biosci Bioeng 120:657–665
Ur-Rehman S, Mushtaq Z, Zahoor T, Jamil A, Murtaza MA (2015) Xylitol: a review on bioproduction, application, health benefits, and related safety issues. Crit Rev Food Sci Nutr 55:1514–1528
van Maris AJ, Winkler AA, Kuyper M, de Laat WT, van Dijken JP, Pronk JT (2007) Development of efficient xylose fermentation in Saccharomyces cerevisiae: xylose isomerase as a key component. Adv Biochem Eng Biotechnol 108:179–204
Yoon SH, Mukerjea R, Robyt JF (2003) Specificity of yeast (Saccharomyces cerevisiae) in removing carbohydrates by fermentation. Carbohydr Res 338:1127–1132
Zhang M, Eddy C, Deanda K, Finkelstein M, Picataggio S (1995) Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science 267:240–243
Zhou L, Zuo ZR, Chen XZ, Niu DD, Tian KM, Prior BA, Shen W, Shi GY, Singh S, Wang ZX (2011) Evaluation of genetic manipulation strategies on D-lactate production by Escherichia coli. Curr Microbiol 62:981–989
Zhou L, Niu DD, Tian KM, Chen XZ, Prior BA, Shen W, Shi GY, Singh S, Wang ZX (2012) Genetically switched D-lactate production in Escherichia coli. Metab Eng 14:560–568
Acknowledgements
This research was supported financially by The Raising Program of Innovation Team for Tianjin Universities (TD12-5002).
Supporting information
Supplementary Table 1—Primers used in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sun, J., Wang, J., Tian, K. et al. A novel strategy for production of ethanol and recovery of xylose from simulated corncob hydrolysate. Biotechnol Lett 40, 781–788 (2018). https://doi.org/10.1007/s10529-018-2537-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-018-2537-0