Abstract
Mucin-type O-glycosylation is initiated by UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases (ppGalNAc-Ts or PGANTs), attaching GalNAc to serine or threonine residue of a protein substrate. In the insect model from Lepidoptera, silkworm (Bombyx mori), however, O-glycosylation pathway is totally unexplored and remains largely unknown. In this study, as the first report regarding protein O-glycosylation analysis in silkworms, we verified the O-glycan profile that a common core 1 Gal (β1-3) GalNAc disaccharide branch without terminally sialylated structure is mainly formed for a baculovirus-produced human proteoglycan 4 (PRG4) protein. Intriguingly, functional screenings in cultured silkworm BmN4 cells for nine Bmpgants reveal that Bmpgant2 is the solo functional BmPGANT for PRG4, implying that Bmpgants may have unique cell/tissue or protein substrate preferences. Furthermore, a recombinant BmPGANT2 protein was successfully purified from silkworm-BEVS and exhibited a high ability to transfer GalNAc for both peptide and protein substrates. Taken together, the present results clarified the functional BmPGANT2 in cultured silkworm cells, providing crucial fundamental insights for future studies dissecting the detailed silkworm O-glycosylation pathways and productions of glycoproteins with O-glycans.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glycosylations are found typically for extracellular and membrane-bound targets, and it has been estimated that half of the whole proteins hold glycans (Apweiler et al. 1999). There are two commonly detected types of glycosylation, referred to as N- and O-linked glycosylations (Stanley 2011). Both N- and O-glycans have been reported to serve a variety of structural and functional roles in membrane-bound and secreted proteins (Taylor and Drickamer 2007; van Kooyk and Rabinovich 2008; Kulkarni et al. 2010; Rillahan and Paulson 2011). It is known that O-linked glycans are typically attached to Ser, Thr, or Tyr residues, with no fixed consensus sequence, which poses a challenge to fully characterize O-glycan in terms of site occupancy (Lairson et al. 2008). Although still only poorly understood, recent reports implicate roles for O-glycoproteins like fetuin, proteoglycan, and mucin, in protein storage/transportation, diseases, and development in higher eukaryotes, and moreover, aberrant expression of O-glycans, e.g., sialylated Tn (STn), is considered as a hallmark of many malignant tumors (Munkley 2016).
The predominant form of O-linked glycosylation is O-GalNAc or mucin-type glycosylation (hereafter referred to as O-glycosylation), which is initiated by a large family of Golgi resident glycosyltransferases (GTs) named UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase (ppGalNAc-T in mammals or pgant in fruit fly), transferring of GalNAc directly to an S/T from a polypeptide chain to create the Tn antigen (GalNAcαSer/Thr) structure (Bennett et al. 2012). Subsequently, elaboration of O-linked linkages could be achieved by the transfer of additional saccharide units in a stepwise fashion to the Tn antigen backbone, employing sets of GTs to generate common core O-glycan structures, such as core 1 (Galβ1-3GalNAcαSer/Thr) and core 2 (GlcNAcβ1-6(Galβ1-3)GalNAcαSer/Thr), etc. (Hang and Bertozzi 2005; Fig. 1).
Canonical biosynthesis of mucin-type O-glycans. Biosynthesis of O-glycans starts from the transfer of N-acetylgalactosamine (GalNAc) to Ser or Thr (S/T) of membrane-bound or secreted protein targets, forming the Tn antigen (Tn Ag). The addition of other sugars in a gradual manner results in the formation of the common core O-glycan structures, core 1 [T antigen (TAg)], core 2, core 3, and core 4, respectively. Additional extensions of linear or branched structures to the O-glycans by further adding, e.g., negative charged sugar sialic acid or galactose, are typically observed in mammals. The number of silkworm orthologs (also listed in Table 1) responsible for each structure was noted in red (e.g., nine isoforms of Bmpgant were found in silkworms). Abbreviations: ppGalNAc-Ts or Pgants, UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases; C1GalT1 or T-syn, core 1 β1,3-galactosyltransferase; β3Gn-T6, β1,3-N-acetylglucosaminyltransferase 6; β6GlcNAc-T, β1,6-N-acetylglucosaminyltransferase
To date, lots of glycozymes and cofactors have been identified to participate in N- and O-glycan biosynthesis pathways. The GTs involved in the O-glycosylation biosynthesis pathway have been largely investigated in mammals such as human and mouse, while most of the information on insect glycobiology comes from the model organism, Drosophila melanogaster (Schwientek et al. 2002; Tran and Ten Hagen 2013). In another insect model from Lepidoptera, silkworm (Bombyx mori), however, O-glycosylation pathway is totally unexplored and remains elusive. Recently, emerging evidence indicates that silkworm serves as a wonderful insect model for fundamental research (Xia et al. 2014) and as a promising host for the baculovirus expression vector system (BEVS) to engineer glycoproteins with mammalian-like PTMs in a large-scale manner (Kato et al. 2010; Lee et al. 2012a, 2012b). It would be essential to draw a fine map of O-glycosylation pathway in the silkworm, B. mori, to uncover the functions of relevant O-glycosylation-related proteins, e.g., glycosyltransferases. To this end, as the first report in terms of protein O-glycosylation analysis in silkworms, we confirmed the O-glycan profile of a silkworm-BEVS-produced human proteoglycan 4 (PRG4, also known as lubricin) with a majority of common disaccharide Gal (β1-3) GalNAc (core 1) branch structure, indicating that the conserved and simplified O-glycosylation pathway exists in silkworms (Table 1). Subsequently, a functional screening for nine Bmpgant isoforms was executed by employing advanced soaking RNAi-BEVS platform in cultured silkworm BmN4-SID1 cells (Mon et al. 2012). Intriguingly, only BmPGANT2 is recognized as a functional BmPGANT in the ovary-derived cells. Furthermore, a recombinant BmPGANT2 protein from silkworm-BEVS was purified, and it exhibited a high ability to transfer GalNAc specifically from UDP-GalNAc to the acceptor peptide and protein substrates. Taken together, the present results clarified the functional BmPGANT2 in mucin-type O-glycosylation in cultured silkworm cells, providing crucial fundamental insights for the following studies in dissecting detailed silkworm O-glycosylation pathways and glycoprotein engineering.
Materials and methods
Animals and cultured cells
All the silkworm larvae, a60, d17, f38, g32, n21, and w17 strains, were provided by the silkworm stock center of Kyushu University, which was supported by the National BioResource Project (NBRP). The silkworms were reared routinely with mulberry leaves in silkworm-rearing rooms under controlled conditions at 25–27 °C. The B. mori cultured cell lines, BmN-BD (a gift from Dr. Hisanori Bando, Hokkaido University), BmN-FK (Funakoshi Inc.), BmN4 (a gift from Dr. Chisa Aoki, Kyushu University), BmN4-SID1 (Mon et al. 2012), Bm5 (Thermo Fisher Scientific), Bme21 and Bmc140 (a gift from Dr. Chisa Aoki, Kyushu University; Lee et al. 2012a, 2012b), and BmO2 (NIAS-Bm-oyanagi2, a gift from Dr. Shigeno Imanashi, National Institute of Agrobiological Sciences, Japan) were maintained at 27 °C in the Insect Genome Science Laboratory (Kyushu University) on IPL-41 medium (Sigma-Aldrich, St. Louis, MO, USA), supplemented with 10% fetal bovine serum (Life Technologies).
Virus
The recombinant bacmid BmNPV-polh-HsPRG4-TEV-His8 for expression of human PRG4 protein (Nagata et al. 2012), BmNPV-polh-30K6G-PSTBP3-TEV-His8 for pufferfish saxitoxin- and tetrodotoxin-binding protein 3 (Nagata et al. 2013; Xu et al. 2013; TrPSTBP3: Takifugu rubripes PSTBP3; TnPSTBP3: Takifugu niphobles PSTBP3), BmNPV-polh-30K6G–Bip-EPO-TEV-His8 for human erythropoietin (Nagata et al. 2013), BmNPV-polh-30K6G-SERPING1-TEV-His8-STREP for expression of human plasma protease C1 inhibitor (SERPING1, UniProtKB-P05155), and BmNPV-polh-30K6G-Bmpgant2-deltaTM-TEV-His8-STREP for expression of silkworm BmPGANT2 protein in BEVS were generated following the manufacturer’s instruction (Life Technologies), respectively. The BmO2 cells were utilized for transfection (FuGENE HD from Promega, Madison, WI) of 2 μg Bacmid per 6-well plate to generate recombinant baculovirus. The high P3 viral stocks were obtained by serial infections and kept in the dark at 4 °C until use. All the viral titers were determined by the end-point dilution method (O’Reilly et al. 1992). As for the expression verifications in cultured cells, the recombinant baculovirus at a multiplicity of infection (MOI) of 1 was used in this study to infect indicated cells. The fifth instar larvae at day 3 were carefully injected with the recombinant baculovirus at 1 × 105 plaque-forming unit (PFU) per larva using a microliter syringe with a 30-gauge needle (Hamilton Co., USA). The culture medium from infected cells at 3 days postinfection (dpi) or hemolymph from infected silkworm larvae at 5 dpi was collected and subjected to Western blot analysis or purification process.
In silico identification of target genes
Using amino acid sequences of Gal/GalNAc-T motif from either HsGalnt1 (LPVRTPTMAGGLFSIDRDYFQEIGTYDAGMDIWGGENLEISFRIWQCGGTLEIVTCSHVGHVFR) or pgant5 (APLRTPTMAGGLFSIDKDYFYEIGSYDEGMDIWGGENLEMSFRIWQCGGILEIIPCSHVGHVFR) as queries, BlastP was performed against the silkworm genome database (SilkDB, http://silkworm.genomics.org.cn/or http://silkbase.ab.a.u-tokyo.ac.jp/), with an E-value threshold of 10−5. The nine output protein sequences, termed BmPGANT1 to BmPGANT9, were further utilized for the domain analysis via the Pfam motif database (http://www.genome.jp/tools/motif/) to verify whether or not it has typical and integral domains of GalNAc-T enzymes. Meanwhile, these sequences were also employed for multiple alignments with human (Homo sapiens), mouse (Mus musculus), and fruit fly (D. melanogaster) pgants and for phylogenetic analyses using CLC sequence viewer and MEGA software. Phylogenetic trees were constructed using the neighbor-joining (NJ) method with 1000 bootstrap replicates (Tamura et al. 2011).
Tissue and cultured cell expression profiles and microarray analysis
Total RNAs were isolated from various silkworm tissues including the head, fat body, silk gland, midgut, testis, and ovary on day 3 of the fifth instar larvae; from eggs on indicated days (day 1 to day 10) after oviposition; and from different cultured silkworm tissue-derived cell lines including BmN-BD (ovary-derived), BmN-FK (ovary-derived), BmN4 (ovary-derived), Bm5 (ovary-derived), Bme21 (embryo-derived), and BmO2 (embryo-derived) by using ISOGEN (Nippon Gene, Japan). Subsequently, 1 μg RNA was subjected to first-strand cDNA synthesis by ReveTra Ace cDNA synthesis kit according to the manufacturer’s instructions (TOYOBO). Then PCR was then performed by gene-specific primers listed in Table 2 using KOD-Plus-Neo polymerase (TOYOBO). The expression of housekeeping gene BmGAPDH was employed as an internal control. As for the expression analysis of all Bmpgants, microarray data representing genome-wide gene expression in silkworm multiple tissues on day 3 of the fifth instar were taken from the SilkDB database and visualized in HemI software.
Soaking RNA interference and gene overexpression
For double-stranded RNA (dsRNA) synthesis, all the fragments were amplified from the first-strand cDNA of either silkworm fat body or cultured silkworm BmN4 cells using the gene-specific oligos including an additional T7 promoter sequence at both the 5′ and 3′ terminus (listed in Table 2). The dsRNAs were synthesized by in vitro transcription with T7 RNA polymerase (Roche). Then soaking RNAi assay was performed in BmN4-SID1 cells expressing Caenorhabditis elegans SID-1 (CeSID-1) transmembrane protein by protocols established previously (Mon et al. 2012). In this study, we use 100 ng dsRNA/gene/24-well (premixed dsRNA with culture medium) to knock down genes of interest for 5 days before viral infection. At 3 dpi of rBmNPV, the culture medium was then harvested for Western blot to detect expressed glycosylated rPRG4 or SERPING1.
As for the overexpression of BmPGANT2 or BmGOLGIN84 (BGIBMGA010211) in BmN4-SID1 cells, the coding sequence (CDS) of Bmpgant2 or Bmgolgin84 was cloned to the C-terminus EGFP or DsRED modification of pENTR™11 vector (Invitrogen, Carlsbad, CA) and subcloned (Gateway LR reaction, Thermo Fisher Scientific) to the piggyBac-based transposition destination vector pBac-IE2-Puro (Li et al. 2012; Chen et al. 2016), respectively. Subsequently, the resulting plasmid pBac-IE2-Bmpgant2-cEGFP-Puro was co-transfected with a piggyBac helper plasmid into BmN4-SID1 cells, followed by selection with the antibiotic puromycin to establish stable cell lines. Soaking RNAi and rescue assays were performed in BmN4-SID1 and BmN4-SID1-Bmpgant2-cEGFP cells, respectively. The expression of stably transformed Bmpgant2 was confirmed by observing the green EGFP signals under a fluorescence microscope (Keyence). To verify the subcellular co-localization of BmPGANT2 with BmGOLGIN84, the BmN4 cells transfected with pBac-IE2-Bmpgant2-cEGFP-Puro and pBac-IE2-Bmgolgin842-cDsRed-Puro were directly harvested and washed with ice-cold PBS twice. Subsequently, cells were prepared by a cytospinner (cytospin4; Thermo Scientific) on a slide fixed in methanol at − 20 °C for 30 min. The fixed cells were then stained with DAPI (0.1 μg/ml; Invitrogen) for visualizing the nucleus. The mounted cells (Vector Labs) were subjected to fluorescence microscopy (Keyence) for detecting green EGFP and DsRED signals of expressed BmPGANT2 and BmGOLGIN84 proteins.
Expression and purification of recombinant proteins
The BmNPV-sensitive silkworm f38 (Hino et al. 2016; Chen et al. 2016) and d17 (Lee et al. 2012b) larvae were utilized to purify recombinant HsPRG4 and BmPGANT2, respectively, according to our previously published procedures with slight modifications (Masuda et al. 2015; Morokuma et al. 2017). At 5 dpi, 10 ml serum from 20 infected larvae was harvested and centrifuged at 10,000g to remove cell debris. The supernatants were then diluted in buffer A (20 mM Tris–HCl pH 7.4, 0.5 M NaCl, 1 mM PMSF) and filtered through a 0.45-μm filter (Millipore, USA). The clear solution containing soluble proteins was then subjected to a nickel affinity chromatography with 1-ml HisTrap excel column (GE Healthcare Bioscience, USA) and eluted by 500 mM imidazole solution buffer. The eluted samples were verified by SDS-PAGE and further concentrated by ultrafiltration using Amicon 30K filters (Millipore, USA) in an exchange PBS buffer (pH 7.5). As for the purification of rBmPGANT2, a two-step purification protocol was performed based on the terminal tandem His and Strep tags. Briefly, after the first purification using 5-ml HisTrap excel column (GE Healthcare Bioscience, USA), the elution was buffer-changed in a binding buffer B (100 mM Tris–HCl pH 8.4, 150 mM NaCl, 1 mM EDTA). Subsequently, the resulting solution was applied to a 5-ml StrepTrap HP column (GE Healthcare Bioscience, USA) and eluted by elution buffer with 2.5 mM desthiobiotin. Finally, all elution was buffer-exchanged against Tris–HCl (pH 8.0) buffer and concentrated by ultrafiltration using Amicon 30K filters (Millipore, USA). The purified proteins were quantified by ImageJ software using bovine serum albumin (BSA) as standard.
Western blot and lectin blot analysis
The SDS-PAGE and Western blotting procedures were described previously (Xu et al. 2013). Concisely, all protein samples were electrophoresed on 12% SDS-PAGE under the reducing condition and further transferred to the polyvinylidene difluoride (PVDF) membrane (Millipore, Milford, MA). The membrane was blocked in 5% skim milk (Wako, Tokyo, Japan), followed by incubation with HisProbe-HRP (1: 20,000; Thermo Scientific, Rockford, IL) or Streptavidin-HRP (1: 20,000; Thermo Scientific, Rockford, IL) for 1 h at room temperature. Lectin blot was performed essentially as Western blot, except the antibody was HRP-conjugated Peanut Agglutinin (PNA, Vector Laboratories, Burlingame, CA), which binds to nonsialylated Galβ1-3GalNAcα1-Ser/Thr (core 1) in O-glycans. The protein bands were then visualized using the Super Signal West Pico Chemiluminescent Substrate (Thermo Scientific Rockford, IL).
MALDI-TOF mass spectrometry
One microgram of the purified rHsPRG4 alkylated by 17.5 mM IAA was digested by 400 units of trypsin (sequencing grade, Roche) at 37 °C for 1 h. After heating at 90 °C for 5 min, the peptide was deglycosylated by 50 units of O-glycosidase (NEB) at 37 °C overnight. The released O-glycans were purified and labeled with N-[(aminooxy) acetyl] tryptophanyl arginine methyl ester (aoWR) using BlotGlyco (Sumitomo Bakelite) according to the manufacturer’s protocol. The aoWR-labeled O-glycans were analyzed by using MALDI-TOF MS on AXIMA-CFR Plus (Shimadzu, Japan). The exact mass of glycans was calculated by the following formulation: [observed m/z (numbers at peak)] + [H2O (18.01)] − [aoWR (447.22)] − [H (1.00)]. The peaks less than m/z 550 were ignored and considered as contaminant peaks derived from the isolation reagents.
Glycosyltransferase assay
The activity of rBmPGANT2 was assayed using the glycosyltransferase activity kit (EA001, R&D Systems, UK) based on the release of phosphate during transfer of GalNAc from the donor UDP-GalNAc to the acceptor peptide EA2 (AnaSpec Inc., Catalog #63841; PTTDSTTPAPTTK) by rBmPGANT2. The detailed protocols were also described in Wu et al. (2011). The standard curve was generated in a microreader plate according to the manufacturer’s instructions, and the assay was conducted in the conditions described as follows: rBmPGANT2 (0, 0.005, 0.01, 0.02, 0.03, 0.04, 0.05 μg), coupling phosphatase I (0.2 μg), EA2 peptide (0.1 nM), and UDPGalNAc/UDP-Gal/UDP-Fuc/CMP-Sia (0.5 mM). All the components were diluted if needed in the assay buffer (50 mM Tris, 2.5 mM MnCl2, 1 mM CaCl2, pH 8.0). As for the control groups, assay buffer only (blank control), 50 ng human rB4GalT1 (Morokuma et al. 2017), or 50 ng mouse rMmIL1 (negative control, laboratory manufacture, Hino et al. 2016) was tested, respectively. As for the glycosyltransferase activity using various protein substrates, human alpha-1-antitrypsin (A1AT, Sigma) and fetal bovine fetuin (Sigma) with (heat inactivation afterward) or without O-glycosidase treatments were further employed instead of the EA2 peptide. The optical density was measured with a microplate reader at 594 nm (Bio-Rad). The equation specific activity (SA, pmol/min/μg) = [Phosphate released (nmol) × (1000 pmol/nmol)]/[Incubation time (min) × amount of enzyme (μg)] was employed to calculate the specific activity.
Data analysis and statistics
The NetOGlyc4.0 (Steentoft et al. 2013, http://www.cbs.dtu.dk/services/NetOGlyc/) was used to evaluate O-glycosites of rHsPRG4-cTEV-His8 in the study. Student’s t test was used to determine the statistical significance among different groups. Data are expressed as mean ± SD of three independent experiments. ***P < 0.001, n.s. (not significant), compared to the control.
Results
Genome-wide identification of genes involved in O-glycosylation biosynthesis pathway
As demonstrated in Fig. 1, the biosynthesis of mucin-type O-glycans starts from the transfer of GalNAc to S/T residue to create the Tn antigen (Tn Ag) structure by ppGalNAc-Ts/Pgants. The addition of other sugars to Tn Ag by the corresponding GTs in a gradual manner results in the formation of several common core O-glycan structures, core 1 (T antigen (TAg)), core 2 (GlcNAcβ1-6(Galβ1-3)GalNAcαSer/Thr), core 3 (GlcNAcβ1-3GalNAcαSer/Thr), and core 4 (GlcNAcβ1-6(GlcNAcβ1-3)GalNAcαSer/Thr), respectively. In mammals, additional extensions of linear or branched structures to the O-glycans by further adding, e.g., negative charged sugar sialic acid or galactose, are typically observed (Varki 2008). To dig the relevant GTs in silkworms, we performed a genome-wide BLAST with amino acid sequences of GTs reported from human or fruit fly. As shown in Fig. 1, the number of silkworm orthologs responsible for each GT was indicated in red. Briefly, nine silkworm orthologs for ppGalNAc-Ts [initial GTs: SilkDB Nos.: BGIBMGA005279 (Bmpgant1), BGIBMGA008904 (Bmpgant2), BGIBMGA005280 (Bmpgant3), BGIBMGA008895 (Bmpgant4), BGIBMGA010523 (Bmpgant5), BGIBMGA001064 (Bmpgant6), BGIBMGA000733 (Bmpgant7), BGIBMGA001365 (Bmpgant8), and BGIBMGA014556 (Bmpgant9)] and one single core 1 β1,3-galactosyltransferase [C1GALT1, aka core 1 synthase, BGIBMGA014130 (Bmc1galt1)] were recorded, while no acetylglucosaminyltransferase such as β1,3-galactosyltransferase (β3GnT for core 3) and β1,6-N-acetylglucosaminyltransferase (β6GlcNAc-T, for core 2 and core 4) was found in the silkworm genome database, suggesting that insects such as silkworm and fruit fly might be unable to synthesize core 2/3/4 types of O-glycans as mammals do (Tian and Ten Hagen 2008). It is known in human and mouse that C1GALT1-specific chaperone 1 (C1GALT1C1 or COSMC) is required for the full function of C1GALT1 (Ju and Cummings 2002) in order to elongate glycans beyond the initial Tn structure. Interestingly, same to the C1GalT1 (CG9520) from D. melanogaster, ortholog of C1GALT1C1 is also absent in the silkworm genome, indicating that both insects do not require this chaperone for the folding, stability, and/or full activity of the C1GalT1, and it is possible that some other unknown factor(s) exists in insects to stable C1GalT1 for its function.
Silkworm Bmpgants demonstrate overlapping tissue-specific expression profile
Given the expression levels of pgants significantly differ in tissues and cells from mammals and fruit fly (Schwientek et al. 2002; Lairson et al. 2008; Walski et al. 2017), we then analyzed the expression levels in different tissues of the fifth instar larvae using the genome-wide microarray data, which are publicly available in SilkDB (Xia et al. 2007). As shown in Supplemental Fig. S1, the overall expression levels of Bmpgants were relatively limited (in blue) in silkworm tissues, except that Bmpgant6 (BGIBMGA001064), an ortholog of HsGalNAcT-10, and pgant6, have broad expressions in various tissues and are found substantially expressed in the head and midgut tissues, indicating that Bmpgants express in a tissue-specific fashion and BmPGANT6 may be comprehensively involved in O-glycosylation in silkworms. Attractively, among all tissues analyzed, we noticed that additional Bmpgants (Bmpgant-2, Bmpgant-3, Bmpgant-5, Bmpgant-6, Bmpgant-8) are overlapped and expressed more in the silkworm testis, implying that those BmPGANTs may work in a collaborative model to support O-glycosylated proteins in the testis. On the other hand, however, normal or very weak mRNA expression levels are observed for most Bmpgants in the ovary, suggesting that more Bmpgants are potentially indispensable for the testis but not for the ovary.
Silkworm Bmpgants have corresponding orthologs in D. melanogaster
As studied by Ten Hagen et al. (2003), up to 14 pgants present in the fruit fly genome and 9 pgants (pgant1–8 and pgant35a) were identified as functional peptide and/or glycopeptide transferases against different substrates (Table 1). To verify whether or not silkworm Bmpgants have corresponding orthologs in the fruit fly D. melanogaster, a phylogenetic tree based on amino acid sequences (Supplemental Fig. S2) from 14 DmPGANTs and 9 BmPGANTs was constructed, and the results showed that each BmPGANT matches single pgant, indicating that silkworm has similar PGANT family members with fruit fly. It is interesting to recognize that fruit fly has several novel pgant homologs that are absent in the silkworm genome which raises questions on the functional variations among these PGANTs against substrates and in different tissues of silkworm and fruit fly.
BmPGANT2 initiates core 1 O-glycosylation in cultured silkworm BmN4 cells
Together with our previous results (Nagata et al. 2012), both silkworm larvae (Fig. 3a) and cultured silkworm cells (Fig. 3b) produce O-glycosylated protein, human PRG4 (Fig. 2a, b). To answer which O-glycans are attached to the rHsPRG4, we then purified the rHsPRG4 protein from baculovirus-infected cultured silkworm Bme21 cells (Lee et al. 2012a) and silkworm strain f38 due to the promising expression level found among cultured cells and silkworm strains for secreted proteins (Imai et al. 2015; Hino et al. 2016). As demonstrated in Fig. 2c, Bme21-derived rHsPRG4 was also sensitive to O-glycosidase and it resulted in a broad range of the de-O-glycosylated band. Then PNA lectin was employed to detect the nonsialylated core 1 structure (Galβ1-3GalNAcα1-Ser/Thr), and the target band for rHsPRG4 disappeared with O-glycosidase treatment. These results indicated that core 1 and/or core 2 O-glycans presented in silkworms. Results from MALDI-TOF MS (Fig. 2d) revealed that only a major disaccharide structure (HexHexNAc) peak specifying the core 1 structure of O-glycans was confirmed on rHsPRG4. Since there are no silkworm orthologs of β3Gn-T6 (for core 3) and β6GlcNAc-T (for core 2/4), we then proposed that similar to D. melanogaster (Aoki et al. 2008) and Caenorhabditis elegans (Ju et al. 2006), silkworm has the ability to synthesize O-glycans up to core 1 on protein substrates, e.g., HsPRG4 in this study.
A previously established (Nagata et al. 2009) recombinant partial human PRG4 (rHsPRG4) protein with a C-terminal His8 tag was employed in this study as an O-glycosylation reporter (a). SMB: somatomedin B domain; PEX: haemopexin domain; TEV: tobacco etch virus (TEV) protease cleavage site. Putative 55 O-glycosylated sites (S/T) were predicted through NetOGlyc 3.1 Server (b). The rHsPRG4 protein was purified from Bme21-BEVS and digested with O-glycosidase (*). Then lectin blot was performed using HRP-conjugated Peanut Agglutinin (PNA), which binds to nonsialylated Galβ1-3GalNAcα1-Ser/Thr in O-glycans. The deglycosylation status was confirmed by Western blot (His-Probe, left panel) or by lectin blot (PNA, right panel). Arrow indicates the fully glycosylated form of rHsPRG4 protein (c). Subsequently, O-linked glycans released from rHsPRG4 by O-glycosidase was analyzed by MALDI-TOF MS (d). The yellow square and green circle represent GalNAc and galactose, respectively. The numbers at the peak represent molecular masses of the labeled glycans
Subsequently, a soaking RNAi-BEVS platform was performed to investigate which BmPGANT initiates O-glycan synthesis in cultured silkworm cells (Mon et al. 2012; Xu et al. 2013; Nagata et al. 2013; Imai et al. 2015; Chen et al. 2016; Fig. S3a for effective RNAi validated by RT-PCR). It is clearly determined that shifted bands were revealed only when dsRNA against Bmpgant2 was presented in soaking RNAi (Fig. S3b: lane 3; Fig. 3c: lane 3; Fig. S3b: lane 2; Fig. S3c: lanes 2–8, 10, 11). As expected, no shifted bands were seen when there were several negative proteins of interest (PoTBT-BP1, PoTBT-BP3, PTnPSTBP3, and Bip-EPO) without putative O-glycosylation sites (Fig. 3d, e). It is shown in Fig. 3f that there were similar deglycosylated patterns from RNAi and O-glycosidase treatment. To our surprise, we also observe an O-glycosidase-resistant type of PTM, indicating silkworms generate heterogeneous products and might also have novel PTMs rather than core 1 O-glycans on rHsPRG4. To fully clarify the RNAi effects of Bmpgant2, we also designed and performed a rescue assay (Fig. 3g, h) in the stably transformed cells (BmN4-SID-BmPGANT2-EGFP). It is demonstrated in Fig. 3i, j that depletion of endogenous BmPGANT2 in BmPGANT2-EGFP overexpression cells has less rHsPRG4 deglycosylated. In other words, the introduced BmPGANT2 significantly rescued the function of RNAi-depleted endogenous Bmpgant2, suggesting that BmPGANT2 is the initial enzyme in cultured BmN4 cells when rHsPRG4 was used as a reporter.
Functional screening of Bmpgants in cultured silkworm BmN4-SID1 cells. Using a recombinant baculovirus BmNPV-rHsPRG4, O-glycosylation status of expressed reporter (secreted form) from either serum of various silkworm larvae strains (a: d17, a60, g32, w17, and n21) or medium of cultured silkworm cells (b: BmN4-SID1, BmN4, Bm5, Bme21 BmO, and Bmc140) was investigated by Western blot, respectively. Soaking RNAi against EGFP (control) or every nine ppGalNAc-T candidates was performed in BmN4-SID1 cells followed by infection with BmNPV-rHsPRG4 viruses (MOI = 5). As an initial screening for the active Bmpgants, we employed single (single dsRNA), triple (dsRNAs from three different Bmpgants), octagon (dsRNAs from eight different Bmpgants), and nonagon (all nine dsRNAs together) RNAi in BmN4-SID1 cells (Supplemental Fig. S2). Then the expressed rHsPRG4 proteins from culture media were investigated by Western blot (c). Likewise, as negative controls, N-glycoprotein candidates without putative O-glycosylation sites, PoTBT-BP1, PoTBT-BP3, TnPSTBP3, and Bip-EPO were tested after elimination for each Bmpgant in the BmN4-SID1/BmNPV system (d, e). Culture medium from EGFP dsRNA-treated cells was treated with O-glycosidase (NEB) to validate the deglycosylated patterns (f) according to the protocols provided by the manufacturer (see “Materials and methods”). To fully clarify the RNAi effects of Bmpgant2, we also performed a rescue assay by introducing plasmid-derived Bmpgant2 coding region (IE2-EGFP-PGANT2) after eliminating endogenous Bmpgant2 by soaking RNAi with dsRNA against CDS or 3′UTR (g) in BmN4-SID1 cells. Soaking RNAi efficiency for all dsRNAs and rescue assay was validated by evaluating deglycosylation of rHsPRG4 through Western blot (h, i). All density values from ImageJ were nominalized to the control (dsegfp) full-glycosylated rHsPRG4 protein (100%) to give a percentage of O-glycosylated rHsPRG4 (j). Data were shown as the mean ± SD of three independent experiments, ***P < 0.001, n.s. (not significant), compared to the control
Bmpgant2 is a resident protein in the Golgi apparatus
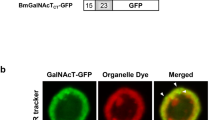
As for the Bmpgant2 which we focused on later in this study, as listed in Table 1, it is a homolog of human GalNAcT1 or fruit fly pgant5, with a high similarity of 58 and 67%, respectively. Motif analysis indicates that Bmpgant2 is composed of several typical conserved domains, which was plotted in Fig. 4a, b. Detailed amino acid alignment, motif, and phylogenetic tree analysis for those PGANTs are shown in Supplemental Figs. S4 and S5. As shown in Fig. 4c, Bmpgant2 expresses ubiquitously in silkworm tissues and cultured cells and during development, which was validated by semiquantitative RT-PCR. These results indicated that it is essential for silkworm development, like its ortholog pgant5 in the fruit fly, D. melanogaster (Tran and Ten Hagen 2013). Compared with the microarray results (Supplemental Fig. S1) from the Chinese silkworm strain Dazao, the p50T strain (WT strain in Japan) showed different expression patterns in several tissues such as fat body and head, indicating that there might not be identical in O-glycosylation status in those two WT strains. To determine the subcellular localization of BmPGANT2, we employed a C-terminus EGFP-fused BmPGANT2 and DsRed-fused BmGOLGIN84 (Golgi marker protein) to be co-expressed in BmN4 cells. As shown in Fig. 4d, the expressed EGFP-fused BmPGANT2 exhibited punctate morphology distributions co-localized with BmGOLGIN84 in the cytoplasm, which is a typical Golgi subcellular localization in cultured insect cells like Drosophila S2 cells and silkworm BmN cells (Rabouille et al. 1999; Cherry et al. 2006; Wang et al. 2013).
Expression profile and subcellular localization of BmPGANT2. The BmPGANT2 is an ortholog to Dmpgant5 and mammalian galnt1/13 (a). Motif analysis indicates that Bmpgant2 is composed of several typical conserved domains: cytoplasmic domain (Cyt.), stem region (SR), two catalytic domains [glycosyltransferase 1 (GT1) motif, Gal/GalNAc glycosyltransferase (Gal/GalNAc-T) motif], and a C-terminal Ricin motif. Representative alignments of amino acids from BmPGANTs, DmPGANT5 (#AAQ56702), and human GALNT1 (HsGALNT1) (b). Semiquantitative RT-PCR was performed to investigate the expression level of Bmpgant2 in different tissues and cultured silkworm cells (c, upper panel), which are normalized to the expression of Bmgapdh. Developmental expression pattern of Bmpgant2 during embryogenesis was examined until day 10 after oviposition (c, lower panel). Specific primers for Bmpgant2 and Bmgapdh genes were listed in Table 2. As for testing the subcellular distribution of BmPGANT2, C-terminal EGFP-fused BmPGANT2 (PGANT2-EGFP) and C-terminal DsRED-fused BmGOLGIN84 were co-expressed in cultured silkworm BmN4 cells. At 72 h after transfection, cells were fixed, and nuclei were labeled with DAPI. A representative merged image was shown in d. Scale bar, 20 μm
Purified rBmPGANT2 by silkworm-BEVS is active for transferring GalNAc
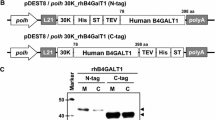
To explore if BmPGANT2 has actual functional activity as a GalNAc transferase, we then expressed and purified rBmPGANT2 by silkworm-BEVS. A schematic representation (Fig. 5a) explained the design of a secreted rBmPGANT2 with an N-terminal His8-Strep-TEV tandem fusion tag. As plotted in Fig. 5b, the rBmPGANT2 protein was highly expressed in d17 strains, and either HRP-His or HRP-Strep antibody was employed for validation of purified rBmPGANT2 protein by Western blot, which was shown in Fig. 5c that a single and sharp band was detected. Strikingly, we were able to obtain ~ 5.8 mg rBmPGANT2 from 10 ml silkworm hemolymph (~ 20 infected silkworms) with a satisfactory purity. Subsequently, its ability to transfer GalNAc from UDP-GalNAc (donor) to peptide EA2 (receptor) was assessed by glycosyltransferase assay (Wu et al. 2011). It is shown in Fig. 5d, e that rBmPGANT2 (0, 0.005, 0.01, 0.02, 0.03, 0.04, 0.05 μg), assay buffer only (blank control), 50 ng human rB4GalT1 (Morokuma et al. 2017, positive control, purified from silkworm hemolymph), or 50 ng mouse rMmIL1 (negative control, purified from silkworm hemolymph) was tested for GalNAc-specific transfer activity, respectively. As a result, it is clear to present here in Fig. 6 that there was no color alternation for the blank and negative controls, and when increasing amount of rBmPGANT2 was added, the optical density was also improved, indicating secreted rBmPGANT2 is an active GalNAc transferase. An equation for the specific activity (SA) was utilized to estimate the activity of the rBmPGANT2 (see “Materials and methods” for detailed procedures). Meanwhile, we also tested several common nucleotide sugars including UDP-Fuc, UDP-Gal, and CMP-Sia to evaluate the specificity of rBmPGANT2. According to our assay conditions, the purified rBmPGANT2 has an SA of ~ 2507 pmol/min/μg. When using fetuin as the acceptor, rBmPGANT2 exhibits an SA of ~ 562 pmol/min/ng, suggesting rBmPGANT2 is functional for both peptide and glycopeptide/protein. Compared to human GALNT-1 with an SA of ~ 1967 pmol/min/μg (Wu et al. 2011), we claimed that silkworm-purified rBmPGANT2 enzyme in this study is an active GalNAcT in vitro.
Function evaluation of purified recombinant BmPGANT2. Schematic representation of constructed recombinant BmPGANT2 with N-terminal His8-Strep-TEV tandem fusion tags. 30K6G is a secretion signal peptide (SP) from silkworm 30K6G protein (a). Purification of rBmPGANT2 from rBmNPV infected silkworm hemolymph (10 ml from ~20 infected silkworms). Subsequently, a two-step purification protocol was employed for His8 and STREP tags (see “Materials and methods” for detailed procedures). M, protein markers; FT, flow-through; lanes 2–5, nickel affinity chromatography; lanes 6–8, Strep-Tactin affinity chromatography; lane 4, elution fraction by 250 mM imidazole; lane 8, elution fraction by 2.5 mM desthiobiotin. All samples were resolved in 12% SDS-PAGE and visualized by CBB staining (b). Purified rBmPGANT2 was further verified by Western blot using either His-Probe or Strep-Probe. M, a protein marker for Western blot. Arrowhead indicates the correct band for rBmPGANT2 (c). Then the purified rBmPGANT2 was utilized for glycosyltransferase assay using glycosyltransferase activity kit (Catalog # EA001) according to the manufacturer’s instructions. Its ability to transfer GalNAc from UDP-GalNAc, but not UDP-Fuc/UDP-Gal/CMP-Sia, to peptide EA2 was investigated (d, e). Phosphate standard was prepared for generating the standard curve in a microreader plate, and 50 ng rBmPGANT2, assay buffer only (blank control), 50 ng human rB4GalT1, or 50 ng mouse rMmIL1 (negative control) was tested for GalNAc-specific transfer activity, respectively. The optical density was measured at 594 nm. An equation Specific Activity (SA, pmol/min/μg) = [Phosphate released (nmol) × (1000 pmol/nmol)]/[Incubation time (min) × amount of enzyme (μg)] was employed to calculate specific activity. According to our assay conditions, the purified rBmPGANT2 has an SA of ~ 2507 pmol/min/μg (e)
Application of soaking RNAi-BEVS system for BmPGANT2 using human SERPING1 as the O-glycoprotein reporter (a). The glycosylated and deglycosylated rSERPING1 was indicated in black arrow and gray arrow, respectively. b The commercial bovine fetuin and human A1AT as acceptors in the phosphatase-coupled glycosyltransferase assay. Standard reaction was performed according to the manufacturer’s instructions (b, upper panel). The green color was only developed with the addition of rBmPGANT2 for both protein accepters (b, lower panel). The fold change was based on the optical density measured at 594 nm
Subsequently, to further validate the RNAi effect of Bmpgant2 for other protein substrates, we expressed and investigated another O-glycoprotein from human, Plasma protease C1 inhibitor (SERPING1). As plotted in Fig. 6a, similar to the results from rPRG4, only depletion of Bmpgant2 caused the shifted bands, indicating that Bmpgant2 is the solo functional initial enzyme for O-glycosylation. Finally, to figure out whether rBmPGANT2 could initiate the O-glycosylation for native O-glycoproteins, we utilized commercial bovine fetuin and human A1AT as acceptors in the phosphatase-coupled glycosyltransferase assay (Wu et al. 2011). As summarized in Fig. 6b, the green color was only developed with the addition of rBmPGANT2 for both protein acceptors, indicating the purified silkworm rBmPGANT2 could be further applied for protein engineering for the O-glycosylation of proteins of interest.
Discussion
To date, O-glycosylations have been extensively studied in the fly D. melanogaster (Tran and Ten Hagen 2013; Harvey et al. 2016) but not in the lepidopteran insects like the silkworm, B. mori. Although only limited literature was found, the identified insect mucin-type glycoproteins (GPs), such as mucin-D, and invertebrate intestinal mucin (IIM) were proved to be sensitive to O-glycosidase treatment and carry core 1 Gal (β1-3) GalNAc disaccharide O-glycans (Kramerov et al. 1997; Wang and Granados 1997; Zhong et al. 2012). Until now, a total of four major glycans have been confirmed from the O-linked profile of embryos (OreR) in D. melanogaster that core 1 disaccharide is the predominant glycan, together with Tn antigen, glucuronylated core 1 disaccharide, and O-Fuc trisaccharide (Aoki et al. 2008). In our previous work, using BEVS, we also have proved that silkworm larvae-expressed rHsPRG4 protein was sensitive to O-glycosidase treatment (Nagata et al. 2012). In this study, the O-glycan profile from silkworm-produced rHsPRG4 was investigated by lectin blot and MALDI-TOF MS, showing that silkworm assigns a majority of core 1 disaccharide structure (Galβ1-3GalNAcα1-Ser/Thr) to the rHsPRG4 protein. These results were consistent with the evidence that there were nine Bmpgants for initial GalNAc transfer and one BmC1GALT1 for Gal transfer, allowing silkworm to be able to synthesize core 1 O-glycans. It is known that both dipteran Drosophila and lepidopteran Bombyx hold potentials to sialylate N-/O-glycans since both insects encode putative α2,6-sialyltransferases, DmST6Gal (Koles et al. 2004) and BmST (Kajiura et al. 2015), respectively. Both in vitro purified DmST6Gal and BmST proteins exhibited weak/no activities toward Galβ1-3GalNA or Galβ1-3GalNAc-O-Ser/Thr from mucin-D glycoprotein glycans (Koles et al. 2004; Kajiura et al. 2015). Our results suggested that BmST contributes inefficiently to the terminal sialylation of O-glycoproteins in the silkworm. Besides, it is worth noting that there is an O-glycosidase-resistant product from the silkworm-produced rHsPRG4 (Nagata et al. 2012; Fig. 3). It would be interesting to further investigate the heterogeneity of the products to uncover which decorations negatively affect this glycoside hydrolases since there is no counterpart to PNGase-F enzyme for O-linked glycoproteins (Hang et al. 2003).
In terms of the initial enzymes which we focused on later in this study, as many as nine BmPGANTs were identified in the silkworm by this study, suggesting that the complexity of mucin-type glycosylated protein expressions also exists in silkworms. It is known that a high degree of sequence similarities exists between mouse and human ppGalNAc-Ts (Bennett et al. 2012), and we showed that the amino acid sequences of nine BmPGANTs also display high similarities with each DmPGANT ortholog from D. melanogaster (Supplemental Fig. S2). Taken together with the expression profile in various silkworm tissues (Supplemental Fig. S1), the different numbers of PGANT homologs in silkworm and fruit fly may suggest the complexity and diversity in their functional distributions in these two insect models. Intriguingly, only BmPGANT2 is identified as a functional BmPGANT out of nine homolog candidates in the ovary-and embryo-derived cells, indicating that BmPGANTs may have unique cell/tissue or protein substrate preferences (Zhang and Ten Hagen 2010). In Drosophila, loss function of pgant4, pgant5, pgant7, CG30463, or pgant35 resulted in significant lethality, indicating that O-linked glycans are essential during development (Tran and Ten Hagen 2013). Although no phenotype was detected after depletion of Bmpgant(s) in cultured BmN4 cells, it would be of great interest to investigate if significant lethality occurs after depletion of silkworm BmPGANTs. Further investigations are required to decipher the mechanisms of the involvement of Bmpgants during development and found out the direct downstream GP targets in silkworms.
In this study, recombinant BmPGANT2 protein from silkworm-BEVS was purified, and it exhibited a substantial ability to transfer GalNAc specifically from UDP-GalNAc to both acceptor peptide EA2 and purified proteins, such as fetuin and A1AT. To our best knowledge, this is the first functional PGANT purified from silkworm and lepidopteran insects. It is known that PGANTs, such as human GLANT5 and fruit fly PGANT35A, display preferences on different substrates (Ten Hagen and Tran 2002; Gerken et al. 2006; Raman et al. 2008). In a future study, all BmPGANTs should be tested for their potential substrate preferences in vitro and in vivo. Moreover, recent evidence indicated that, when employing the BEVS, silkworms could serve as a promising insect bioreactor for the mass production of target proteins including glycoproteins of interest (Kato et al. 2010; Lee et al. 2012a, 2012b), offering many advantages in exploring the glycan structure and function. Together with our previous work (Nagata et al. 2012), we concluded that both silkworm larvae and cultured cells are capable of decorating O-glycans to the GPs, suggesting that silkworm should be a promising host for producing with potential O-glycosylation sites since silkworm-BEVS often results in satisfactory yields of proteins of interest. Our soaking RNAi-BEVS platform could allow us to produce GPs with different N-/O-glycans, which potentially has a bright future in applied glycoprotein bioengineering for pharmaceutical purposes (Nagata et al. 2013; Xu et al. 2013; Chavez-Pena and Kamen 2018). Taken together, the present results clarified the functional BmPGANT2 in mucin-type O-glycosylation in cultured silkworm cells, providing crucial fundamental insights for the following studies in dissecting detailed silkworm O-glycosylation pathways and productions of glycoproteins with different O-glycans. The purified rBmPGANT2 in this study also allows us to initiate O-glycosylation synthesis by decorating proteins of interest with GalNAc in vitro.
References
Aoki K, Porterfield M, Lee SS, Dong B, Nguyen K, McGlamry KH, Tiemeyer M (2008) The diversity of O-linked glycans expressed during Drosophila melanogaster development reflects stage- and tissue-specific requirements for cell signaling. J Biol Chem 283:30385–30400. https://doi.org/10.1074/jbc.M804925200
Apweiler R, Hermjakob H, Sharon N (1999) On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta 1473:4–8. https://doi.org/10.1016/S0304-4165(99)00165-8
Bennett EP, Mandel U, Clausen H, Gerken TA, Fritz TA, Tabak LA (2012) Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. Glycobiology 22:736–756. https://doi.org/10.1093/glycob/cwr182
Chavez-Pena C, Kamen AA (2018) RNA interference technology to improve the baculovirus-insect cell expression system. Biotechnol Adv 36:443–451. https://doi.org/10.1016/j.biotechadv.2018.01.008
Chen J, Xu J, Hino M, Yamashita M, Hirata K, Patil AA, Tatsuke T, Mon H, Banno Y, Kusakabe T, Lee JM (2016) Co-expression of silkworm allatostatin-C receptor BNGR-A1 with its cognate G protein subunits enhances the GPCR display on the budding baculovirus. J Asia Pac Entomol 19:753–760. https://doi.org/10.1016/j.aspen.2016.07.007
Cherry S, Kunte A, Wang H, Coyne C, Rawson RB, Perrimon N (2006) COPI activity coupled with fatty acid biosynthesis is required for viral replication. PLoS Pathog 2:e102. https://doi.org/10.1371/journal.ppat.0020102
Gerken TA, Raman J, Fritz TA, Jamison O (2006) Identification of common and unique peptide substrate preferences for the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases T1 and T2 derived from oriented random peptide substrates. J Biol Chem 281:32403–32416. https://doi.org/10.1074/jbc.M605149200
Hang HC, Bertozzi CR (2005) The chemistry and biology of mucin-type O-linked glycosylation. Bioorg Med Chem 13:5021–5034. https://doi.org/10.1016/j.bmc.2005.04.085
Hang HC, Yu C, Kato DL, Bertozzi CR (2003) A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proc Natl Acad Sci U S A 100:14846–14851. https://doi.org/10.1073/pnas.2335201100
Harvey BM, Rana NA, Moss H, Leonardi J, Jafar-Nejad H, Haltiwanger RS (2016) Mapping sites of O-glycosylation and fringe elongation on Drosophila notch. J Biol Chem 291:16348–16360. https://doi.org/10.1074/jbc.M116.732537
Hino M, Kawanami T, Xu J, Morokuma D, Hirata K, Yamashita M, Karasaki N, Tatsuke T, Mon H, Iiyama K, Kamiya N, Banno Y, Kusakabe T, Lee JM (2016) High-level expression and purification of biologically active human IL-2 using silkworm-baculovirus expression vector system. J Asia Pac Entomol 19:313–317. https://doi.org/10.1016/j.aspen.2016.03.014
Imai S, Kusakabe T, Xu J, Li Z, Shirai S, Mon H, Morokuma D, Lee JM (2015) Roles of silkworm endoplasmic reticulum chaperones in the secretion of recombinant proteins expressed by baculovirus system. Mol Cell Biochem 409:255–262. https://doi.org/10.1007/s11010-015-2529-5
Ju T, Cummings RD (2002) A unique molecular chaperone Cosmc required for activity of the mammalian core 1 beta 3-galactosyltransferase. Proc Natl Acad Sci U S A 99:16613–16618. https://doi.org/10.1073/pnas.262438199
Ju T, Zheng Q, Cummings RD (2006) Identification of core 1 O-glycan T-synthase from Caenorhabditis elegans. Glycobiology 16:947–958. https://doi.org/10.1093/glycob/cwl008
Kajiura H, Hamaguchi Y, Mizushima H, Misaki R, Fujiyama K (2015) Sialylation potentials of the silkworm, Bombyx mori; B. mori possesses an active α2,6-sialyltransferase. Glycobiology 25:1441–1453. https://doi.org/10.1093/glycob/cwv060
Kato T, Kajikawa M, Maenaka K, Park EY (2010) Silkworm expression system as a platform technology in life science. Appl Microbiol Biotechnol 85:459–470. https://doi.org/10.1007/s00253-009-2267-2
Koles K, Irvine KD, Panin VM (2004) Functional characterization of Drosophila sialyltransferase. J Biol Chem 279:4346–4357. https://doi.org/10.1074/jbc.M309912200
van Kooyk Y, Rabinovich GA (2008) Protein-glycan interactions in the control of innate and adaptive immune responses. Nat Immunol 9:593–601. https://doi.org/10.1038/ni.f.203
Kramerov AA, Mikhaleva EA, Rozovsky YM, Pochechueva TV, Baikova NA, Arsenjeva EL, Gvozdev VA (1997) Insect mucin-type glycoprotein: Immunodetection of the O-glycosylated epitope in Drosophila melanogaster cells and tissues. Insect Biochem Mol Biol 27:513–521. https://doi.org/10.1016/S0965-1748(97)00026-X
Kulkarni AA, Weiss AA, Iyer SS (2010) Glycan-based high-affinity ligands for toxins and pathogen receptors. Med Res Rev 30:327–393. https://doi.org/10.1002/med.20196
Lairson LL, Henrissat B, Davies GJ, Withers SG (2008) Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem 77:521–555. https://doi.org/10.1146/annurev.biochem.76.061005.092322
Lee JM, Kawakami N, Mon H, Mitsunobu H, Iiyama K, Ninaki S, Maenaka K, Park EY, Kusakabe T (2012a) Establishment of a Bombyx mori nucleopolyhedrovirus (BmNPV) hyper-sensitive cell line from the silkworm e21 strain. Biotechnol Lett 34:1773–1779. https://doi.org/10.1007/s10529-012-0971-y
Lee JM, Mon H, Banno Y, Iiyama K, Kusakabe T (2012b) Bombyx mori strains useful for efficient recombinant protein production using a baculovirus vector. J Biotechnol Biomater s9. https://doi.org/10.4172/2155-952X.S9-003
Li Z, Cheng D, Mon H, Tatsuke T, Zhu L, Xu J, Lee JM, Xia Q, Kusakabe T (2012) Genome-wide identification of polycomb target genes reveals a functional association of Pho with Scm in Bombyx mori. PLoS One 7:e34330. https://doi.org/10.1371/journal.pone.0034330
Masuda A, Xu J, Mitsudome T, Nagata Y, Morokuma D, Mon H, Banno Y, Kusakabe T, Lee JM (2015) Mass production of an active peptide-N-glycosidase F using silkworm-baculovirus expression system. Mol Biotechnol 57:735–745. https://doi.org/10.1007/s12033-015-9866-1
Mon H, Kobayashi I, Ohkubo S, Tomita S, Lee J, Sezutsu H, Tamura T, Kusakabe T (2012) Effective RNA interference in cultured silkworm cells mediated by overexpression of Caenorhabditis elegans SID-1. RNA Biol 9:40–46. https://doi.org/10.4161/rna.9.1.18084
Morokuma D, Xu J, Hino M, Mon H, Merzaban JS, Takahashi M, Kusakabe T, Lee JM (2017) Expression and characterization of human β-1, 4-galactosyltransferase 1 (β4GalT1) using silkworm–baculovirus expression system. Mol Biotechnol 59:151–158. https://doi.org/10.1007/s12033-017-0003-1
Munkley J (2016) The role of sialyl-Tn in cancer. Int J Mol Sci 17:275. https://doi.org/10.3390/ijms17030275
Nagata Y, Sakashita K, Imanishi S, Lee JM, Kusakabe T (2012) Expression of glycosylated mucin-like domain using baculovirus expression system in silkworm, Bombyx mori. J Fac Agr Kyushu Univ 57:83–86
Nagata Y, Lee JM, Mon H, Imanishi S, Hong SM, Komatsu S, Oshima Y, Kusakabe T (2013) RNAi suppression of β-N-acetylglucosaminidase (BmFDL) for complex-type N-linked glycan synthesis in cultured silkworm cells. Biotechnol Lett 35:1009–1016. https://doi.org/10.1007/s10529-013-1183-9
O’Reilly DR, Miller LK, Luckow VA (1992) Baculovirus expression vectors. In: Freeman WH (ed) A laboratory manual. Oxford University Press, New York, pp 139–166
Rabouille C, Kuntz DA, Lockyer A, Watson R, Signorelli T, Rose DR, van den Heuvel M, Roberts DB (1999) The Drosophila GMII gene encodes a Golgi alpha-mannosidase II. J Cell Sci 112(Pt 19):3319–3330
Raman J, Fritz TA, Gerken TA, Jamison O, Live D, Liu M, Tabak LA (2008) The catalytic and lectin domains of UDP-GalNAc:polypeptide-N-acetylgalactosaminyltransferase function in concert to direct glycosylation site selection. J Biol Chem 283:22942–22951. https://doi.org/10.1074/jbc.M803387200
Rillahan CD, Paulson JC (2011) Glycan microarrays for decoding the glycome. Annu Rev Biochem 80:797–823. https://doi.org/10.1146/annurev-biochem-061809-152236
Schwientek T, Bennett EP, Flores C, Thacker J, Hollmann M, Reis CA, Behrens J, Mandel U, Keck B, Schäfer MA, Haselmann K, Zubarev R, Roepstorff P, Burchell JM, Taylor-Papadimitriou J, Hollingsworth MA, Clausen H (2002) Functional conservation of subfamilies of putative UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferases in Drosophila, Caenorhabditis elegans, and mammals. One subfamily composed of l(2)35Aa is essential in Drosophila. J Biol Chem 277:22623–22638. https://doi.org/10.1074/jbc.M202684200
Stanley P (2011) Golgi glycosylation. Cold Spring Harb Perspect Biol 3. https://doi.org/10.1101/cshperspect.a005199
Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KTBG, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, Gupta R, Bennett EP, Mandel U, Brunak S, Wandall HH, Levery SB, Clausen H (2013) Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J 32:1478–1488. https://doi.org/10.1038/emboj.2013.79
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739. https://doi.org/10.1093/molbev/msr121
Taylor ME, Drickamer K (2007) Paradigms for glycan-binding receptors in cell adhesion. Curr Opin Cell Biol 19:572–577. https://doi.org/10.1016/j.ceb.2007.09.004
Ten Hagen KG, Tran DT (2002) A UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase is essential for viability in Drosophila melanogaster. J Biol Chem 277:22616–22622. https://doi.org/10.1074/jbc.M201807200
Ten Hagen KG, Tran DT, Gerken TA, Stein DS, Zhang Z (2003) Functional characterization and expression analysis of members of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family from Drosophila melanogaster. J Biol Chem 278:35039–35048. https://doi.org/10.1074/jbc.M303836200
Tian E, Ten Hagen KG (2008) Recent insights into the biological roles of mucin-type O-glycosylation. Glycoconj J 26:325–334. https://doi.org/10.1007/s10719-008-9162-4
Tran DT, Ten Hagen KG (2013) Mucin-type O-glycosylation during development. J Biol Chem 288:6921–6929. https://doi.org/10.1074/jbc.R112.418558
Varki A (2008) Sialic acids in human health and disease. Trends Mol Med 14:351–360. https://doi.org/10.1016/j.molmed.2008.06.002
Walski T, De Schutter K, Van Damme EJM, Smagghe G (2017) Diversity and functions of protein glycosylation in insects. Insect Biochem Mol Biol 83:21–34. https://doi.org/10.1016/j.ibmb.2017.02.005
Wang P, Granados RR (1997) An intestinal mucin is the target substrate for a baculovirus enhancin. Proc Natl Acad Sci U S A 94:6977–6982
Wang Q, Shen B, Zheng P, Feng H, Guo Y, Cao W, Chen L, Liu X, Zhao G, Xu S, Shen W, Chen J, Teng J (2013) BmCREC is an endoplasmic reticulum (ER) resident protein and required for ER/Golgi morphology. J Biol Chem 288:26649–26657. https://doi.org/10.1074/jbc.M113.463018
Wu ZL, Ethen CM, Prather B, Machacek M, Jiang W (2011) Universal phosphatase-coupled glycosyltransferase assay. Glycobiology 21:727–733. https://doi.org/10.1093/glycob/cwq187
Xia Q, Cheng D, Duan J, Wang G, Cheng T, Zha X, Liu C, Zhao P, Dai F, Zhang Z, He N, Zhang L, Xiang Z (2007) Microarray-based gene expression profiles in multiple tissues of the domesticated silkworm, Bombyx mori. Genome Biol 8:R162. https://doi.org/10.1186/gb-2007-8-8-r162
Xia Q, Li S, Feng Q (2014) Advances in silkworm studies accelerated by the genome sequencing of Bombyx mori. Annu Rev Entomol 59:513–536. https://doi.org/10.1146/annurev-ento-011613-161940
Xu J, Nagata Y, Mon H, Li Z, Zhu L, Iiyama K, Kusakabe T, Lee JM (2013) Soaking RNAi-mediated modification of Sf9 cells for baculovirus expression system by ectopic expression of Caenorhabditis elegans SID-1. Appl Microbiol Biotechnol 97:5921–5931. https://doi.org/10.1007/s00253-013-4785-1
Zhang L, Ten Hagen KG (2010) Dissecting the biological role of mucin-type O-glycosylation using RNA interference in Drosophila cell culture. J Biol Chem 285:34477–34484. https://doi.org/10.1074/jbc.M110.133561
Zhong X, Zhang L, Zou Y, Yi Q, Zhao P, Xia Q, Xiang Z (2012) Shotgun analysis on the peritrophic membrane of the silkworm Bombyx mori. BMB Rep 45:665–670. https://doi.org/10.5483/BMBRep.2012.45.11.261
Acknowledgments
We thank Dr. Aoki (Kyushu University Graduate School) for providing the Bm5/Bme21 cell lines.
Funding
This work was supported by JSPS KAKENHI Grant Number JP15H04612.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 5561 kb)
Rights and permissions
About this article
Cite this article
Xu, J., Morio, A., Morokuma, D. et al. A functional polypeptide N-acetylgalactosaminyltransferase (PGANT) initiates O-glycosylation in cultured silkworm BmN4 cells. Appl Microbiol Biotechnol 102, 8783–8797 (2018). https://doi.org/10.1007/s00253-018-9309-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-018-9309-6