Abstract
Enterobacteriaceae is one kind of harmful microorganisms commonly presented in raw fish products, and detection of Enterobacteriaceae plays a very important role in evaluating microbial contamination. This work was carried out to exploit the potential of emerging hyperspectral imaging technique to determine the Enterobacteriaceae contamination of salmon flesh during cold storage. The spectral information ranging from 900 to 1700 nm (239 wavelengths) was extracted to relate to the Enterobacteriaceae loads (recorded as log 10 CFU/g) using partial least square (PLS) regression, developing a PLS model with correlation coefficient of prediction (r P) of 0.94 and root mean square error of prediction (RMSEP) of 0.53 as well as residual predictive deviation (RPD) of 2.97. By applying successive projection algorithm (SPA), eight wavelengths at 924, 931, 964, 1068, 1262, 1373, 1628 and 1668 nm among the 239 wavelengths were selected as informative wavelengths to reduce the information redundancy and optimise the PLS model. With the eight informative wavelengths, a simplified PLS model defined as SPA-PLS was established with r P of 0.95, RMSEP of 0.47 and RPD of 3.23. To visualise the contamination degree of salmon flesh caused by Enterobacteriaceae, the SPA-PLS model was transferred to each pixel of images, and colourful distribution maps were produced with different colour represented different numbers of Enterobacteriaceae colonies. The results showed that hyperspectral imaging operating in 900–1700 nm is promising in evaluating Enterobacteriaceae contamination of salmon products. More studies are still required to further refine the multispectral imaging system to achieve online application.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
With the development of optical technology, a novel technique called hyperspectral imaging has emerged as a smart and non-destructive technique to create, record and display the images at a large number of discrete, contiguous spectral bands (ElMasry et al. 2012a). Because of its powerful ability to provide spectral and spatial information simultaneously, hyperspectral imaging technique has attracted tremendous interests and attentions both in academia and industry. In principle, a hyperspectral image of a sample is composed of a series of congruent three-dimensional “datacube” (x, y, λ), in which (x, y) and (λ) represent spatial and spectral information, respectively. The hyperspectral image can be described either as separate spatial image (x, y) at each individual wavelength (λ) or as a spectrum (λ) at each pixel (x, y) (Wu and Sun 2013a). Spectral data combined with spatial images provide a large amount of information, which can be analysed to characterise a target object more objectively and reliably. Moreover, imaging function, as an obvious advantage of hyperspectral imaging, provides a direct and simple way to visualise the spatial variation of quality attributes of an object, which cannot be done by conventional spectroscopic technology and other destructive methods. On the other hand, although imaging or computer vision is often used for quality assessment by providing spatial images of food samples (Valous et al. 2009; Jackman et al. 2009, 2008; Sun 2004; Wang and Sun 2002), just like hyperspectral imaging does by its visualisation feature, it cannot obtain the quality-related spectral information at every pixel. Hence, it can be said that hyperspectral imaging is a more advanced technique, and its potential should be exploited to guarantee food quality, thus satisfying the increasing consumer demands.
Up to now, many applications on the use of hyperspectral imaging for quality evaluation and assessment have been reported in various kinds of agri-food products, such as pork (Dissing et al. 2012; Barbin et al. 2012), lamb (Kamruzzaman et al. 2011; 2012a), beef (Wu et al. 2012a; ElMasry et al. 2011b, 2012b), turkey ham (ElMasry et al. 2011c), poultry (Feng and Sun 2013), fruits and vegetables (Rajkumar et al. 2012; Taghizadeh et al. 2009), eggs (Abdel-Nour and Ngadi 2011), milk (Qin et al. 2012) and cereals (Shahin et al. 2013). Besides, aquatic products have also been evaluated in terms of physical and chemical attributes, involving texture profile analysis (Wu and Sun 2013b; He et al. 2014a), moisture and drip loss distribution (He et al. 2013, 2014b) and colour measurement (Wu et al. 2012b). Among aquatic products, salmon is especially worth mentioning because of its preferable nutritional value and economic importance. Being rich in many valuable nutritive elements including high-quality lipids, tocopherol and astaxanthin, salmon becomes one of the most popular fish species (Johnston et al. 2006). Salmon farming has been one of the most successful aquaculture industries with a production substantially larger than the total aquaculture production in recent decades (Asche et al. 2013). In fish market, salmon fillets are always sold on ice and exposed directly to the air without any protection from microbial contamination. On the other hand, salmon fillets are so perishable because of excellent environment (moist and nutritious flesh) for growth of spoilage microorganisms (Wu and Sun 2013c). Therefore, it is very necessary to inspect and control emerging microorganism contamination in order to ensure salmon fillets being safely consumed.
Enterobacteriaceae as a large group of harmful microorganisms mainly contains Escherichia coli, Shigella, Salmonella and Yersinia. They can transform carbohydrate (sugar) into acids and gas during the process of fermentation (Feng et al. 2012). The output of the transformation could cause serious public health concerns to consumers (Pitout and Laupland 2008). In food and food products, the number of Enterobacteriaceae colonies, which is Enterobacteriaceae loads, is often used as a useful indicator for food safety evaluation (Zaragozá et al. 2012; Tosukhowong et al. 2011). Conventional techniques for Enterobacteriaceae detection commonly include standard plate colony-counting method (Botsoglou et al. 2010), immunological method (enzyme-linked immunosorbent assay, ELISA) (Dwivedi and Jaykus 2011), molecular method (polymerase chain reaction, PCR) (Liu et al. 2013) as well as PCR-ELISA method (Kuo et al. 2010). However, these methods are time-consuming, labour-intensive, tedious, inefficient, destructive and thus not suitable for a rapid, non-destructive and high-efficient inspection application. Therefore besides methods and techniques such as drying (Sun and Byrne 1998; Sun and Woods 1997; Delgado and Sun 2002a, b), refrigeration (Sun 1997; Sun et al. 1996; McDonald and Sun 2001; Kiani and Sun 2011) and edible coating (Xu et al. 2001) are often required to ensure food quality and safety, there is a urgent need for efficient and effective assessment methods.
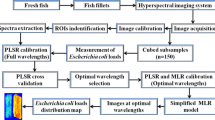
Hyperspectral imaging that emerged as a promising technique provides a possible way to conduct quality evaluation in fish products. In recent years, hyperspectral imaging has been used as an analytical tool for quality assessment of fish products such as flesh tenderness evaluation (He et al. 2014a), fat determination (ElMasry and Wold 2008), water-holding capacity measurement (Wu and Sun 2013d; Zhu et al. 2012) and freshness identification (Sone et al. 2012). Besides, hyperspectral imaging has also been used for microbial evaluation in meat (Peng et al. 2009, 2011; Tao et al. 2012; Teena et al. 2013), contamination in fruits (Liu et al. 2007) and parasite detection in fish fillets (Sivertsen et al. 2011). In this study, the main objective of this study was to explore the potential of hyperspectral imaging for rapid determination of Enterobacteriaceae in farmed salmon flesh during cold storage. The contamination degree of salmon samples caused by Enterobacteriaceae was quantified based on the analysis of hyperspectral images and then further visualised using distribution maps. The specific aims of this study were to (1) acquire hyperspectral images of salmon samples in the wavelength range of 900–1700 nm at different storage time, (2) identify the regions of interest (ROI) within the acquired hyperspectral images and extract the spectral information from the ROIs, (3) develop a mathematical model to correlate the extracted spectra of samples with their Enterobacteriaceae loads measured by the standard pour plate method, (4) select informative wavelengths for the reduction of redundant information among the full wavelength range, (5) establish optimised models based on the selected informative wavelengths and evaluate their performance in terms of correlation coefficient (r) and root mean square error (RMSE) as well as residual predictive deviation (RPD), and (6) transfer a better optimised model into the pixels of images of samples to generate colour maps for Enterobacteriaceae load visualisation.
Materials and Methods
Fillet Preparation and Sampling
Thirty fresh farmed Atlantic salmon fillets (Salmo salar) originated from Norway were supplied by local seafood supermarkets in Dublin, Ireland. The fillets were vacuum-packed and transported to the laboratory of Food Refrigeration and Computerised Food Technology (FRCFT), University College Dublin, Ireland, and then used for sampling and experiment. Each fillet was cut into several cubes (approximately 10 g) with the size of 3 cm × 3 cm × 1 cm (length × width × thickness), obtaining a total of 94 samples from the thirty fillets. All the cubed samples were then packed using cling film (Tesco Stores Ltd, UK), labelled and stored at 4 °С in a digital refrigerator for 0, 3, 6, 8, 10 and 13 days.
Hyperspectral Imaging System
In this work, a line scan pushbroom near-infrared hyperspectral imaging system was used for the acquisition of hyperspectral images of the cubed samples in reflectance mode. The main components of the system are presented in Fig. 1. In detail, the hyperspectral imaging system is composed of an imaging spectrograph (Specim ImSpector N17E, Spectral Imaging Ltd., Oulu, Finland), a CCD camera (SUI Goodrich SU320M-1.7RT, a 12-bit high performance of 320 spatial × 256 spectral), a lens (Xeva 992, Xenics Infrared Solutions, Belgium), a pair of tungsten lamps (V-light, Lowel Light Inc, USA), a translation stage (MSA15R-N, AMT-Linearways, SuperSlides & Bushes Corp., India) and a computer installed with an image acquisition software (SpectralCube, Spectral Imaging Ltd., Oulu, Finland). The spectrograph covers a broad near-infrared spectral range from 897 to 1753 nm (256 spectral bands) with an increment of 3.34 nm between every two wavebands. Due to the low signal-to-noise ratio of the ranges within 897–900 and 1700–1753 nm, only the wavelength range of 900–1700 nm with 239 spectral wavebands was used for further data analysis.
Image Acquisition and Data Extraction
At each test day, about 15 samples were taken out of the refrigerator and placed on the translation stage of the hyperspectral imaging system. Scanning was performed line by line to acquire hyperspectral images of the samples, with the moving speed of translation stage of 2.8 cm/s to obtain the same resolution in the horizontal and vertical directions. The acquired images were the combination of congruent two-dimensional sub-images (x, y) at the 239 wavelength bands (λ) that spanned from 900 to 1700 nm. In fact, the raw hyperspectral images collected by the CCD camera attached in the hyperspectral imaging system reflected the signal intensity rather than the reflectance spectra. Therefore, image calibration was required to correct the raw hyperspectral images into reflectance images. Two reference images, white and black, were needed during the calibration process. The white image (I White) was obtained by scanning a white board with about 99.9 % reflectance. The black image (I Black) was acquired by turning off the light source and covering the camera lens completely, making the black image with about 0 % reflectance. The raw hyperspectral images (I Raw) were finally calibrated (I Calibrated) using a formula shown below:
With the regions of interest (ROI) function of ENVI v4.6 software (Research Systems Inc., Boulder, CO, USA), the region that had the same shape of the corresponding sample in the calibrated hyperspectral image was isolated from the background. The spectra of all pixels within the isolated ROI were extracted and averaged to obtain one spectrum representing the ROI. As a result, 94 mean spectra corresponding to the 94 samples were obtained as the basis for further spectral analysis.
Microbiological Analysis
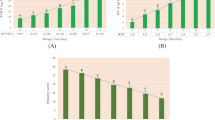
Once the image acquisition and calibration were completed, the salmon samples were immediately used to measure their reference numbers of Enterobacteriaceae colonies according to the method described by Sallam (Sallam 2007). Enterobacteriaceae presented in each sample were incubated in a pour plate of Violet Red Bile Glucose Agar (VRBGA, CM0485, Oxoid, Basingstoke, UK) at 37 °С and then enumerated after 24 h. The Enterobacteriaceae loads were recorded as colony-forming units (CFU) and logarithmically transformed. The final results are listed in Table 1, and the growing trend of Enterobacteriaceae loads of salmon samples is presented in Fig. 2. To avoid bias in the selection of calibration and prediction data, the 94 samples were sorted from small to large according to their values of Enterobacteriaceae loads. One of every four samples was selected into the prediction group. Finally, 71 samples among the 94 samples were selected for model calibration, and the rest of the samples (23 samples) were used for prediction.
Chemometric Analysis
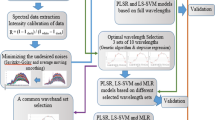
To establish the quantitative relationship between the spectra extracted from the salmon samples and the measured Enterobacteriaceae loads, chemometric analysis was conducted to mine the spectral data using partial least square (PLS) regression. PLS is a linear algorithm and often applied to model in the situation where the number of predictor variables (e.g. wavelengths) is larger than that of response variables, and there is a high correlation or colinearity among the predictor variables at the same time (Barbin et al. 2013). In this study, by reducing the original wavelengths to a set of unrelated wavelengths, optimal latent variables (LVs) were found in the PLS procedure to predict Enterobacteriaceae loads from the 239 spectral variables.
The performance of established regression models was examined in terms of correlation coefficient of calibration (r C), root mean square error of calibration (RMSEC), correlation coefficient of prediction (r P), root mean square error of prediction (RMSEP) and residual predictive deviation (RPD). Generally, a good model should have high correlation coefficients (r C and r P), high RPD and low root mean square errors (RMSEC and RMSEP) (ElMasry and Wold 2008; Gómez et al. 2006). In addition, an absolute value between RMSEC and RMSEP, which is |RMSEC−RMSEP|, was also calculated to evaluate model robustness.
Informative Wavelength Selection
In general, hyperspectral images containing hundreds or thousands of spectral variables are always characterized by high dimensionality, which requires more time to do information processing (Kamruzzaman et al. 2012b). Moreover, the contiguous wavelengths often exhibit similar spectral information, which produces a problem of multicollinearity, leading to information redundancy. In addition, irrelevant or useless information might hide in the spectra, which will weaken the performance of predictive model. The elimination of uninformation variables (wavelengths) is helpful to predigest calibration and improve prediction accuracy (Wu et al. 2012c). Therefore, the most useful wavelengths, that is, informative wavelengths, are preferable to be selected for data analysis. As claimed by Keskin et al. (Keskin et al. 2004), the wavelengths holding the most useful information should be recommended while the wavelengths carrying irrelevant information should be eliminated. To sum up, the implementation of informative wavelength selection will not only contribute to the reduction of information dimensionality and multicollinearity, but also simplify the modelling process and accelerate the data analysis.
It has been reported that regression coefficients of PLS model can be used for informative wavelength selection (ElMasry et al. 2011). In this method, wavelengths corresponding to the large values of regression coefficients (regardless of sign) are regarded as informative wavelengths and carry the most important spectral information related to specific quality parameter. Successive projection algorithm (SPA) is another variable selection method proposed to solve the collinearity problems of spectral wavelengths (Wu et al. 2014). In SPA procedure, there are two steps required to select the important wavelengths for Enterobacteriaceae contamination prediction. First, the candidate subsets of variables including spectra (X) and reference values of Enterobacteriaceae loads (Y) were constructed and arranged in a matrix. Second, the constructed candidate subsets were evaluated according to the performance of the calibrated model. Informative wavelength selection by regression coefficients of PLS model and SPA was operated with software Unscrambler v9.7 (CAMO, Oslo, Norway) and Matlab R2010b (The Mathworks, Inc., Natick, MA, USA), respectively.
Visualisation of Enterobacteriaceae Distribution
Visualisation of quality attributes is an advanced function of hyperspectral imaging over traditional spectroscopy. Such function enables hyperspectral imaging to visualise the spatial variation of quality attributes in the target sample (He et al. 2015; Sun 2010). In practice, it is very useful to show the microbial change of food products using a visual image (spatial distribution), especially many harmful microbes that cannot be observed by naked eyes. A quantitative prediction model in tandem with a spatial image will be more powerful and effective in evaluating microorganism contamination in food products, like Enterobacteriaceae present in salmon flesh.
In this study, there were two steps required to carry out the Enterobacteriaceae load visualisation in salmon samples. First, different optimised models established with the informative wavelengths selected by regression coefficients of PLS model and SPA were, respectively, evaluated and then compared in terms of predictive ability. Second, a model with better accuracy was chosen and then transferred to each pixel of images with the help of an image processing algorithm. The distribution map of Enterobacteriaceae loads was at last created to exhibit the Enterobacteriaceae load variation from sample to sample and spot to spot within the same sample. The whole visualisation process was performed with the Matlab R2010b software.
Results and Discussion
Spectral Profiles of Salmon Samples
The typical spectral profiles extracted from the ROI of hyperspectral images of tested salmon samples with different Enterobacteriaceae loads are shown in Fig. 3. It was observed that there are several obvious absorption peaks emerged in the whole 900–1700-nm range, which were associated with overtone and combination vibrations of hydrogen-containing functional groups, such as C–H, O–H, N–H and S–H (ElMasry et al. 2011). In specific, absorption maxima that appeared at around 975 nm was assigned to the O–H stretching second overtone of water (Ritthiruangdej et al. 2011). The absorption peak around 1210 nm was due to the C–H stretching second overtone (presence of fat) (Morales-Sillero et al. 2011). The absorption peak at 1450 nm was related to the O–H stretching first overtone (presence of water) (Cozzolino et al. 2013). In general, the spectral profiles mainly exhibited the information of chemical molecules such as water and fat, which are the main components of salmon flesh. As shown in Fig. 3, the three reflectance spectra curves had similar trends and that may be due to the similar chemical compositions of salmon samples. On the other hand, small differences between each curve were observed, which may be caused by the changing of the major chemical compositions in salmon sample during the microbial contamination process.
Although no typical feature peaks of Enterobacteriaceae were found in the near-infrared spectral region, spectral information concerning these chemical components could be mined using appropriate chemometrics method for the determination of Enterobacteriaceae loads in salmon flesh.
PLS Analysis Based on Full Range Spectra
In the PLS calibration, two matrixes containing the extracted full range spectral data and the measured Enterobacteriaceae loads were obtained for PLS model calibration and prediction, respectively. The first matrix that had the size of 71 × 239 (sample × wavelength) was used for the model calibration, while the other matrix with the size of 23 × 239 (sample × wavelength) was used as an independent set to evaluate the performance of the calibrated PLS model. After executing PLS algorithm in software Unscrambler, a PLS model based on the full 900–1700-nm range spectra was developed, and its performance is shown in Table 2. The PLS model exhibited a high ability for the prediction of Enterobacteriaceae loads of salmon flesh, with r P of 0.94 and RMSEP of 0.53. Moreover, the |RMSEC−RMSEP| value was 0.10, indicating a good robustness of the PLS model. The nearer the value of |RMSEC−RMSEP| approximates to zero, the better robustness PLS model has. Besides, the PRD value in this study was 2.81, which is over 2.5 and that indicated an excellent prediction accuracy of the PLS model (Nicolaï et al. 2007).
Wavelength Selection and Model Optimisation
Although the predictive ability of the PLS model for Enterobacteriaceae load prediction was good, the number of the full 900–1700-nm wavelengths was still large, which would need more time for data processing and model calibration. It was expected that the number of full wavelengths could be reduced, and a new model with similar performance could be developed based on the reduced wavelengths, compared with the original PLS model. In view of this, wavelength selection was conducted by the regression coefficients of PLS model and SPA, respectively, to select informative wavelengths from the whole 900–1700-nm range.
Informative Wavelengths Selected by Regression Coefficients
In the process of selecting the informative wavelengths by regression coefficients, the wavelengths among the full 900–1700-nm range corresponding to the large values of regression coefficients (regardless of sign) of the PLS model were considered as the informative wavelengths for the PLS model optimisation. As shown in Fig. 4, 11 individual wavelengths at 921, 931, 964, 1008, 1105, 1145, 1252, 1366, 1628, 1658 and 1685 nm were selected as the informative wavelengths.
Based on the selected wavelengths, a new matrix with the dimension of 94 × 11 (sample × wavelength) was obtained and used for PLS model optimisation. In the new matrix, the first 71 lines of the matrix were used for calibration, and the remaining 23 lines were used for prediction. PLS algorithm was applied with the new matrix to establish an optimised PLS model (RC-PLS model for short), which had the r C of 0.95 with RMSEC of 0.47 and r P of 0.95 with RMSEP of 0.47 (Table 2). Although the number of wavelength variables was reduced from 239 to 11, the performance of the RC-PLS model in predicting Enterobacteriaceae loads of salmon flesh was similar to the original PLS model. Moreover, the |RMSEC−RMSEP| value in RC-PLS model was 0.01, 90 % less than that of PLS model, which indicated that the RC-PLS model had better robustness than the PLS model. Besides, the RPD value of 3.11 in RC-PLS model was about 11 % higher than that (2.81) of PLS model, showing a better ability of RC-PLS model in predicting Enterobacteriaceae loads. In general, use of regression coefficients of PLS model for informative wavelength selection was helpful to improve the model predictive accuracy and robustness. Based on the regression coefficients of RC-PLS model, a formula was obtained and expressed as follows:
Informative Wavelengths Selected by SPA
By running SPA program in software Matlab, RMSEV plots were obtained to select the optimal number of informative wavelengths. As shown in Fig. 5a, the formed RMSEV curve exhibited a rise trend as the number of selected informative wavelengths (variables) was increased from one to three, then followed by a sharp fall when the variable number increased from three to seven and at last a gradual fall when the variable number increased from seven to thirteen. The eighth plot was picked and marked with a square marker as shown in Fig. 5a. The particular informative eight wavelengths of 924, 931, 964, 1068, 1262, 1373, 1628 and 1668 nm were identified and exhibited with square markers in Fig. 5b.
With the eight important wavelengths selected by SPA, the original matrix used for PLS model development was reduced to 94 × 8 (sample × wavelength), with 71 × 8 submatrix and 23 × 8 submatrix used for a new model calibration and prediction, respectively. With the reduced matrix, the PLS model was simplified, and an optimised model defined as SPA-PLS model was developed for Enterobacteriaceae load determination. As shown in Table 2, although the wavelength number was reduced by 97 % (239 vs 8), the performance of the SPA-PLS model was comparable to the original PLS model. The |RMSEC−RMSEP| value of 0.08 in SPA-PLS model was reduced by 20 % (0.10) compared with the PLS model, which showed that the SPA-PLS model had a better robustness than the PLS model. Besides, the RPD value of SPA-PLS model was 3.23, about 15 % higher than that of PLS model, which indicated a better predictive ability of SPA-PLS model over the PLS model in predicting Enterobacteriaceae loads. According to the regression coefficients of the SPA-PLS model, an equation for the prediction of Enterobacteriaceae loads of salmon samples was obtained and is shown below:
It was observed from Table 2 that the performance of the SPA-PLS model was similar to that of the RC-PLS model in predicting Enterobacteriaceae loads, although the wavelength number in SPA-PLS model was less than that of RC-PLS model (8 vs 11). The wavelength selection results showed that SPA and regression coefficients of PLS model were all effective in select informative wavelengths for PLS model optimisation. In practice, the SPA-PLS model was more suitable for further development of multispectral imaging system for online application, because of its less wavelength number.
Distribution of Enterobacteriaceae Loads
Besides chemometric analysis of spectra to develop quantitative model for Enterobacteriaceae contamination prediction, Enterobacteriaceae loads distributed in the salmon samples were also spatially visualised by the near-infrared hyperspectral imaging technique. The aim of visualising the distribution of Enterobacteriaceae loads was to show the unvisible changes of Enterobacteriaceae from sample to sample or spot to spot within the same sample in pseudocolour images, achieving and enhancing a better understanding of the Enterobacteriaceae contamination process when salmon flesh was stored in cold conditions. Such visualisation is generally the final important step in the whole analysis of the acquired hyperspectral images and also an advantageous feature of hyperspectral imaging over the traditional spectroscopic technology.
In this study, the Enterobacteriaceae load visualisation was realized by transferring the SPA-PLS model to each pixel of images. The dot product was calculated between the regression coefficients of SPA-PLS model shown in Eq. (3) and the spectrum of each pixel in the hyperspectral images at the selected eight informative wavelengths, resulting in the predicted Enterobacteriaceae loads of every pixel within the ROI of salmon samples. A distribution map of Enterobacteriaceae loads was then generated based on the predicted values of Enterobacteriaceae loads and their pixel positions within the ROI. In the generated map, pixels having similar spectral features produced similar predicted values of Enterobacteriaceae loads, which were then visualised in similar colour in the distribution map.
In Fig. 6, several distribution maps of salmon samples were displayed as examples to illustrate the spatial variation of Enterobacteriaceae loads during the contamination process. In the maps, a linear colour scale on the right side of samples was assigned from blue to red to indicate the Enterobacteriaceae loads in the salmon samples from low to high. The colour scale was used to exhibit the contamination process of salmon samples, with blue, green and red colour representing the low, middle and high degree of Enterobacteriaceae contamination, respectively. As shown in Fig. 6, a general trend of the increment of contamination degree of salmon samples was observed with the average Enterobacteriaceae loads that increased from low to high. At early stage of contamination, the distribution maps of Enterobacteriaceae loads were almost in blue colour (e.g. the sample with the mean Enterobacteriaceae loads of 2.643). At the middle stage, green and red colour accounted for the most of the distribution maps of Enterobacteriaceae loads, like the distribution map of the sample with the average Enterobacteriaceae loads of 4.741. At the later stage, red was the overwhelming majority colour (e.g. distribution map with mean Enterobacteriaceae loads being 6.601), demonstrating the serious contamination caused by Enterobacteriaceae.
Combination of spectral and spatial information enabled the hyperspectral imaging technique to determine the Enterobacteriaceae contamination of salmon flesh in a more objective and reliable way. In addition to mining spectra related to the reference Enterobacteriaceae loads, spatial image was also produced to form a distribution map for visualising the spatial changes of Enterobacteriaceae loads during the process of contamination. The results consisted of quantitative models and colourful distribution maps indicating that hyperspectral imaging is a more useful and powerful technique in evaluating the Enterobacteriaceae infection of salmon fillets.
Conclusions
Rapid determination of harmful microorganisms such as Enterobacteriaceae is very necessary in monitoring food quality, especially perishable foods like salmon products. Emergence of hyperspectral imaging integrating spectroscopy and imaging techniques provides a potential way to evaluate food quality rapidly and non-destructively. The results presented in current study indicated that near-infrared hyperspectral imaging has a great potential for the evaluation of Enterobacteriaceae contamination in salmon flesh during cold storage. Near-infrared hyperspectral images of examined salmon samples were successfully acquired at different storage times. Spectral and spatial information within the ROIs of the hyperspectral images were extracted, and the spectral data were analysed by PLS algorithm. Eleven informative wavelengths (921, 931, 964, 1008, 1105, 1145, 1252, 1366, 1628, 1658 and 1685 nm) were selected by regression coefficients of PLS model and eight ones (924, 931, 964, 1068, 1262, 1373, 1628 and 1668 nm) were selected by SPA to, respectively, optimise the PLS model developed with the full spectra of 900–1700-nm range. Two optimised models, namely RC-PLS and SPA-PLS, were developed, with RC-PLS model using 11 wavelengths obtained from regression coefficient method and SPA-PLS model using eight wavelengths obtained from SPA method, respectively. By comparison, the RC-PLS model and the SPA-PLS model were all comparable to the original PLS model in predicting Enterobacteriaceae contamination. The RC-PLS model and the SPA-PLS model had similar abilities to predict Enterobacteriaceae loads of salmon flesh. In addition, the distribution of Enterobacteriaceae loads was visualised by generating colourful maps after the transformation of the SPA-PLS model. The degree of Enterobacteriaceae contamination of salmon flesh was indicated and exhibited with the extension of storage times. The imaging feature would further enhance the understanding of quality changes of salmon products in cold condition. Near-infrared hyperspectral imaging showed its great promise for rapid and non-contact Enterobacteriaceae inspection in salmon flesh. More studies are still required to refine the multispectral imaging system for further online/offline application.
References
Abdel-Nour N, Ngadi M (2011) Int J Food Sci Nutr 62:418–422
Asche F, Roll KH, Sandvold HN, Sørvig A, Zhang D (2013) Aquac Econ Manag 17:322–339
Barbin DF, ElMasry G, Sun D-W et al (2012) Predicting quality and sensory attributes of pork using near-infrared hyperspectral imaging. Anal Chim Acta 719:30–42
Barbin DF, ElMasry G, Sun D-W, Allen P (2013) Food Chem 138:1162–1171
Botsoglou E, Govaris A, Christaki E, Botsoglou N (2010) Food Chem 121:17–22
Cozzolino D, Roumeliotis S, Eglinton J (2013) J Food Eng 114:545–549
Delgado AE, Sun D-W (2002a) Desorption isotherms for cooked and cured beef and pork. J Food Eng 51(2):163–170
Delgado AE, Sun D-W (2002b) Desorption isotherms and glass transition temperature for chicken meat. J Food Eng 55(1):1–8
Dissing B, Papadopoulou O, Tassou C, Ersbøll B, Carstensen J, Panagou E, Nychas G-J (2012) Food Bioprocess Technol 6:2268–2279
Dwivedi HP, Jaykus L-A (2011) Crit Rev Microbiol 37:40–63
ElMasry G, Wold JP (2008) J Agric Food Chem 56:7672–7677
ElMasry G, Sun D-W, Allen P (2011a) Food Res Int 44:2624–2633
ElMasry G, Sun D-W, Allen P (2011b) Non-destructive determination of water-holding capacity in fresh beef by using NIR hyperspectral imaging. Food Res Int 44(9):2624–2633
ElMasry G, Iqbal A, Sun D-W et al (2011c) Quality classification of cooked, sliced turkey hams using NIR hyperspectral imaging system. J Food Eng 103(3):333–344
ElMasry G, Barbin DF, Sun D-W, Allen P (2012a) Crit Rev Food Sci Nutr 52:689–711
ElMasry G, Sun D-W, Allen P (2012b) Near-infrared hyperspectral imaging for predicting colour, pH and tenderness of fresh beef. J Food Eng 110(1):127–140
Feng Y-Z, Sun D-W (2013) Talanta 105:244–249
Feng Y-Z, ElMasry G, Sun D-W, Walsh D, Morcy N (2012) Food Chem 138:1829–1836
Gómez AH, He Y, Pereira AG (2006) J Food Eng 77:313–319
He H-J, Wu D, Sun D-W (2013) Innovative Food Sci Emerg Technol 18:237–245
He H-J, Wu D, Sun D-W (2014a) J Food Eng 126:156–164
He H-J, Wu D, Sun D-W (2014b) Food Chem 156:394–401
He H-J, Wu D, Sun D-W (2015) Crit Rev Food Sci Nutr 55:864–886
Jackman P, Sun D-W, Du C-J et al (2008) Prediction of beef eating quality from colour, marbling and wavelet texture features. Meat Sci 80(4):1273–1281
Jackman P, Sun D-W, Du C-J et al (2009) Prediction of beef eating qualities from colour, marbling and wavelet surface texture features using homogenous carcass treatment. Pattern Recogn 42(5):751–763
Johnston IA, Li X, Vieira VLA, Nickell D, Dingwall A, Alderson R, Campbell P, Bickerdike R (2006) Aquaculture 256:323–336
Kamruzzaman M, ElMasry G, Sun D-W et al (2011) Application of NIR hyperspectral imaging for discrimination of lamb muscles. J Food Eng 104(3):332–340
Kamruzzaman M, ElMasry G, Sun D-W, Allen P (2012a) Anal Chim Acta 714:57–67
Kamruzzaman M, Barbin D, ElMasry G, Sun D-W, Allen P (2012b) Innovative Food Sci Emerg Technol 16:316–325
Keskin M, Dodd RB, Han YJ, Khalilian A (2004) Appl Eng Agric 20:851–860
Kiani H, Sun D-W (2011) Water crystallization and its importance to freezing of foods: a review. Trends Food Sci Technol 22(8):407–426
Kuo J-T, Cheng C-Y, Huang H-H, Tsao C-F, Chung Y-C (2010) J Ind Microbiol Biotechnol 37:237–244
Liu Y, Chen Y-R, Kim MS, Chan DE, Lefcourt AM (2007) J Food Eng 81:412–418
Liu C-C, Yeung C-Y, Chen P-H, Yeh M-K, Hou S-Y (2013) Food Chem 141:2526–2532
McDonald K, Sun D-W (2001) The formation of pores and their effects in a cooked beef product on the efficiency of vacuum cooling. J Food Eng 47(3):175–183
Morales-Sillero A, Fernández-Cabanás V-M, Casanova L, Jiménez M-R, Suárez M-P, Rallo P (2011) J Food Eng 107:99–106
Nicolaï BM, Beullens K, Bobelyn E, Peirs A, Saeys W, Theron KI, Lammertyn J (2007) Postharvest Biol Technol 46:99–118
Peng Y, Zhang J, Wu J, Hang H (2009) 73150Q–73150Q. doi: 10.1117/12.819424
Peng Y, Zhang J, Wang W, Li Y, Wu J, Huang H, Gao X, Jiang W (2011) J Food Eng 102:163–169
Pitout JDD, Laupland KB (2008) Lancet Infect Dis 8:159–166
Qin J, Chao K, Kim MS (2012) Food Chem 138:998–1007
Rajkumar P, Wang N, ElMasry G, Raghavan GSV, Gariepy Y (2012) J Food Eng 108:194–200
Ritthiruangdej P, Ritthiron R, Shinzawa H, Ozaki Y (2011) Food Chem 129:684–692
Sallam KI (2007) Food Chem 101:592–600
Shahin MA, Symons SJ, Hatcher DW (2013) Food Bioprocess Technol 7:224–234
Sivertsen AH, Heia K, Stormo SK, Elvevoll E, Nilsen H (2011) J Food Sci 76:S77–S83
Sone I, Olsen RL, Sivertsen AH, Eilertsen G, Heia K (2012) J Food Eng 109:482–489
Sun D-W (1997) Thermodynamic design data and optimum design maps for absorption refrigeration systems. Appl Therm Eng 17(3):211–221
Sun D-W (2004) Computer vision - an objective, rapid and non-contact quality evaluation tool for the food industry. J Food Eng 61(1):1–2
Sun D-W (2010) Hyperspectral imaging for food quality analysis and control. Academic Press/Elsevier, San Diego
Sun D-W, Eames IW, Aphornratana S (1996) Evaluation of a novel combined ejector-absorption refrigeration cycle .1. Computer simulation. Int J Refrig 19(3):172–180
Sun D-W, Woods JL (1997) Simulation of the heat and moisture transfer process during drying in deep grain beds. Dry Technol 15(10):2479–2508
Sun D-W, Byrne C (1998) Selection of EMC/ERH isotherm equations for rapeseed. J Agric Eng Res 69(4):307–315
Taghizadeh M, Gowen A, O’Donnell C (2009) Sens & Instrumen Food Qual 3:219–226
Tao F, Peng Y, Li Y, Chao K, Dhakal S (2012) Meat Sci 90:851–857
Teena M, Manickavasagan A, Mothershaw A, El Hadi S, Jayas D (2013) Food Bioprocess Technol 6:1621–1634
Tosukhowong A, Visessanguan W, Pumpuang L, Tepkasikul P, Panya A, Valyasevi R (2011) Food Chem 129:846–853
Valous NA, Mendoza F, Sun D-W et al (2009) Colour calibration of a laboratory computer vision system for quality evaluation of pre-sliced hams. Meat Sci 81(1):132–141
Wang HH, Sun D-W (2002) Melting characteristics of cheese: analysis of effect of cheese dimensions using computer vision techniques. J Food Eng 52(3):279–284
Wu D, Sun D-W (2013a) Innovative Food Sci Emerg Technol 19:1–14
Wu D, Sun D-W (2013b) Food Chem 145:417–426
Wu D, Sun D-W (2013c) Talanta 111:39–46
Wu D, Sun D-W (2013d) Talanta 116:266–276
Wu D, Wang S, Wang N, Nie P, He Y, Sun D-W, Yao J (2012a) Food Bioprocess Technol 6:2943–2958
Wu D, Sun D-W, He Y (2012b) Innovative Food Sci Emerg Technol 16:361–372
Wu D, Nie P, He Y, Bao Y (2012c) Food Bioprocess Technol 5:1402–1410
Wu D, Chen X, Cao F, Sun D-W, He Y, Jiang Y (2014) Food Bioprocess Technol 7:1555–1569
Xu SY, Chen XF, Sun D-W (2001) Preservation of kiwifruit coated with an edible film at ambient temperature. J Food Eng 50(4):211–216
Zaragozá P, Fuentes A, Fernández-Segovia I, Vivancos J-L, Rizo A, Ros-Lis JV, Barat JM, Martínez-Máñez R (2012) Food Chem 138:1374–1380
Zhu F, Zhang D, He Y, Liu F, Sun DW (2012) Food Bioprocess Technol 6:2931–2937
Acknowledgments
Hong-Ju He thanks the Chinese Scholarship Council for supporting his PhD study (under UCD-CSC funding programme).
Conflict of Interest
Hong-Ju He declares that he has no conflict of interest. Da-Wen Sun declares that he has no conflict of interest.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
He, HJ., Sun, DW. Selection of Informative Spectral Wavelength for Evaluating and Visualising Enterobacteriaceae Contamination of Salmon Flesh. Food Anal. Methods 8, 2427–2436 (2015). https://doi.org/10.1007/s12161-015-0122-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12161-015-0122-x