Abstract
The peptide-N 4-(N-acetyl-β-d-glucosaminyl) asparagine amidase F (PNGase F) catalyzes the cleavage of N-linked oligosaccharides between the innermost GlcNAc and asparagine residues of high mannose, hybrid and complex oligosaccharides from glycoproteins. The PNGase F has broad substrate specificity and thus is extensively used for the structural and functional studies of the glycoproteins. In this study, we tried to produce active recombinant PNGase F as secreted and intracellular-expressed forms using baculovirus expression vector system (BEVS) through silkworm larvae or cultured cells. PNGase F itself contains potential N-linked glycosylation sites and we found that it was N-glycosylated when PNGase F secreted from silkworm cells. Intriguingly, the secreted recombinant PNGase F has the lower catalytic activity and self-digests its N-linked glycans and therefore this secreted form of this enzyme produced from BEVS is not appropriate for carbohydrate chain analysis. Instead, we successfully mass-produced (2.1 mg/20 silkworm larvae) and purified active recombinant PNGase F as an intracellular protein without N-glycosylations. Besides, we confirmed by directed mutagenesis that several amino acid residues are crucial for the function of PNGase F. Our results provide an alternative method for the mass production of active enzymes involved in the study of glycoproteins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
N-Linked glycosylation is one of the post-translational modifications of protein that is critical for its correct folding, stability, and biological activity [1]. Proteins in secretion pathway are usually modified by specific glycotransferases with N-linked and/or O-linked oligosaccharides in endoplasmic reticulum (ER) and/or Golgi apparatus. N-glycans are modified at asparagine (Asn) residues of a consensus sequence Asn-X-Ser/Thr (where X can be any amino acid except proline). Glycoproteins are widely spread among the eukaryotic organisms, and it has been predicted that more than half of the eukaryotic proteome is glycosylated with about 90 % of these glycoproteins are likely to be N-glycosylated [2]. Hence, it is essential to elucidate the biological and physiological meanings of the protein glycosylation. In spite of the significance of carbohydrate chains, their detail structures and functions have not yet been well understood due to their remarkable heterogeneity.
Carbohydrate chain-cleaving enzymes are used for investigating oligosaccharide structures, structure–function relationships, and glycoprotein biosynthesis by virtue of their applicability under physiological conditions. Most common N-linked carbohydrates can be removed by peptide-N 4-(N-acetyl-β-d-glucosaminyl) Asn amidase F (PNGase F, EC3.5.1.52). The PNGase F is a 34.8 kDa (314 amino acids) enzyme secreted by the Gram-negative bacterium Flavobacterium meningosepticum [3]. PNGase F catalyzes the cleavage of N-linked oligosaccharides between the innermost GlcNAc and Asn residues of high mannose, hybrid and complex oligosaccharides from N-linked glycoproteins [4]. During the removal of intact glycans, the modified Asn residue is converted to aspartic acid (Asp), and the ammonia is liberated. Despite being active against most N-linked glycans, it cannot cleave the substrate when the innermost GlcNAc is linked to an α-(1,3)-fucose, most commonly found in plant and some insect glycoproteins ([5, 6], also see Supplemental Fig. 1). PNGase F has very broad substrate specificity and is widely used in proteomic research, biotechnology, and glycobiology.
It is known that purification of native PNGase F is a time-consuming, multi-step process with a frustrated low yield: 0.1–0.5 mg of pure protein per liter of F. meningosepticum culture, which significantly is a human opportunistic pathogen [7, 8]. To increase the amount of protein and to avoid the large culture of F. meningosepticum, PNGase F has been cloned and heterologously expressed in various protein expression systems, such as Escherichia coli and Pichia pastoris. The recombinant PNGase F (rPNGase F) expressed in E. coli remained cell membrane anchored instead of being secreted into periplasm [9]. Loo et al. improved the production and enzyme activity of rPNGase F in E. coli by removing the native signal sequence, and obtained 8 mg of protein from 1 L culture medium [10]. Recently, rPNGase F was expressed in the P. pastoris, and the yield was 800 mg/L [11]. The P. pastoris expression system has been reported as an excellent secretor of heterologous proteins into the culture medium, and thus the expressed rPNGase F might avoid the intracellular accumulation. However, the rPNGase F purified from P. pastoris was aberrantly N-glycosylated for the presence of multiple potential glycosylation sites. These undesirable N-linked oligosaccharides might interfere with the accurate analysis of N-glycan structures.
The baculovirus expression vector system (BEVS) is widely utilized and often expressed the recombinant proteins at extremely high levels, due to being driven by the strong polyhedrin promoter. It also has great advantages for expressing eukaryotic proteins because the insect cells can add the mammalian-like posttranslational modifications into proteins of interest [12, 13]. In this paper, we reported the expression, purification, and characterization of secreted and intracellular-expressed rPNGase F from the BEVS. The rPNGase F secreted into cultured silkworm cell medium was N-glycosylated and the enzymatic activity was diminished to a low level, whereas intracellular rPNGase F was not N-glycosylated and exhibited the activity equivalent to commercial native PNGase F. In addition, we introduced three mutations (R61G, W120C, and N243I), and compared their activities with each other using the purified enzymes.
Materials and Methods
Cells and Silkworms
The NIAS-Bm-oyanagi2 (kindly provided from Dr. Imanishi) and Bme21 cells were cultured in IPL-41 medium (Sigma, St. Louis, MO) supplemented with 10 % fetal bovine serum (Gibco, Grand Island, NY). The silkworm strains used in this study were obtained from the Institute of Genetic Resources, Kyushu University Graduate School. The larvae were reared on mulberry leaves at 25–27 °C.
Construction of Expression Vectors
An insect codon adapted coding sequence of the F. meningosepticum PNGase F (DDBJ accession No. LC004726) were chemically synthesized (Genscript, Tokyo, Japan) and cloned into pUC57 vector (Fig. 1a). To construct gateway-based entry clones, open-reading frame (ORF) of PNGase F was amplified by polymerase chain reaction (PCR) with KOD-Plus-Neo DNA polymerase (TOYOBO, Tokyo, Japan) using the specific primers PNGaseF-5 (5′-gctcccgctgacaacactgtgaacatcaag-3′) and PNGaseF-3-XhoI (5′-ccccctcgagtttgtgacgactggggctgag-3′). The amplified PCR product without native signal peptide was digested with XhoI, and further inserted into the EcoRV and XhoI double-digested site of the modified pENTR11 (pENTR11L2130KTEVH8STREP) vector [14] by DNA Ligation Kit Ver.2.1 (TaKaRa, Tokyo, Japan). The resulting construct was named pENTRL2130K-PNGaseF-TEVH8STREP. The 30K signal peptide sequences were removed by inverse PCR using the primer pairs L21-ATG-3 (5′-catggtggcggttttttaggagttgcctgc-3′) and PNGaseF-5 and termed pENTRL21NoSP-PNGaseF-TEVH8STREP. These vectors were subsequently used as templates to generate the seven mutations (R61G, W120C, N243I, R61G/W120C, R61G/N243I, W120C/N243I, and R61G/N243I/W120C) by inverse PCR. The following primers were used for introducing the base substitutions, R61 Ga: 5′-aacgtttacgtgaagaacaagacaaccg-3′, R61 Gb: 5′-ggcgtatccatcccattcgtcacaagtc-3′, W120Ca: 5′-gctaagggcagggagtactccgtggacttc-3′, W120Cb: 5′-caagcaggtttctgtgtagatcttgagctc-3′, N243Ia: 5′-gctcctgacagggctggatggtgtccagg-3′, N243Ib: 5′-ccagataccagggctctggttgttgatagg-3′.
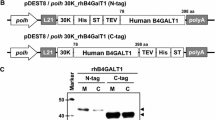
Schematic representation of the chemically synthesized coding sequences of PNGase F and expression constructs in this study. a The nucleotide sequences of an insect codon adapted PNGase F with deduced amino acid sequences. Nucleotide and amino acid residue numbers are shown on the left. For comparison, the amino acid residues mutated in this study were shown in bold italic letters, with substituted amino acid residues underneath. b Schematic representation of recombinant BmNPV/T3. The plasmid pDEST8/30K-PNGaseF-TEVH8STREP and pDEST8/NoSP-PNGaseF-TEVH8STREP were generated from its corresponding pENTR and pDEST8 vector by Gateway reaction. All destination vectors were under the control of polyhedrin (Polh) promoter and followed by SV40 polyadenylation signal (polyA). B1 and B2, represent recombination sites for Gateway cloning. L21 leader sequence for enhancing translation efficiency, 30K signal peptide of silkworm 30 kDa protein, PNGaseF peptide-N-glycosidase F, TEV tobacco etch virus protease cleavage site, H8: 8× histidine tag, STREP strep-tag
Generation of Recombinant BmNPVs for rPNGase F
The transfer plasmids for generating recombinant baculovirus were subsequently constructed using gateway LR reaction between the pENTRL2130K-PNGaseF-TEVH8STREP or pENTRL21NoSP-PNGaseF-TEVH8STREP and pDEST8 vector (Invitrogen, Carlsbad, California) according to the manufacture’s protocol (Fig. 1b). Recombinant baculoviruses were generated using BmNPV/T3 bacmid system as described previously [15]. The 200 ng bacmid DNAs were transfected into 1 × 105 NIAS-Bm-oyanagi2 cells in 24-well plates using FuGENE HD transfection reagent (Promega, Madison, WI). Then 4 days after incubation at 27 °C, the cell culture medium was harvested and centrifuged at 1000×g for 10 min at 4 °C. The supernatant containing recombinant P1 viruses were collected and stored at 4 °C in the dark. High-titer virus (P3) stock was prepared by serial infection following the protocols recommended in the manufacturer’s manual (Invitrogen). On the other hand, the cell pellets suspended in ice-cold PBS were subjected to Western blotting analysis.
All samples were electrophoresed on a 10 % sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under the reducing condition and transferred to polyvinylidene difluoride (PVDF) membrane (Millipore, Milford, MA), and further blocked in 5 % skim milk (Wako, Tokyo, Japan) followed by incubation with HisProbe-HRP (1:2000; Thermo Scientific, Rockford, IL). The protein bands were visualized using the Super Signal West Pico Chemiluminescent Substrate (Thermo Scientific).
Expression of rPNGase F in Cultured Silkworm Bme21 Cells and in Silkworm Larvae
Bme21 cells (1 × 105 cells/1 mL) in 32 mL IPL-41 insect cell culture medium were infected with the recombinant virus at an MOI of 10. At 4 days post infection (DPI), the samples were recovered by centrifugation at 1000×g (7780II Kubota, Tokyo, Japan) for 10 min. The extracellular PNGase F was prepared from the supernatant and the intracellular PNGase F was prepared from the cell pellet by freeze–thaw treatment. The supernatant was diluted in buffer A (20 mM Tris–HCl pH 7.4, 0.5 M NaCl), and the cell pellet suspended in buffer A was frozen at −80 °C overnight and then the thawed sample was rotated (2 rpm, RT-5 Taitec, Tokyo, Japan) for 30 min at 4 °C. The samples containing rPNGase F were then centrifuged at 52,500×g for 30 min at 4 °C, and further filtered with a 0.45-μm filter (Millipore).
As for the expression in silkworm larvae, the recombinant virus (1 × 105 plaque-forming unit per larva) was injected into the 5th instar silkworm larvae (day 3) of each strain. 4 days after inoculation, the silkworm was collected and stocked at −80 °C. Before purification, fat body from 20 freeze-thawed larvae was collected and suspended into buffer A containing 20 mM 1-phenyl-2-thiourea and 1 mM PMSF. To extract the rPNGase F, the protein sample was rotated (2 rpm, RT-5 Taitec) for 30 min at 4 °C and centrifuged at 52,500×g for 30 min at 4 °C. The supernatant was filtered using a 0.45-μm filter (Millipore) and subjected to the purification process.
Purification of rPNGase F
For the purification of rPNGase F, two-step purification protocol was performed based on the presence of the C-terminal His8-tag and STREP-tag. The samples containing the rPNGase F were purified by nickel affinity chromatography using HisTrap excel column (GE Healthcare Bioscience, Piscataway, NJ), and eluted by 500 mM imidazole solution buffers. After concentrated by ultrafiltration using Amicon 10 K filters (Millipore) in binding buffer B (100 mM Tris–HCl pH 8.4, 150 mM NaCl, 1 mM EDTA), the rPNGase F was applied to StrepTrap HP column (GE Healthcare), and then eluted by the buffer B containing 2.5 mM desthiobiotin.
The purified fractions were then visualized with Coomassie Brilliant Blue (CBB) R-250 after separation through 10 % SDS-PAGE. The purified rPNGase F was concentrated again and the buffer was exchanged to the storage buffer (20 mM Tris–HCl pH 7.5, 50 mM NaCl, 5 mM Mg2EDTA, 50 % Glycerol). The purified rPNGase F was quantified by ImageJ software using bovine serum albumin (BSA) as standard.
Deglycosylation Activity Assay
The deglycosidase assay was performed in reaction buffer (50 mM sodium phosphate buffer, pH 7.5 and 1 % NP-40) using RNase B (New England Biolabs, Beverly, MA) as substrate. The native PNGase F purified from F. meningosepticum (NEB) was used as a positive control. In general, 1 unit of PNGase F is defined as the amount of enzyme that digests 95 % of the N-glycans from 10 μg of RNase B when reacted at 37 °C for 1 h. 2.5–10 μg of RNase B or 5 μg of fetuin (Sigma, St. Louis, MO) or ovalbumin (Sigma, St. Louis, MO) were denatured with Glycoprotein Denaturing Buffer (NEB) at 100 °C for 10 min and 1.5–300 ng of native and rPNGase F were mixed with the reaction buffer and incubated at 7–37 °C for 1 h. The self-digestion analysis was performed by incubating 2.5 μg of non-denatured PNGase F from the medium of Bme21 cells. All the reactions were terminated by rapid heating to 95 °C for 5 min, and samples were resolved on 10 or 15 % SDS-PAGE and visualized by CBB R-250 staining.
N-Linked Glycan Analysis Using MALDI-TOF MS Spectrometry
40 μg of RNase B (NEB) alkylated by 123 mM IAA was digested by approximately 1 μg of trypsin (Roche) for 1 h at 37 °C. After heating at 90 °C for 5 min, RNase B was deglycosylated by 22.5 ng of NoSP-PNGase F (purified from silkworm larvae) for 12 h. The released N-linked glycans were purified and labeled with N-[(aminooxy)acetyl]tryptophanylarginine methyl ester (aoWR) using BlotGlyco (Sumitomo Bakelite, Tokyo, Japan) and analyzed using matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) performed on AXIMA-CFR Plus (Shimadzu, Japan).
Screening of Silkworm Strains for Efficient Production of rPNGase F
Seventeen silkworm strains were used for the screening of rPNGase F expressions. The virus expressing intracellular form of rPNGase F was injected into each silkworm strain. Then each silkworm was dissected at 4 days post infection (DPI) and 100 mg of fat body was suspended into 1 mL buffer A containing 20 mM 1-phenyl-2-thiofurea and 1 mM PMSF. After the centrifugation at 8500 rpm for 20 min, 2 μL of the supernatants was resolved on 10 % SDS-PAGE. The expression levels of randomly selected three samples of each infected strain were analyzed by Western blotting.
The expression levels were compared using the fat bodies from 20 infected larvae of p50 and p53 strains. The fat bodies were suspended to 100 mL buffer A and centrifuged at 52,500× g for 30 min, and 2 μL of the supernatants was analyzed by Western blotting. The band intensities were quantified with ImageJ software and further normalized to the controls from CBB staining.
Results and Discussion
Construction of Expression Vectors
PNGase F is a secreted protein from the gram-negative soil bacterium F. meningosepticum. Thus, in the present work, we utilized the 30K signal peptide (Fig. 1b) from silkworm 30 kDa protein, which is one of the most efficient signal peptides in recombinant secretory protein production in the BEVS [16]. The previous studies reported that the rPNGase F expressed in P. pastoris was aberrantly N-glycosylated [11]. Bacteria express proteins without N-glycans in the native host. When expressed in eukaryotic hosts, many proteins may become glycosylated if containing potential glycosylation sites and may result in lower function. In the case of glycosidase, this abnormal N-linked oligosaccharide is most likely to interfere the enzymatic activity. Thus, we also generated the expression construct without any signal peptide (Fig. 1b). To improve the purity of proteins of interest, 8× His-tag and Strep-tag were added to the C-terminus of PNGase F for the efficient two-step purification.
PNGase F is one of the well-characterized glycosidases and the three-dimensional high-resolution-structure was determined [8, 17]. Several amino acid residues were identified that are essential to its activity using site-directed mutagenesis and crystallographic analysis [18]. However, the exact catalytic mechanism has not been described so far. Interestingly, we obtained four PNGase F sequences from UniProtKB/Swiss-Prot (Accession No. P21163: wild-type) or DDBJ/GenBank (Accession Nos. M57237: 61G, J05411: 120C, J05449: 243I, respectively) with conflicting amino acid substitutions [9, 19, 20]. To determine whether or not these substitutions could affect its activity, three site-specific mutations, R61G, W120C, and N243I were introduced into PNGase F as described under the “Materials and Methods” section (see Fig. 1a). In addition, the vectors bearing double or triple mutations were also generated. Taken together, we made 16 mutant constructs of PNGase F for recombinant baculoviruses.
Expression of rPNGase F in BmNPV-Infected Cultured Silkworm Cells
The 16 bacmid DNAs were transfected into the NIAS-Bm-oyanagi2 cells, and 4 days post transfection, the cells were subjected to western blotting analysis. As shown in Fig. 2a, all the NoSP-PNGase F proteins were detected as a clear single band and their expression levels were similar among the mutants. Unfortunately, it is difficult to detect the 30K-PNGase F in the cell culture media of NIAS-Bm-oyanagi2 (data not shown). The 30K-PNGase F proteins from cell lysates, however, generate the ladder-like bands, and it is noteworthy that the band patterns were distinct among mutants, suggesting that the N-glycosylated PNGase F proteins were accumulated in ER or Golgi apparatus, but hardly secreted because of some redundant modifications. Since the potential N-glycosylation sequences Asn-Lys-Thr (53-55) and Asn-Lys-Ser (152-154) were not located in the mutation sites, the differences observed in the band patterns were attributed to the structural differences of N-glycans. It is possible that the N-glycans aberrantly appended to the 30K-PNGase F proteins were self-digested to some extent during the secretion pathway. If this is true, the increased band shift is correlated to loss of self-digestion activities.
Expression and purification of rPNGase F from cultured silkworm cells. a PNGase F expression in NIAS-Bm-oyanagi2 cells infected with the recombinant BmNPV (wild-type, R61G, W120C, N243I, R61G/W120C, R61G/N243I, W120C/N243I, and R61G/W120C/N243I). 4 days post transfection, rPNGase F proteins accumulated inside the cells were detected by Western blotting using HisProbe-HRP. The upper panel shows the NoSP-PNGase F mutants, and the lower panel shows 30K-PNGase F mutants. b Purification of wild-type NoSP-PNGase F from the pellets of silkworm e21 cells and c 30K-PNGase F from the Bme21 cell cultured medium through the two-step affinity chromatography. Lanes 1–4 nickel affinity chromatography, lanes 5–8 strep-tag affinity chromatography, M molecular markers, IP input, FT flow-through, WS wash fraction, lane 4 elution fraction by 500 mM imidazole, lane 8 elution fraction by 2.5 mM desthiobiotin. The samples were analyzed by Coomassie-stained 10 % SDS-PAGE. The arrows indicate the bands of rPNGase F
Purification of Wild-Type PNGase F Proteins from Silkworm Bme21 Cells
Because of the low secretion amount of the PNGase F from NIAS-Bm-oyanagi2 cells, we then used the BmNPV hypersensitive Bme21 cells to produce PNGase F [21] in intracellular or secreted forms. Both PNGase F proteins were purified in high purity as observed in the SDS-PAGE (Fig. 2b, c). Unlike the NIAS-Bm-oyanagi2 cells, Bme21 cells successfully secreted high yield of rPNGase F with different molecular weight forms. Deglycosylation treatment by commercial PNGase F confirmed that the secreted recombinant 30K-PNGase F contains N-linked glycans (data not shown).
Deglycosylation Activity
To confirm our hypothesis on the band shift observed in Fig. 2a, the self-digestion analysis was performed. As shown in Fig. 3a, the ladder-like bands of secreted PNGase F were shifted to a single sharp band after 1-h incubation with reaction buffer. This result clearly indicates that the contamination of N-glycan derived from secreted PNGase F is inevitable, and hence secreted-type PNGase F is not suitable for the structural analysis of glycoconjugates. To compare the enzymatic activity, the amount of 30K-PNGase F was measured after digestion of N-glycan and utilized the following experiments.
Enzymatic activities of rPNGase F proteins. a Self-digestion of secreted PNGase F. 2.5 μg of secreted PNGase F was incubated in the reaction buffer for indicated times at 37 °C, and analyzed by 10 % SDS-PAGE. b Enzymatic activities of native and recombinant PNGase F proteins. RNase B was incubated with Native PNGase F (lane 2–4), NoSP-PNGase F (lane 5–7) and 30K-PNGase F (lane 8–10) with indicated amount of enzyme, and subjected to 15 % SDS-PAGE and Coomassie staining. Lane 1 (mock) represents RNase B without enzyme treatment
PNGase F activity can be detected by the mobility shift of glycoproteins due to the removal of the glycans. To analyze the activities of purified rPNGase F proteins, RNase B, a high-mannose glycoprotein carrying a single N-glycan was employed as substrate for deglycosylation reaction.
As shown in Fig. 3b, the deglycosylation activity of NoSP-PNGase F appears to be identical to that of the commercial PNGase F and is not affected by the H8-STREP-tag. On the other hand, the secreted PNGase F showed significantly low activity. The aberrant N-glycan attached to the 30K-PNGase F is assumed to be the reason to the decreased enzymatic activity. To confirm the effect of N-glycan, the enzymatic activity was compared between 30K-PNGase F before and after self-deglycosylation. Whether or not the 30K-PNGase F possesses N-glycan, the enzymatic activity was equal and lower than that of NoSP-PNGase F (Supplemental Fig. 2). When 30K-PNGase F digests its own N-glycan, the Asn is converted to Asp (see Supplemental Fig. 1), which is likely to be responsible for the low enzymatic activity.
Screening of Silkworm Strains for Large-Scale Expression of rPNGase F
As mentioned above, intracellular form is more valuable than secreted PNGase F for glycobiology research. However, the yield of NoSP-PNGase F purified from Bme21 cells was low: 30 μg of pure protein from 32 mL culture medium. To achieve a mass production of rPNGase F, silkworm-BEVS was employed. The protein expression using silkworm or its pupae is approximately 10 to 100-fold higher than that from cultured silkworm cells, and the silkworm expression platform is inexpensive and easy to scale-up for mass production [22, 23].
Then firstly, we screened the silkworm strains maintained in Kyushu University to approve whether there is a suitable strain for the mass production of PNGase F. In general, the d17 strain is highly permissive to BmNPV and suitable for large-scale expression of foreign proteins [14, 24]. As shown in Fig. 4a, several strains show relatively lower NoSP-PNGase F expression, there was no strain showed significantly higher expression level than that of d17. This is somewhat surprising, since many proteins tested in our laboratory exhibited different amounts of production among the strains [14, unpublished data]. In addition to BmNPV-susceptibility, the total amount of fat body may contribute to the yield of recombinant proteins. To validate this, total fat bodies from 20 silkworm larvae were collected and the expression amount was compared. However, there was no significant difference between wild-type strain p50 [BmNPV low-permissive strain, 24] and p53 strain which shows high level expression of other recombinant proteins (Fig. 4b) [14, unpublished data].
Screening of silkworm strains for mass production of rPNGase F. a 2 mg of fat bodies from the silkworms infected with recombinant BmNPV/NoSP-PNGase F were resolved in 10 % SDS-PAGE, and the PNGase F was detected by Western blotting using HisProbe-HRP. Three individual samples were prepared from each strain, and the expression from d17 strain fat body was used as the control. b The comparison of expression levels between p50 and p53 silkworm strains. Crude extracts (5 μL) from 20 silkworm fat bodies was resolved in 10 % SDS-PAGE and the PNGase F was detected by Western blotting using HisProbe-HRP or visualized by Coomassie Brilliant Blue R-250 staining. The expression levels of the NoSP-PNGase F were quantitated by ImageJ software and normalized to the expression level from p50 strain
Purification and Enzymatic Characterization of rPNGase F from Silkworm Larvae
At 4 DPI, the fat bodies from infected silkworm larvae were harvested. After serial purifications through nickel affinity and Strep-tag affinity chromatography, approximately 2.1 mg of PNGase F was purified from 20 p53 silkworm larvae in high purity (Fig. 5a), and this is much more higher than that from Bme21 cells (30 μg/32 mL culture medium, Supplemental Fig. 3). Our results suggested that the silkworm-BEVS could be applicable for mass production of the PNGase F.
Purification and characterization of PNGase F from silkworm fat bodies. a The purification of NoSP-PNGase F through two-step affinity chromatography. Lanes 1–4 nickel affinity chromatography, lanes 5–8 strep-tag affinity chromatography, M molecular markers, IP input, FT flow-through, WS wash fraction, lane 4 elution fraction by 500 mM imidazole, lane 8 elution fraction by 2.5 mM desthiobiotin. The black and gray arrows indicate the recombinant and native PNGase F, respectively. Comparison of deglycosylation performance between native and recombinant PNGase F. b 10 μg of RNase B was incubated with 1.5–150 μg Native PNGase F (lane 2–4) or NoSP-PNGase F (lane 5–7) for 1 h at 37 °C, and c 2.5 μg of RNaseB was incubated with 3 ng of native (lane 2–6) and NoSP-PNGae F (lane 7–11) for 1 h at the temperature indicated above. d Fetuin (lane 1–3) and ovalbumin (lane 4–6) were digested with Native PNGase F (lane 2, 5) and NoSP-PNGase F (lane 3, 6) and analyzed by 10 % SDS-PAGE
Using the purified NoSP-PNGase F protein, we compared its deglycosylation activity with commercial native enzyme using RNase B as substrate. As shown in Fig. 5b, 15 ng of the NoSP-PNGase F (~10 units) was sufficient for the complete deglycosylation in 1 h that is comparable to the PNGase F from NEB. Therefore, approximately 1,400,000 units of rPNGase F were purified from 20 silkworm fat bodies. As shown in Fig. 5d, the purified rPNGase F was highly active at 24–37 °C (Fig. 5c) and could digest complex- and hybrid-type N-glycans from bovine fetuin [25] and chicken ovalbumin [26, 27]. These results indicated that the activity of rPNGase F was comparable to those of commercial PNGase F.
To validate the substrate specificity of NoSP-PNGase F, the N-linked glycans released from RNase B by rPNGase F were subjected to MALDI-TOF MS. The mass of each N-linked glycan was measured as the additional mass (477.22) due to labeling with N-[(aminooxy)acetyl]tryptophanylarginine methyl ester (aoWR) for high sensitivity detection. Oligosaccharides from bovine pancreas RNase B have been characterized to be Man5GlcNAc2 through Man9GlcNAc2 ([28], also see Supplemental Fig. 4). The MALDI-TOF MS measurement for the aoWR-labeled glycans digested by NoSP-PNGase F were detected as five major peaks corresponding to Man5GlcNAc2-(aoWR), Man6GlcNAc2-(aoWR), Man7GlcNAc2-(aoWR), Man8GlcNAc2-(aoWR), and Man9GlcNAc2-(aoWR), respectively (Fig. 6). This result clearly proved that NoSP-PNGase F cleaves of N-linked oligosaccharides between the innermost GlcNAc and Asn residue.
Effects of R61G, W120C, and N243I Mutations on the Enzymatic Activity of rPNGase F
To validate the influence of point mutations in rPNGase F, wild-type and mutants of intracellular-expressed PNGase F (R61G, W120C, and N243I) were purified from silkworm larvae infected with each recombinant BmNPV. The yields of mutants were almost the same as wild-type rPNGase F (data not shown). To compare the enzymatic activities, the RNase B was deglycosylated by 1.5–150 ng of wild-type or mutant PNGase F. As shown in Fig. 7, the enzymatic activity was significantly reduced in the all of the mutant enzymes: R61G, W120C, and N243I. The site-directed mutagenesis and crystallographic analysis by Kuhn et al. suggested that D60 accepts an H+ from water molecule to activate bound water acting as the nucleophile, thus R61 is presumably important for generation of hydrophilic environment in the active site. It also suggested the E118 is important for substrate binding and recognition, thus W120 is most likely to relate to the substrate binding [18]. These results on enzymatic activity reinforce that the ladder-like bands in Fig. 2a are correlated with the catalytic activity of each mutant. Moreover, our results demonstrated here indicated that other amino acid residues also affect the function of PNGase F as a glycosidase. However, further efforts, e.g., crystal structure determination, should be made to interpret the detailed mechanisms in lower function of mutant proteins.
The comparison of enzymatic activity among wild-type and mutant PNGase F proteins. RNase B (2.5 μg) was incubated with 3–300 ng of each NoSP-PNGase F, wild-type (lane 1–3), R61G (lane 4–6), W120C (lane 7–9), and N243I (lane 10–12), for 1 h at 37 °C were analyzed by CBB-stained 15 % SDS-PAGE. The arrow indicates the rPNGase F. The glycosylated or deglycosylated proteins are indicated the right. M molecular mass markers
In summary, we successfully produced rPNGase F as secreted and intracellular-expressed forms using BEVS through silkworm larvae or cultured cells. About 2.1 mg of pure and fully active PNGase F was obtained from 20 silkworm larvae. Secreted rPNGase F was redundantly N-glycosylated contributing to its significant lower catalytic activity and could self-deglycosylated. Besides, we confirmed by mutagenesis that several amino acid residues are crucial for the function of PNGase F. The silkworm-BEVS is easy to scale-up for the larger amount of PNGase F, which enables us to perform N-glycomics analysis.
References
Imperiali, B., & O’Connor, S. E. (1999). Effect of N-linked glycosylation on glycopeptides and glycoprotein structure. Current Opinion in Chemical Biology, 3, 643–649.
Apweiler, R., Hermjakob, H., & Sharon, N. (1999). On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochimica et Biophysica Acta, 1473, 4–8.
Plummer, T. H, Jr, Elder, J. H., Alexander, S., Phelan, A. W., & Tarentino, A. L. (1984). Demonstration of peptide: N-glycosidase F activity in endo-beta-N-acetylglucosaminidase F preparations. The Journal of Biological Chemistry, 259, 10700–10704.
Elder, J. H., & Alexander, S. (1982). Endo-beta-N-acetylglucosaminidase F: Endoglycosidase from Flavobacterium meningosepticum that cleaves both high-mannose and complex glycoproteins. Proceedings of the National Academy of Sciences of the United States of America, 79, 4540–4544.
Chu, F. K. (1986). Requirements of cleavage of high mannose oligosaccharides in glycoproteins by peptide N-glycosidase F. The Journal of Biological Chemistry, 261, 172–177.
Tretter, V., Altmann, F., & März, L. (1991). Peptide-N 4-(N-acetyl-beta-glucosaminyl)asparagine amidase F cannot release glycans with fucose attached alpha 1-3 to the asparagine-linked N-acetylglucosamine residue. European Journal of Biochemistry, 199, 647–652.
Mussar, K. J., Murray, G. J., Martin, B. M., & Viswanatha, T. (1989). Peptide: N-glycosidase F: studies on the glycoprotein aminoglycan amidase from Flavobacterium meningosepticum. Journal of Biochemical and Biophysical Methods, 20, 53–68.
Norris, G. E., Stillman, T. J., Anderson, B. F., & Baker, E. N. (1994). The three-dimensional structure of PNGase F, a glycosylasparaginase from Flavobacterium meningosepticum. Structure, 2, 1049–1059.
Barsomian, G. D., Johnson, T. L., Borowski, M., Denman, J., Ollington, J. F., Hirani, S., et al. (1990). Cloning and expression of peptide-N 4-(N-acetyl-beta-D-glucosaminyl) asparagine amidase F in Escherichia coli. The Journal of Biological Chemistry, 265, 6967–6972.
Loo, T., Patchett, M. L., Norris, G. E., & Lott, J. S. (2002). Using secretion to solve a solubility problem: high-yield expression in Escherichia coli and purification of the bacterial glycoamidase PNGase F. Protein Expression and Purification, 24, 90–98.
Hua, L., Gao, X., Yang, X., Wan, D., He, C., Cao, J., et al. (2014). Highly efficient production of peptides: N-Glycosidase F for N-glycomics analysis. Protein Expression and Purification, 97, 17–22.
O’Reilly, D. R., Miller, L., & Luckow, V. A. (1992). Baculovirus expression vectors: A laboratory manual (pp. 216–234). New York: Oxford University Press.
Kato, T., Kajikawa, M., Maenaka, K., & Park, E. Y. (2010). Silkworm expression system as a platform technology in life science. Applied Microbiology and Biotechnology, 85, 459–470.
Mitsudome, T., Xu, J., Nagata, Y., Masuda, A., Iiyama, K., Morokuma, D., et al. (2014). Expression, purification, and characterization of endo-β-N-acetylglucosaminidase H using baculovirus-mediated silkworm protein expression system. Applied Biochemistry and Biotechnology, 172, 3978–3988.
Ono, C., Nakatsukasa, T., Nishijima, Y., Asano, S., Sahara, K., & Bando, H. (2007). Construction of the BmNPV T3 bacmid system and its application to the functional analysis of BmNPV he65. Journal of Insect Biotechnology and Sericology, 76, 161–167.
Soejima, Y., Lee, J., Nagata, Y., Mon, H., Iiyama, K., Kitano, H., et al. (2013). Comparison of signal peptides for efficient protein secretion in the baculovirus-silkworm system. Central European Journal of Biology, 8, 1–7.
Kuhn, P., Tarentino, A. L., Plummer, T. H, Jr, & Van Roey, P. (1994). Crystal structure of peptide-N 4-(N-acetyl-beta-D-glucosaminyl) asparagine amidase F at 2.2-A resolution. Biochemistry, 33, 11699–11706.
Kuhn, P., Guan, C., Cui, T., Tarentino, A. L., Plummer, T. H., & Van Roey, P. (1995). Active site and oligosaccharide recognition residues of peptide-N 4-(N-acetyl-beta-D-glucosaminyl)asparagine amidase F. The Journal of Biological Chemistry, 270, 29493–29497.
Lemp, D., Haselbeck, A., & Klebl, F. (1990). Molecular cloning and heterologous expression of N-glycosidase F from Flavobacterium meningosepticum. The Journal of Biological Chemistry, 265, 15606–15610.
Tarentino, A. L., Quinones, G., Trumble, A., Changchien, L. M., Duceman, B., Maley, F., et al. (1990). Molecular cloning and amino acid sequence of peptide-N 4-(N-acetyl-beta-D-glucosaminyl)asparagine amidase from Flavobacterium meningosepticum. The Journal Biological Chemistry, 265, 6961–6966.
Lee, J., Kawakami, N., Mon, H., Mitsunobu, H., Iiyama, K., Ninaki, S., et al. (2012). Establishment of a Bombyx mori nucleopolyhedrovirus (BmNPV) hyper-sensitive cell line from the silkworm e21 strain. Biotechnology Letters, 34, 1773–1779.
Motohashi, T., Shimojima, T., Fukagawa, T., Maenaka, K., & Park, E. Y. (2005). Efficient large-scale protein production of larvae and pupae of silkworm by Bombyx mori nuclear polyhedrosis virus bacmid system. Biochemical and Biophysical Research Communications, 326, 564–569.
Mon, H., Lee, J., Fukushima, M., Nagata, Y., Fujii, M., Xu, J., et al. (2013). Production and characterization of the celery mismatch endonuclease CEL II using baculovirus/silkworm expression system. Applied Microbiology and Biotechnology, 97, 6813–6822.
Kawakami, N., Lee, J., Mon, H., Kubo, Y., Banno, Y., Kawaguchi, Y., et al. (2008). Efficient protein expression in Bombyx mori larvae of the strain d17 highly sensitive to B. mori nucleopolyhedrovirus. Molecular Biotechnology, 40, 180–185.
Green, E. D., Adelt, G., Baenziger, J. U., Wilson, S., & Van Halbeek, H. (1988). The asparagine-linked oligosaccharides on bovine fetuin. Structural analysis of N-glycanase-released oligosaccharides by 500-megahertz 1H NMR spectroscopy. The Journal of Biological Chemistry, 263, 18253–18268.
Corradi Da Silva, M. L., Stubbs, H. J., Tamura, T., & Rice, K. G. (1995). 1H NMR characterization of a hen ovalbumin tyrosinamide N-linked oligosaccharide library. Archives of Biochemistry and Biophysics, 318, 465–475.
Harvey, D. J., Wing, D. R., Küster, B., & Wilson, I. B. (2000). Composition of N-linked carbohydrates from ovalbumin and co-purified glycoproteins. Journal of American Society for Mass Spectrometry, 11, 564–571.
Fu, D., Chen, L., & O’Neill, R. A. (1994). A detailed structural characterization of ribonuclease B oligosaccharides by 1H NMR spectroscopy and mass spectrometry. Carbohydrate Research, 261, 173–186.
Acknowledgments
The NIAS-Bm-oyanagi2 cell line for propagation of recombinant BmNPVs was kindly provided by Dr. Imanihi (National Institute of Agrobiological Sciences, Japan). We also thank Dr. Chisa Aoki (Kyushu University Graduate School) for providing the Bme21 cell line for the expression of recombinant protein. The MALDI-TOF MS was kindly supported by Center for Advanced Instrumental and Educational Supports (Faculty of Agriculture, Kyushu University).
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1
Diagram of substrate specificities of PNGase F. The reaction results in release of the oligosaccharide and the aspartic acid-containing polypeptide. PNGase F is not able to cleave N-linked glycans from glycoproteins when the innermost GlcNAc residue is linked to an α(1-3) fucose residue. Black arrow represents the cleavage site of PNGase F. Gray circles: mannose, Black squares: GlcNAc, Black triangle: fucose, X: any sugar. Supplementary material 1 (JPEG 420 kb)
Supplemental Fig. 2
Comparison of the enzymatic activity of 30K-PNGase F before and after the self-digestion. 30K-PNGase F (self-digested) was generated by incubating 30K-PNGase F under the reaction condition at 37 °C for 1 h. 200 ng of 30K-PNGase F (with N-glycan) (lane 2, 3), 30K-PNGase F (self-digested) (lane 4, 5) and NoSP-PNGase F (lane 6, 7) which were incubated with denatured 15 μg fetuin (lane 3, 5, 7) at 37 °C for 1 h and that before the incubation (lane 2, 4, 6) were analyzed by CBB-stained 10 % SDS-PAGE. The arrows indicate the bands of rPNGase F. Mock (lane 1): fetuin without enzyme treatment. M: molecular mass markers. Supplementary material 2 (JPEG 525 kb)
Supplemental Fig. 3
Purification of rPNGase F by nickel affinity chromatography. The NoSP-PNGase F proteins purified from (a) the Bme21 cells, and (b) the silkworm fat bodies by nickel affinity chromatography. M molecular mass markers, IP input, FT flow-through, WS wash fraction, lanes 1–9 elution fractions by 500 mM imidazole. All samples were analyzed by Coomassie-stained 10 % SDS-PAGE, and the arrows indicate the bands of rPNGase F. Supplementary material 3 (JPEG 652 kb)
Supplemental Fig. 4
Structure of N-linked oligosaccharides of RNase B. The gray circles and black squares represent mannose and GlcNAc, respectively. Black arrow represents the cleavage site of PNGase F. Supplementary material 4 (JPEG 524 kb)
Rights and permissions
About this article
Cite this article
Masuda, A., Xu, J., Mitsudome, T. et al. Mass Production of an Active Peptide-N-Glycosidase F Using Silkworm-Baculovirus Expression System. Mol Biotechnol 57, 735–745 (2015). https://doi.org/10.1007/s12033-015-9866-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-015-9866-1