Abstract
Colorectal cancer (CRC) is the third highest frequent malignancy and ultimate critical source of cancer-associated mortality around the world. Regardless of latest advances in molecular and surgical targeted medicines that have increased remedial effects in CRC patients, the 5-year mortality rate for CRC patients remains dismally low. Evidence suggests that microRNAs (miRNAs) execute an essential part in the development and spread of CRC. The miRNAs are a type of short non-coding RNA that exhibited to control the appearance of tumor suppressor genes and oncogenes. miRNA expression profiling is already being utilized in clinical practice as analytical and prognostic biomarkers to evaluate cancer patients' tumor genesis, advancement, and counteraction to drugs. By modulating their target genes, dysregulated miRNAs are linked to malignant characteristics (e.g., improved proliferative and invasive capabilities, cell cycle aberration, evasion of apoptosis, and promotion of angiogenesis). This review presents an updated summary of circulatory miRNAs, tumor-suppressive and oncogenic miRNAs, and the potential reasons for dysregulated miRNAs in CRC. Further we will explore the critical role of miRNAs in CRC drug resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cancer-related pathways that promote tumor aggressiveness and metastasis, like angiogenesis, vasculogenic mimicry, and neovascularization, are controlled mainly by non-coding RNAs and communicate in a complicated manner. This cross-talk maintains the equilibrium of various neural networks. There is increasing evidence to suggest that non-coding RNAs (ncRNAs), which include microRNAs and long-noncoding RNAs (lncRNAs), interact with each other directly, fine-tuning the effects of their control, in addition to influencing mRNA translation through separate processes [1]. Recent studies have concentrated on these lncRNAs, which act as competing endogenous RNAs (ceRNAs) to control gene expression by sponging miRNAs via common miRNA response domains. miRNAs are short non-coding RNAs found to control gene expression by blocking and subjecting the degradation of target mRNAs. Cancer development may be linked to their aberrant expression [2,3,4,5]. Argonaute proteins, lipids, and microvesicles form complexes with miRNAs to preserve them from degradation and make them relatively long-lasting in storage [6,7,8,9]. So they should be utilized as biomarkers. There have been several studies on serum or plasma miRNAs as diagnostic biomarkers for CRC [10, 11]. Lin4, the earliest miRNA, was found in Caenorhabditis elegans (C. elegans) in 1993 [12]. Reinhart et al. published the first mammalian miRNA, let-7, in 2000. They observed that let-7 suppresses the heterochronic gene lin-41's expression by engaging with its mRNA’s 3′-UTRs through a sequence-specific RNA-RNA interaction [13, 14]. Later in 2002, Dr Croce’s team was the first to show that miRNA has a role in human cancer genesis. The team also discovered that miR-16-1 and miR-15a on the 13q14 chromosome are commonly disrupted in B-cell chronic lymphocytic leukemia (CLL) [15]. Many research investigations have highlighted the roles of miRNAs in numerous diseases, including cancer, throughout the last decade [16].
Colon cancer is a type of common malignant tumor that develops at the junction of the rectum and sigmoid colons. CRC has been identified as the third most frequent and second most lethal cancer [17]. In 2018, CRC ranked second in mortality and fourth in occurrence [18]. People suffering from CRC showed a five-year survival rate of 65% [19]. In CRC, miRNA dysregulation causes broad changes in gene expression patterns, leading to apoptosis, abnormal cell proliferation, metabolism, epithelial-mesenchymal transition (EMT), and treatment hindrance [20, 21]. Based on accumulating data, miRNAs may be promising indicators for prognosis, diagnosis, and therapeutic targets in CRC [22]. This article contains a brief discussion of the deregulation of microRNAs in CRC and how microRNAs function as oncogenes or tumor suppressors to manage the growth of CRC by ameliorating its various critical targets. In addition, we investigate the analytical, prognostic, and clinical prospects of miRNAs in colorectal cancer. In CRC patients, metastasis is strongly linked to poor prognoses [23]. Metastasis is a crucial process that can disclose the malignant transformation of neoplasms and the clinical phases of malignant tumor’s to some extent [24]. Cancer metastasis has been the subject of several investigations. Epithelial-mesenchymal transition (EMT) [25], angiogenesis [26], hypoxia [27], and the tumor microenvironment (TME) are the most important variables influencing cancer spread. TME transformation is required for CRC carcinogenesis [28, 29]. Molecules that interact with the microenvironment (MET) may also have a role in metastasis [28]. EMT is controlled by a network of intricate molecular pathways that include microRNAs, epigenetic and posttranslational regulators, and alternative splicing processes [25]. These pathways are responsible for modulating EMT (Figs. 1 and 2).
An overview of the miRNA biogenesis and its functional mechanism. Series of events occurs during process (A) Drosha, the first nuclear ribonuclease III, recognizes pri-miRNA and cuts the double-stranded RNA freeing a pre-miRNA. (B) Pre-miRNA hairpin is exported from the nucleus in a process involving the nucleocytoplasmic protein Exportin-5 (RAN GTPase). (C) In the cytoplasm, the pre-miRNA hairpin is cleaved by the RNase III enzyme Dicer and produce sense and anti-sense strands, approximately 20 nucleotides in length, the effective strand called anti-sense and known as mature miRNA and short-lived complementary sequence called passenger strand (miR*). (D) The anti-sense stranded miRNA is combined into RISC, which then targets it to the target 30 untranslated region mRNA sequence. (E) Mature miRNA acts either by degrading the mRNA target or by regulating gene expression which further leads to Cancer development, Cell proliferation, and apoptosis
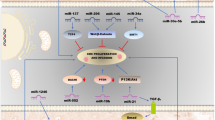
Colorectal cancer was induced through miRNA pathways (RAS/MAPK, EMT, Notch, Wnt/β Catenin, TGF-β). Tumor suppressor genes (APC, TP53) are downregulated, resulting in K-RAS mutation and MAPK activation. Several miRNAs have a role in cancer development and progression, with miRNA 103 being therapeutically addressed
PubMed, Web of Science, and EMBASE databases were used for this study for this review. Our search terms included “colorectal cancer” and its variants, “CRC” and its variants, “colon cancer” and its variants, and “rectal cancer” and its variants, as well as “metastasis,” “miRNA,” “microRNA,” and their variants”, miRNA signaling in CRC. After that, we reviewed the most recent developments in miRNA research in the metastatic phase of CRC and various signaling pathways. We also discussed their potential as therapeutic targets or biomarkers in treating CRC. We hope this review will contribute to better-comprehending metastasis, dysregulation, and signaling in CRC and future research in the field.
mi-RNA and cellular progression in colon cancer
MiRNA dysregulation in colon cancer alters crucial cellular mechanisms that promote tumor growth. Identifying treatment targets and establishing new diagnostic and prognostic indicators for colon cancer requires understanding miRNA regulation. Depending on their target genes and malignancy, miRNAs can be oncogenes or tumor suppressors [30, 31]. Modifying particular miRNAs may decrease tumor development, suppress metastasis, and improve colon cancer treatment. Abnormal miRNA expression is linked to various biological changes, including death, cell differentiation, and carcinogenesis [30, 32]. Colon cancer links to several miRNAs; e.g., MiR-21, an oncogene that promotes cell proliferation, inhibits apoptosis, and enhances tumor invasion and metastasis, is often elevated. However, miR-34a is commonly downregulated in colon cancer and suppresses tumors by reducing cell proliferation, cell cycle arrest, and death. MiRNAs regulate epithelial-mesenchymal transition (EMT), a critical tumor metastatic pathway. EMT lets cancer cells invade nearby tissues and spread to other organs. MiR-200 family members and miR-155 regulate colon cancer cell EMT. MiRNAs affect colon cancer cell response to chemotherapy and targeted therapies. 5-fluorouracil (5-FU) resistance links to altered miRNA expression [12, 33,34,35,36,37]. The activities of TS-miRNA and onco-miRNA in connection to cancer characteristics like tumor start, EMT, and metastasis are discussed in the next section.
mi-RNAs and epithelial-mesenchymal transition (EMT) process
Epithelial-mesenchymal transitions (EMTs) are observed in various biological processes, including embryonic development, adult tissue regeneration, and cancer progression [38, 39]. While EMT is tightly regulated in normal development, it becomes dysregulated during tumor growth, particularly in cancer metastasis [40]. EMT is a crucial step in the metastatic cascade, where epithelial cells lose their adherence and acquire a mesenchymal phenotype [41]. Specific miRNAs have been identified as critical players in driving EMT and promoting tumour progression [42]. The molecular pathways involved in EMT, including miRNAs, can vary significantly, and different classification systems have been proposed. Indicators commonly used to assess EMT include the loss of E-cadherin expression and the upregulation of EMT-related transcription factors such as ZEB1 [24, 25]. Recent studies have highlighted the interplay between EMT and miRNAs or tumor-associated immune cells [27]. In colorectal cancer (CRC), miR-141, miR-200b, and miR-200c directly target ZEB1 and ZEB2, activating EMT and metastasis [43]. Additionally, various signaling pathways associated with miRNAs have been implicated in inducing EMT [44]. For example, miR-4775 has been shown to enhance metastasis and EMT by targeting the SMAD7/TGF-b axis [45]. Another direct target of miR-496 is Ras association domain family member 6 (RASSF6), whose hypermethylation can enhance migration and EMT through the Wnt signaling pathway in CRC [46]. Conversely, a subset of miRNAs can suppress metastasis by inhibiting the EMT process. Studies have shown that miR-192 and miR-194 can prevent Snail-induced EMT and metastasis [47]. Similarly, miR-150 targets Snail and Gli1, effectively inhibiting CRC metastasis [25]. Additionally, miR-490-3p inhibits EMT by targeting the Wnt/b-catenin pathway through binding to frequently rearranged advanced T cell lymphoma (FRAT) protein [48]. Specific miRNAs also reduce EMT by attenuating EGFR signaling [49]. For example, miR-612 inhibits AKT2, inhibiting EMT-related processes and reducing CRC metastasis [50]. Upregulation of miR-219-5p inhibits lymphoid enhancer-binding factor 1 (LEF1), resulting in downregulation of the AKT/ERK pathway, repression of EMT, and decreased metastasis incidence [51]. Additionally, miRNAs can target other factors involved in carcinogenesis, ultimately leading to the downregulation of EMT. For instance, miR-185 decreases CRC metastasis by reducing stromal interaction molecule 1 (STIM1), preventing EMT [52]. Moreover, miR-296 modulates S100A4-mediated EMT processes, thereby reducing CRC metastasis [53]. Overall, miRNAs play a crucial role in regulating EMT and the metastatic potential of CRC cells by targeting various signaling pathways and downstream effectors.
miRNAs and angiogenesis
Angiogenesis is a crucial hallmark of cancer, involving forming an abnormal vascular network that facilitates tumor growth, metastasis, and progression [54,55,56]. This process is tightly regulated by a complex interplay of pro-angiogenic and anti-angiogenic factors at each stage [57, 58]. MiRNAs have emerged as important regulators of tumor angiogenesis, capable of exerting both pro-angiogenic and anti-angiogenic effects [57, 58]. They can directly modulate endothelial cell function or indirectly influence angiogenesis by targeting proteins involved in vessel development [58, 59]. Consequently, miRNAs have garnered significant attention as potential targets for novel anti-angiogenic therapies. The manipulation of angiogenesis requires precise regulation of the balance between pro-angiogenic and anti-angiogenic factors [60]. Among the key players in angiogenesis are vascular endothelial growth factor A (VEGFA) and its receptors [27]. Recent studies investigating tumor metastasis have underscored the role of miRNAs in angiogenesis [61]. For instance, miR-1249, activated by p53, negatively regulates angiogenesis by targeting VEGFA and modulating the AKT/mTOR signaling pathway [62]. Additionally, miR-590-5p has been found to target both VEGFA and interleukin enhancer-binding factor 3, exerting downstream effects on angiogenesis [63]. MiR-25-3p enhances angiogenesis by targeting Kruppel-like factor 2 (KLF2) and Kruppel-like factor 4 (KLF4), thereby promoting the expression of VEGF receptor 2 (VEGFR2) [64]. Balancing angiogenesis and anti-angiogenesis mechanisms can prevent cancer cell dissemination [63, 65]. Overall, miRNAs contribute to the intricate regulation of angiogenesis and hold promise as potential therapeutic targets for modulating tumour angiogenesis.
miRNAs and hypoxia
Solid tumours, including CRC, are characterized by the presence of hypoxia, which is a low-oxygen environment [66]. Hypoxia-inducible factor 1a (HIF-1a) and the insulin-like growth factor 1 receptor (IGF-1R) play crucial roles in creating this hypoxic microenvironment [26]. Alterations in genetic programs facilitating cellular adaptation to hypoxia can contribute to more aggressive tumour phenotypes. Recent studies have highlighted the significant involvement of miRNAs in cancer metastasis under hypoxic conditions. For instance, the miR-1792 cluster negatively regulates HIF-1a, inhibiting CRC metastasis [27]. Yin and colleagues demonstrated that miR-145 suppresses CRC metastasis by targeting HIF-1a expression [37]. Similarly, miR-143 inhibits angiogenesis and metastasis by targeting the hypoxia-related IGF-1R [65]. Given the close association between hypoxia, angiogenesis, and metastasis, targeting miRNAs associated with hypoxia holds promising implications for controlling cancer spread.
Interaction between miRNAs and TME
The tumour microenvironment (TME) facilitates aggressive metastasis [28]. Previous research has highlighted the close association between miRNAs and the TME, with miRNAs influencing various genes involved in the TME during cancer metastasis [67]. MiRNAs can act as regulators of immune cells, cancer-associated fibroblasts (CAFs), cancer-associated endothelial cells (CAECs), and tumour cells [67,68,69,70,71]. For instance, miR-let-7c-5p interacts with the TME by affecting the expression of collagen type I alpha 2 chains (COL1A2) in CRC [72]. The balance between matrix metalloproteinases (MMPs) and tissue inhibitors of metalloproteinases-2 (TIMP-2) is crucial for ECM degradation in cancer cells [73]. Makondi et al. reported that miR-20a and miR-495 inhibit CRC metastasis by suppressing matrix metalloproteinase expression, specifically MMP 2 [65, 73].
Exosomes derived from M2 macrophages (MDEs) play a significant role in the TME by upregulating miR-21-5p and miR-155-5p. These miRNAs then target S-ribonuclease binding protein (BRG1), leading to reduced BRG1 levels and increased metastasis rates. Tumour-associated macrophages (TAMs) are also essential components of the TME. During cancer development and progression, TAMs undergo M1- or M2-like polarization and subsequently regulate tumour metastasis [74]. Several miRNAs associated with metastasis play important roles in these processes [28]. For example, direct binding of miR-195-5p to the 3’ UTR of Notch2 inhibits M2-like TAM polarization and metastasis in cancer cells [75]. CD163 + TAMs, on the other hand, contribute to the production of interleukin 6 (IL-6) and suppression of miR-506-3p. In turn, miR-506-3p prevents cancer spread by inhibiting the synthesis of C–C motif chemokine ligand 2 (CCL2), a molecular signal that recruits macrophages [76].
The pro-inflammatory cytokine tumour necrosis factor-alpha (TNF-α) plays a central role in altering the TME and regulating specific miRNAs [77]. Recent research has demonstrated that miR-19a promotes TME alterations by enhancing TNF-α, critical for TNF-α-mediated metastasis [78]. Similarly, miR-105 is involved in TNF-α-induced TME modifications by enhancing the nuclear factor-ƙB (NF-ƙB) subunit 1 signalling pathway in CRC. Therefore, the interaction between miRNAs and the TME should be considered an additional important factor when developing strategies to combat metastasis [79].
Prime mechanisms of miRNA in CRC
Colorectal cancer (CRC) is a complex chronic disease characterized by dysregulation of multiple signaling pathways implicated in its development and progression. There is compelling evidence to suggest that miRNAs play a prominent role in CRC formation and metastasis [79]. Further below, under various subheadings, we will discuss the interactions between miRNAs and other key signaling pathways that dysregulate in CRC, including the Wnt pathway, mitogen-activated protein kinases (MAPKs), epithelial-mesenchymal transition (EMT), and transforming growth factor-beta (TGF-β) [80]. Understanding these miRNA-mediated regulatory mechanisms can provide valuable insights into the pathogenesis of CRC and may contribute to developing novel therapeutic strategies targeting these signalling pathways.
Wnt/β-catenin signaling pathway in CRC
Wnt activation defects have a carcinogenic function in CRC via β-catenin accumulation in the cytoplasm [81, 82]. miR-135b is an essential onco-miRNA in CRC that controls numerous crucial tumour suppressor genes. A recent study found that miR-135b suppress factor inhibiting HIF (FIH) to up-regulate hypoxia-inducible factor 1 (HIF-1alpha) to increase colon cancer cell invasion, metastasis, as well as proliferation [83]. This finding was supported by the fact that miR-135b also suppresses APC, which in turn increases the activity of the downstream Wnt pathway. According to the findings of another study [84], the microRNAs miR-135a and miR-135b are responsible for inhibiting APC in CRC while simultaneously activating the Wnt signaling pathway. The microRNA known as miR-155 is very important in regulating the Wnt/β-Catenin mechanism in CRC. Targeting Axin1 and TCF4 allows miR-155 to allow long-term Wnt/β-Catenin activation in CRC cells [85]. This allows miR-155 to manage both cell proliferation and survival. Casein kinase 1 (CK1), Axin, adenomatous polyposis coli (APC), and glycogen synthase kinase 3 (GSK3) are the enzymes responsible for the degradation of β-catenin under typical conditions. Ma and his colleagues discovered that miR-17–5p directly binds P130 and stimulates the Wnt/β-catenin pathway, resulting in increased carcinogenesis and CRC development [86]. Furthermore, by inhibiting GSK3, miR-224 maintains Wnt/β-catenin signaling and the aggressive behavior of CRC in vivo and in vitro [87]. In CRC, lncRNA H19 acts as a competitive endogenous RNA sponge for miR-141, activating the β-catenin pathway. The latter has been shown to limit the stemness of CRC cells by directly targeting β-catenin. Activated β-catenin, on the other hand, binds to and triggers the promoter of the miR-17/92 cluster [88]. The Wnt signaling pathway increases the production of miR-19a, which inhibits phosphatase and tensin homolog (PTEN), bim, p21, and Fbw7, all of which are implicated in CRC cell invasion, proliferation, and survival [89]. TCF4 regulates miR-21 expression, and there was a positive connection between miR-21 and Wnt/ β-catenin signaling in CRC, signifying that miR-21 may be involved in colon cancer growth via the Wnt/ β-catenin pathway [90]. APC2, RNF43, ZNRF3, dickkopf-related protein 1 (DKK1), and DKK3 are among the five Wnt/-catenin regulatory proteins targeted by miR-100 and miR-125b in CRC [91, 92]. As a result, for CRC patients, targeting miRNAs involved in abnormal Wnt/β-catenin signaling pathway can be an alternate therapy. The list of miRNAs and their mRNA targets in Wnt/β-catenin pathway related with the development, advancement and metastasis of CRC are shown in Table 1 respectively.
RAS/MAPK signaling pathway in CRC
The aberrant expression of RAS molecules is a critical factor in CRC progression [106]. RAS can control numerous cascades by expressing (GTP) guanosine triphosphate binding proteins, including the mitogen-activated protein kinase (MAPK) axis [107]. It has been observed that overactivation of the KRAS protein is related to cancer development, metastasis, and invasive cell characteristics. CRC development is accelerated by oncogenic RAS hyperactivation. RAS activation is often caused by enhanced nucleotide exchange or reduced GTP hydrolysis [108]. Because of its significance in cancer development and therapeutic efficacy, the RAS/MAPK signaling axis is an important therapeutic target. Pathogenesis and carcinogenic characteristics like cancer cell migration, senescence, and differentiation, are caused by mutations in the MAPK signaling pathway [109,110,111,112,113]. In colon cancer, dysregulation of MAPK is typically observed due to mutations in KRAS and BRAF 119. KRAS mutation is among the early steps in CRC development, happening in 30% to 40% of colorectal tumors, according to genetic analysis of oncogenic KRAS [115]. It has been shown that the oncoprotein KRAS plays a substantial involvement in tumor growth by modification of several miRNAs such as the miR-200c, miR-221, and miR-222 in CRC cell lines; HCT11660 and also in DLD-1 cells [116] only in three-dimensional (3D) cultures of the colonic‐crypt model. The protein expression of PTEN as a substrate of related miRNAs was studied to corroborate the stimulatory effect of KRAS in miRNA elevation. Following KRAS overexpression, PTEN expression is upregulated in CRC specimens. Many oncogenic miRs, including miR21, miR32, miR92a, miR181a, and miR494 have a role in CRC carcinogenesis by repressing PTEN [116,117,118]. According to the DIANA mirPath prediction study, increased miR215p is closely related to CRC advancement via RAS/MAPK signaling pathways [119]. According to Lu et al. [120], substantial activation of miR335 in CRC tissues stimulates RAS/MAPK signaling via RASA1 repression. Mir335 levels are highly correlated with tumor size and cell differentiation, providing valid interpretations in tumor progression. Likewise, CCAAT/enhancer-binding protein (C/EBP) induced miR223 promotes RAS protein binding to GTP and elevates pMEK1/2 and pERK proteins in CRC tissues via RASA1 downregulation. In colorectal cancer cell lines, miRNA acts as a target gene of the KRASMAPK signaling pathway [121]. miR31 is also implicated in the inhibition of RASA1 at the posttranscriptional level. RASA1 deficiency promotes CRC cell growth and carcinogenesis by increasing RAS GTP, pERK1/2, and proliferative agents like proliferation cell nuclear antigen and KI-67. Inhibiting oncogenic miR31 reduces tumor growth in the nude mice tumor model, indicating a novel therapy option for CRC [122]. Because the tumor suppressor RAS and RASA1 signaling play critical roles in controlling cellular proliferation, RASA1 targeted miRNA profiling investigations are essential in colon cancer therapy options [123]. Using Western blot analysis, bioinformatics, and luciferase experiments, researchers discovered that the miR 1260b inhibitor directly targets the programmed cell death of genes in CRC. miR650 directs its attention squarely on the tumour suppressor gene known as inhibitor of growth protein 4 (ING4), which is responsible for encouraging the migration, proliferation, and epithelial-mesenchymal transition of tumour cells. In SW620 and SW480 cells, miR650 blocked ING4 increases phosphorylated ERK1/2 and p38 MAPK [124]. Rasmussen et al., demonstrated that miR6253p enhances resistance to oxaliplatin (oxPt) via decreasing apoptosis and the cell cycle regulatory network, which is produced by targeting MAP2K6 (also known as MKK6). MAP2K6 and its substrate, p38 MAPK, are reduced by miR6253p. Overexpression of this protein in tumor cells improves drug resistance and inhibits genotoxic stress signaling mediated by MAP2K6MAPK14, which causes medical issues in colorectal cancer therapy [125]. One study has shown that tumor suppressor miRNAs may interact with oncogenic KRAS and inhibit the RAS/MAPK signaling pathway, contrary to oncogenic miRNAs. MiR-487b has been found to reduce CRC metastasis by decreasing KRAS and its downstream pathways, as demonstrated by Hata et al. [126]
P-38, C-jun N-terminal kinase (JNK), and Extracellular signal-regulated kinase (ERK), are all members of the MAPK signaling cascade. ERK signaling has been extensively explored in the advancement of colorectal cancer [114]. miR-143/145 expression is decreased in CRC, and they act as tumour suppressors by targeting ERK5 [127]. MiR-422a specifically targets the 3′ -UTR of MAPK1, and its silencing activates the Raf/MEK/ERK signaling pathway, hence increasing CRC cell proliferation [128]. The MAPK pathway serves as a downstream effector for several different growth factor receptors, including the epidermal growth factor receptor (EGFR) [14]. The EGFR is a transmembrane protein that functions as a tyrosine kinase receptor (RTK).When it comes to CRC, dysregulation of the Ras/Raf1/MEK/ERK signaling pathway is a common occurrence, and it plays a part in tumour development and progression [129]. The rhomboid domain containing 1 (RHBDD1) suppresses CRC carcinogenesis by targeting the EGFR/Raf/MEK/ERK signaling pathway [130]. MiR-195 inhibition promotes CRC cell proliferation and survival by upregulating the expression of RAF-1 [131]. Additionally, miR-143/145 has been shown to affect EGFR and KRAS directly or indirectly, hence suppressing CRC cell proliferation and tumorigenicity [132, 133]. Additionally, miRNAs modulate the MAPK signaling pathway's downstream targets. miR-873 reduces ETS Like-1 protein (ELK1) production directly in CRC, disrupting the ERK-CyclinD1 pathway [134]. This data imply that miRNAs play a critical role in CRC development via modulating the MAPK signaling pathway. Table 2 shows the oncogenic or tumor-suppressor regulatory microRNAs for the RAS/MAPK signaling pathway in colorectal cancer pathology.
TGF-β signaling pathway in CRC
In CRC, the TGF-signaling pathway has two roles. Downregulated TGF-expression leads to the start and progression of CRC in the early stages [160]. In contrary, significant TGF-expression was identified in late stage CRC, indicating that TGF- has an oncogenic function in late stage CRC [161]. Evidence suggests that miRNAs target the TGF signaling pathway to control cell proliferation and differentiation in CRC cells. Apoptosis and repression of cell proliferation in a CRC mouse model have been shown to be facilitated by the suppression of miR-135b [162]. Furthermore, miR-224 expression is linked to tumour burden and microsatellite stability. Upregulated miR-224 enhances CRC metastasis in vitro and in vivo by inhibiting SMAD4, a TGF- downstream effector [83]. Additionally, miR-4260 acts as an onco-miRNA in CRC by targeting the tumour suppressor SMAD4, indicating a critical role for miRNAs in TGF- signaling [163].
EMT signaling pathway
EMT plays a crucial role in tumor development and metastasis. Transcription factors like SNAIL1/2, ZEB1/2, and TWIST1 have been identified as significant drivers of EMT [164]. In colorectal cancer (CRC), miRNAs have been found to influence EMT by targeting genes involved in this process. Among these miRNAs, the miR-200 family (miR-141, miR-200a, miR-200b, miR-200c, and miR-429) is well-known for its tumour-suppressive properties. The miR-200 family has been demonstrated to decrease metastatic potential in various cancers, including CRC, by inhibiting EMT through targeting ZEB1 and ZEB2 [165, 166]. Specifically in invasive CRC tissues, Hur et al. observed reduced expression of miR-200c. MiR-200c has been shown to significantly contribute to CRC metastasis by directly targeting ZEB1, ETS1, and FLT1 (fms-related tyrosine kinase (1) and indirectly influencing the expression of E-cadherin and vimentin [168]. Furthermore, miR-200c-3p has been found to inhibit CRC proliferation and migration by targeting N-BLR, a novel carcinogenic non-coding RNA specific to CRC, and its interaction with E-cadherin and ZEB1 [169]. Given the crucial roles of miR-200 family members in the EMT process, these miRNAs may serve as biomarkers for CRC metastasis. By suppressing E-cadherin and claudin-1 expression, up-regulated miRNA-155 modulates EMT to increase CRC motility and infiltration [170]. Zinc finger protein 281 (ZNF281) is an EMT regulator. MiR-34-mediated ZNF281 downregulation causes mesenchymal-epithelial transition (MET), whereas EMT generating factor SNAIL directly increases ZNF281 transcription and represses miR-34 to ameliorate ZNF281 mRNA downregulation in CRC [171].
miRNAs in the notch signaling pathway
Stimulation of the Notch signaling pathway is crucial for determining cell fate. The Notch pathway facilitates direct cell-to-cell communication in multicellular animals [172]. Cell growth, differentiation, proliferation, and death cannot occur without a properly functioning Notch system [173,174,175]. Carcinogenesis may therefore be linked to Notch signaling. Cancers such as colorectal cancer have been found to have high levels of Notch signaling in multiple investigations (CRC). Four receptors: Notch1, Notch2, and Notch3, are part of the Notch signaling cascade [176]. Previous research on CRC metastasis has shown that the Notch signaling system is implicated, and further, miRNAs may control it post-transcriptionally [177]. MiR-1280 inhibits metastasis by straightly blocking the Notch signaling pathway activators like Zeb1, jagged canonical Notch ligand 2 (JAG2), Gata1/3, and polycomb repressive complex (Suz12) [178]. MiR-200b has also been shown to be a Notch signaling pathway activator [177]. Also, studies have found that miR-34a attached to the putative 3' untranslated regions of Notch1 and Jagged1 in SW480 cells, inhibiting colon cancer cell motility and invasion, according to in vitro miRNA functional experiments. miR-34a was also discovered to suppress the expression of vimentin and fibronectin via Notch1 and Jagged1. Therefore, findings suggest that miR 34a targets and regulates Notch signaling, inhibiting colorectal cancer spread [178, 179].
miRNA dysregulation in CRC
It is widely documented that miRNAs play a vital part in the genesis and advancement of cancer. As regulators of gene expression, miRNAs are committed to the maintenance of cellular homeostasis in normal tissue. However, in most cancers, miRNAs are greatly dysregulated [180,181,182,183]. Some miRNAs may act as oncogenes (oncomiRs), whereas others act as tumor suppressor genes. OncomiRs are highly expressed in cancer; for instance, oncomiRs involved in enhanced proliferation and apoptosis repression include mir-17, mir-19b, miR-21, mir-92a, and mir-106a. In contrast, tumor suppressor genes, along with miR-18a, miR-143, miR-145, and let-7 [184] are down-regulated in cancer. Furthermore, relying on the cellular context of its target genes in various malignancies, a specific miRNA can perform both tumor-suppressive and oncogenic roles. miRNAs have tissue-specific expression forms, and uncontrolled expression may be induced by genetic mutations in the miRNA gene region, inappropriate epigenetic modifications, erroneous transcriptional control, or miRNA synthesis faults. Aberrant miRNA expression is connected to CRC [185]. miRNA signatures are connected to CRC diagnosis, prognosis, progression, metastasis, and therapeutic resistance. Generally, miRNAs have been linked to signaling pathways implicated in (i) The modulation of miR-18a in MAPK pathway genes has been linked to cell proliferation,(ii) Cell survival pathways are linked to cellular apoptotic activity, as evidenced by miR-29a regulation in the PI3K1/AKT/MDM2/p53 pathways. (iii) DNA damage repair pathways, as with miR-155, that regulate RAD51 activity, an essential protein in DNA repair after ionic radiation and (iv) Cancer cell invasion and migration, as seen with miRNA-29a [179]. Alterations in miRNA expression have also been reported to take place at all phases of carcinogenesis, like tumor initiation, progression, and metastasis. Certain tumour types may have a distinctive miRNA profile that distinguishes them from the normal tissue from which they arose and other cancer types. Some of the implications of changed miRNA expression that contribute to cancer formation [183] include dysregulated cell proliferation, cell motility in carcinogenesis [186], and/or abnormalities in hormonal stress response. Dysregulation of microRNAs has been related to several undesirable outcomes, including continued proliferation, evasion of tumour suppression, avoidance of apoptosis, activation of invasion and metastasis, induction of angiogenesis, and therapy resistance [27, 28]. MiRNAs can serve as oncogenes or tumor suppressors by generating these alterations. Most human miRNAs are related to cancer-specific translocation breakpoints, fragile sites, CpG islands, and repetitive sequences [187]. Some research suggests this link is not clear-cut and depends on the kind of cancer [188]. Polymorphism in single nucleotides (SNPs) is well established, suggesting that SNPs alter miRNA destinations in cancer-associated mechanisms [189]. An SNP increase in function may improve its interaction with miRNA targets, increasing its regulatory activity as a tumour suppressor gene. SNP function loss may boost miRNA production, which functions as an oncogene [190]. SNPs in miRNA target regions can also prevent degradation [191]. All these data show that SNPs regulate the synthesis and functioning of miRNAs and thus is the one among the contributor to dysregulation during cancers. Also, miRNA gene expression, especially around CpG islands, is easily altered by methylation processes [192]; various research groups have explored whether hyper- or hypo-methylation (an initial episode in carcinogenesis) influences miRNA gene activity [192,193,194,195]. Multiple studies have shown that DNA methylation affects miRNA activity. Some examples: A study of colon cancer cell lines found that DNA methylation affected the production of 10% of miRNAs and that incomplete methylation pruning were inadequate for miRNA retrieval [196]. CRC screening demonstrated hyper-methylation of CpG islands suppressed miR-34b and miR-34c and affected miR-9 family genes [197]. MiR-9–1 methylation is connected with lymph node metastasis in CRC cells [198, 199]. Because both the conventional and noncanonical routes involve multiple intermediary components that are closely controlled, defects in miRNA synthesis are one of the molecular ways in which miRNAs can contribute to cancer development. Ras oncogene stress responses, Reactive oxygen species, and phorbol esters for example, reduced Dicer protein production in several cell lines [200]. Low levels of dicer expression and functioning have been linked with cancer development and a faulty prediction in individuals with lung, liver, breast, bladder, and CRC [201, 202]. In ovarian cancer, overexpression of Dicer, Drosha, Ago1 and Ago2 has been found [202]. Exportin-5 was discovered to perform an oncogenic function in CRC, with elevated expression levels linked to the worst clinicopathology and poor patient survival [183, 203]. The list of dysregulated miRNAs in CRC is shown in Table 3 respectively.
Clinical implication of miRNAs as reliable biomarkers for CRC diagnosis and prognosis
Various documented shreds of evidence reported that miRNAs are securely nested in exosomes obtained from various biological fluid samples like cerebrospinal fluid, urine, tears, saliva, breast milk, urine, feces, blood, and seminal fluids [227,228,229]. In addition, the bloodstream, viz. plasma, and serum nurtures ample exosomal miRNAs that might be utilized in clinical applications as a classic biomarker for diagnosis and early predictions as they have the prominent capability to lower various unfavorable conditions like high and low temperature, acidic and basic pH [230]. Therefore, microRNAs can be employed to screen people for cancer since they are stable and reliable, and their traces are in both blood and feces Furthermore, their expression patterns are comparable to those identified in tumors taken from patients with colorectal cancer [229, 231]. In principle, screening tests can identify the presence of miRNAs exclusively observed in individuals with intestinal adenomas or CRC [227]. However, the use of miRNAs to screen for colorectal cancer will never be able to match the preventative efficacy of routine colonoscopy. However, it may give an alternative that is less intrusive and more cost-effective than the screening approaches that are now used. MiRNAs for prediction purposes are also promising, particularly as precision medicine becomes more common in CRC treatment (A technique that considers individual differences in gene expression and tumour characteristics to obtain optimal patient outcomes through personalized therapy) [232]. In fact, in CRC patients, some studies reported suppressed levels of miR-24-2 in serum [233] and enhanced amounts of miR-129 in plasma. [234] Moreover, these reports concluded that miRNAs might be ideal biomarkers for people suffering from CRC as bloodstream samples are critical for detecting CRC. [226, 235, 236]. As recently proven for lung cancer, new advancements to miRNA detection technologies like digital PCR may enable better sensitive approaches for absolute quantification of miRNAs [237,238,239]; Various miRNA and mRNA expression patterns might be employed to create CRC diagnostic and predictive assays [237, 240]. Mir-21 expression is enhanced in CRC tumours, with multiple studies finding a step-wise rise as tumours advance to later stages [241,242,243]. MiR-21 is also altered in CRC patients’ blood and stool, and it may reliably predict the extent of local tumour invasion (T), lymph node involvement (N), and the existence of distant metastases (M)—the TNM stage [244]. In addition, researchers have discovered a correlation between increased amounts of miR-21 in primary CRC tissues and matching blood samples and considerable tumour development and distant metastasis [245]. Enhanced miR-21 expression in malignancies is also linked to poor chemotherapeutic response and lower disease-free survival [168]. Despite this, circulating miR-21 levels drop following CRC tumour excision [241]. These findings show that miR-21 levels in serum and stool mirror those in CRC tumours, suggesting that it might be used as an analytical and prognostic biomarker, predicting TNM stage, probable metastasis, and response to treatment. According to research, the miR-17-92 cluster is also implied in carcinogenesis. It has been discovered that levels of many members of this cluster, including miR-17, miR-20a, miR-92a, and miR-18a, are elevated in CRC tumours as well as serum and plasma, with greater levels being associated with recurrence and a poor outcome. Significantly, serum amounts of the miRNAs miR-18a and miR-92a fall after tumour removal. Tests on isolated colonic epithelial cells from CRC patients’ faeces revealed an increased appearance of the miR-17-92 cluster [246]. People with early-stage colorectal cancer and those with advanced adenomas can be differentiated from one another using the oncomir known as miR-29a, which is shown to be high in both CRC tumours and blood. This miRNA’s comprehensive expression profile may restrict its usefulness as a particular biomarker for CRC, yet, screening for miR-17-92 in CRC patients' blood and stool might be a valuable prognostic signal.
Colon cancer researchers recently concluded a clinical trial in which they used ELF/LEBS for depth-selective (from 30 to hundreds of microns) spectroscopy of live tissue to study two CRC animal models (the AOM-treated rat and the Min-mouse) as well as 190 human subjects. Before adenomas, potential changes in the colon might be detected with ELF/LEBS, and other histological/molecular indications of CRC could be found. ELF/LEBS can identify changes in normal rectal tissue induced by adenomas in any part of the colon. Rectal ELF/LEBS significantly surpassed all other known CRC markers in the first experiments. Therefore, testing for ELF/LEBS in the rectum can provide an accurate risk assessment for colon carcinogenesis without stool preparation or colonoscopy. ELF and LEBS fiber-optic probe prototypes have been constructed in vivo by scientists. One clinical trial using miRNA as a diagnostic tool for CRC stage II is still going, and the expected completion date is 2025. In this study, the miRNA apparatus, including miR-21, miR-20a-5p, miR-103a-3p, miR-106b-5p, miR-143-5p, and miR-215, will be used to evaluate patients. As a result of this tool, all the microRNAs in the human genome can be found. Utilizing qRT-PCR, researchers examine surgical material for the presence of these miRNAs and determine their risk score. Then they will classify patients with a score greater than one as exposed [247,248,249,250].
Discussion
There is mounting evidence that microRNAs are critical molecules in controlling each of the hallmarks of cancer. As a direct consequence, several microRNAs have been linked to colorectal cancer diagnosis, progression, and development of colorectal cancer. Multiple studies have identified either an excess of or a deficiency in the levels of miRNAs in CRC tissue samples. This mutation affects the mortality and differentiation of tumour cells while also promoting the proliferation, progression, metastasis, and angiogenesis of tumour cells. This mutation also promotes invasiveness. Inappropriate modulation of the RAS/MAPK signaling pathway is a characteristic feature of malignancies, and it frequently arises as a consequence of aberrant stimulation of associated receptors or dysregulation of RAS or RAF genes. The RAS/MAPK signaling pathway is responsible for regulating the activities of epithelial cells; however, abnormal regulation of this pathway by miRNAs can change its biological function. The fact that microRNAs have accountability in the growth of colorectal cancer as either tumour suppressors or oncogenes suggest that they have the potential to be exploited as diagnostic and prognostic markers. Therapeutic strategies include using inhibitors to target oncogenic miRNAs or restoring tumour-suppressor miRNAs in the body. It has been hypothesized that combining a therapeutic medicine with onco-miRNA inhibitors would result in more favourable outcomes. This is partly due to the complexity of the settings in which cancer cells are found and the dysregulation of many miRNAs in CRC. The understanding and characterization of miRNAs have shown that they belong to the same class of regulatory RNAs as other miRNAs. However, it has also shown that they may be able to assist in identifying a viable therapy and management plan for colorectal cancer (CRC). Many CRC oncomiRs have been linked to CRC metastasis and liver carcinogenesis when contained in cancer exosomes. These CRC oncomiRs include miR-18a, mir-328, miR-17-5p, and miR-92a. However, to be clinically exploited as reliable biomarkers, further validated studies and calibrated protocols for first processing, production, and normalization of miRNA are necessary.
Data availability
No data was generated during this study.
References
López-Urrutia E, Bustamante Montes L, Ladrón de GuevaraCervantes D, Pérez-Plasencia C, Campos-Parra A. Crosstalk Between Long Non-Coding Rnas Micro-Rnas and Mrnas: deciphering molecular mechanisms of master regulators in cancer. Frontiers in Oncology. 2017. https://doi.org/10.3389/fonc.2019.00669.
Chan J, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19(5):1310.
Qi X, Lin Y, Chen J, Shen B. Decoding competing endogenous RNA networks for cancer biomarker discovery. Brief Bioinform. 2019;21(2):441–57.
Ratti M, Lampis A, Ghidini M, Salati M, Mirchev M, Valeri N, Hahne J. Micrornas (Mirnas) and long non-coding Rnas (Lncrnas) as new tools for cancer therapy: first steps from bench To bedside. Target Oncol. 2020;15(3):261–78.
Guelfi G, Cochetti G, Stefanetti V, Zampini D, Diverio S, Boni A, Mearini E. Next generation sequencing of urine exfoliated cells: an approach of prostate cancer micrornas research. Sci Rep. 2018. https://doi.org/10.1038/s41598-018-24236-y.
Pan Y, Qin J, Sun H, Xu T, Wang S, He B. Mir-485-5P as a potential biomarker and tumor suppressor in human colorectal cancer. Biomark Med. 2020;14(3):239–48.
Mall C, Rocke D, Durbin-Johnson B, Weiss R. Stability of Mirna in human urine supports its biomarker potential. Biomark Med. 2013;7(4):623–31.
Blondal T, Jensby Nielsen S, Baker A, Andreasen D, Mouritzen P, Wrang Teilum M, Dahlsveen I. Assessing sample and mirna profile quality in serum and plasma or other biofluids. Methods. 2013;59(1):S1–6.
Mraz M, Malinova K, Mayer J, Pospisilova S. Microrna isolation and stability in stored RNA samples. Biochem Biophys Res Commun. 2009;390(1):1–4.
Schetter A, Harris C. Plasma micrornas: a potential biomarker for colorectal cancer? Gut. 2009;58(10):1318–9.
Imaoka H, Toiyama Y, Fujikawa H, Hiro J, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Mori T, Kato T, Toden S, Goel A, Kusunoki M. Circulating microrna-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann Oncol. 2016;27(10):1879–86.
Lee R, Feinbaum R, Ambros V, The C. Elegans heterochronic gene lin-4 encodes small Rnas with antisense complementarity to lin-14. Cell. 1993;75(5):843–54.
Reinhart B, Slack F, Basson M, Pasquinelli A, Bettinger J, Rougvie A, Horvitz H, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in caenorhabditis elegans. Nature. 2000;403(6772):901–6.
Pasquinelli A, Reinhart B, Slack F, Martindale M, Kuroda M, Maller B, Hayward D, Ball E, Degnan B, Müller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–9.
Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–9.
Rupaimoole R, Slack F. Microrna therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug discov. 2017;16(3):203–22.
Iwasaki H, Shimura T, Kitagawa M, Yamada T, Nishigaki R, Fukusada S, Okuda Y, Katano T, Horike S, Kataoka H. A novel urinary Mirna biomarker for early detection of colorectal cancer. Cancers. 2022;14(2):461.
Bray F, Ferlay J, Soerjomataram I, Siegel R, Torre L, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin. 2018;68(6):394–424.
Siegel R, Miller K, Fedewa S, Ahnen D, Meester R, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA: Cancer J Clin. 2017;67(3):177–93.
Croce C. 37 causes and consequences of microrna dysregulation in cancer. Eur J Cancer. 2012;48:S8–9.
Jevšinek Skok D, Hauptman N, Boštjančič E, Zidar N. The integrative knowledge base for Mirna-Mrna expression in colorectal cancer. Sci Rep. 2019. https://doi.org/10.1038/s41598-019-54358-w.
Iorio M, Croce C. Microrna dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Molecular Med. 2012;4(3):143–15923.
Siegel R, Miller K, Jemal A. Cancer statistics. CA: Cancer J Clin. 2016;66(1):7–30.
Mani SA, Yang J, Brooks M, Schwaninger G, Zhou A, Miura N, Kutok JL, Hartwell K, Richardson AL, Weinberg RA. Mesenchyme forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci USA. 2007;104:10069–74.
Fan Y, Wang K. Mir-205 suppresses cell migration, invasion and EMT of colon cancer by targeting mouse double minute 4. Mol Med Rep. 2020;22(2):633–42.
Xu Z, Zhu C, Chen C, Zong Y, Feng H, Liu D, Feng W, Zhao J, Lu A. CCL19 suppresses angiogenesis through promoting Mir-206 and inhibiting Met/ERK/Elk-1/HIF-1Α/VEGF-A pathway in colorectal cancer. Cell Death Dis. 2018. https://doi.org/10.1038/s41419-018-1010-2.
Ma H, Pan J, Jin L, Wu J, Ren Y, Chen P, Xiao C, Han J. Microrna-17~92 inhibits colorectal cancer progression by targeting angiogenesis. Cancer Lett. 2016;376(2):293–302.
Yang N, Zhu S, Lv X, Qiao Y, Liu Y, Chen J. Micrornas: pleiotropic regulators in the tumor microenvironment. Front Immunol. 2018;9:2491.
Cătană C, Pichler M, Giannelli G, Mader R, Berindan-Neagoe I. Non-coding Rnas, the trojan horse in two-way communication between tumor and stroma in colorectal and hepatocellular carcinoma. Oncotarget. 2017;8(17):29519–34.
Su J, Lu E, Lu L, Zhang C. Mir-29A-3P suppresses cell proliferation in laryngocarcinoma by targeting prominin 1. FEBS Open Bio. 2017;7(5):645–51.
Truong A, Rengaraj D, Hong Y, Hoang C, Hong Y, Lillehoj H. Differentially expressed JAK-STAT signaling pathway genes and target micrornas in the spleen of necrotic enteritis-afflicted chicken lines. Res Vet Sci. 2017;115:235–43.
Seo H, Moeng S, Sim S, Kuh H, Choi S, Park J. Microrna-based combinatorial cancer therapy: effects of micrornas on the efficacy of anti-cancer therapies. Cells. 2019;9(1):29.
Caritg O, Navarro A, Moreno I, Martínez-Rodenas F, Cordeiro A, Muñoz C, Ruiz-Martinez M, Santasusagna S, Castellano J, Monzó M. Identifying high-risk stage II colon cancer patients: a three-microrna-based score as a prognostic biomarker. Clin Colorectal Cancer. 2016;15(4):e175–82.
Bobowicz M, Skrzypski M, Czapiewski P, Marczyk M, Maciejewska A, Jankowski M, Szulgo-Paczkowska A, Zegarski W, Pawłowski R, Polańska J, Biernat W, Jaśkiewicz J, Jassem J. Prognostic value Of 5-microrna based signature In T2–T3N0 colon Cancer. Clin Exp Metas. 2016;33(8):765–73.
Maierthaler M, Benner A, Hoffmeister M, Surowy H, Jansen L, Knebel P, Chang-Claude J, Brenner H, Burwinkel B. Plasma Mir-122 and Mir-200 family are prognostic markers in colorectal cancer. Int J Cancer. 2016;140(1):176–87.
Gilles M, Slack F. <I>Let-7</I> microrna as a potential therapeutic target with implications for immunotherapy. Expert Opin Ther Targets. 2018;22(11):929–39.
Conti I, Varano G, Simioni C, Laface I, Milani D, Rimondi E, Neri L. Mirnas as influencers of cell-cell communication in tumor microenvironment. Cells. 2020;9(1):220.
Kalluri R, Weinberg R. The basics of epithelial-mesenchymal transition. J Clin Investig. 2009;119(6):1420–8.
Zeisberg M, Neilson E. Biomarkers for epithelial-mesenchymal transitions. J Clin Investig. 2009;119(6):1429–37.
Naxerova K, Reiter J, Brachtel E, Lennerz J, van de Wetering M, Rowan A, Cai T, Clevers H, Swanton C, Nowak M, Elledge S, Jain R. Origins of lymphatic and distant metastases in human colorectal cancer. Science. 2017;357(6346):55–60.
Akinc A, Zumbuehl A, Goldberg M, Leshchiner E, Busini V, Hossain N, Bacallado S, Nguyen D, Fuller J, Alvarez R, Borodovsky A, Borland T, Constien R, de Fougerolles A, Dorkin J, Narayanannair Jayaprakash K, Jayaraman M, John M, Koteliansky V, Manoharan M, Nechev L, Qin J, Racie T, Raitcheva D, Rajeev K, Sah D, Soutschek J, Toudjarska I, Vornlocher H, Zimmermann T, Langer R, Anderson D. A combinatorial library of lipid-like materials for delivery of Rnai therapeutics. Nat Biotechnol. 2008;26(5):561–9.
Jin D, Fang Y, Li Z, Chen Z, Xiang J. Epithelial-mesenchymal transition-associated micrornas in colorectal cancer and drug-targeted therapies (Review). Oncol Rep. 2014;33(2):515–25.
.
Ding M, Zhang T, Li S, Zhang Y, Qiu Y, Zhang B. Correlation analysis between liver metastasis and serum levels Of Mir-200 and Mir-141 in patients with colorectal cancer. Mol Med Rep. 2017;16(5):7791–5.
Zhao S, Sun H, Jiang W, Mi Y, Zhang D, Wen Y, Cheng D, Tang H, Wu S, Yu Y, Liu X, Cui W, Zhang M, Sun X, Zhou Z, Peng Z, Yan D. Mir-4775 promotes colorectal cancer invasion and metastasis via the Smad7/Tgfβ-mediated epithelial to mesenchymal transition. Molecular Cancer. 2017. https://doi.org/10.1186/s12943-017-0585-z.
Wang H, Yan B, Zhang P, Liu S, Li Q, Yang J, Yang F, Chen E. Mir-496 promotes migration and epithelial-mesenchymal transition by targeting RASSF6 in colorectal cancer. J Cell Physiol. 2019;235(2):1469–79.
Przygodzka P, Papiewska-Pająk I, Bogusz-Koziarska H, Sochacka E, Boncela J, Kowalska M. Regulation of mirnas by snail during epithelial-to-mesenchymal transition in HT29 colon cancer cells. Sci Rep. 2019. https://doi.org/10.1038/s41598-019-39200-7.
Zheng K, Zhou X, Yu J, Li Q, Wang H, Li M, Shao Z, Zhang F, Luo Y, Shen Z, Chen F, Shi F, Cui C, Zhao D, Lin Z, Zheng W, Zou Z, Huang Z, Zhao L. Epigenetic silencing Of Mir-490-3P promotes development of an aggressive colorectal cancer phenotype through activation of the Wnt/Β-catenin signaling pathway. Cancer Lett. 2016;376(1):178–87.
Vu T, Datta P. Regulation Of EMT in colorectal cancer: a culprit in metastasis. Cancers. 2017;9(12):171.
Sheng L, He P, Yang X, Zhou M, Feng Q. Mir-612 negatively regulates colorectal cancer growth and metastasis by targeting AKT2. Cell Death Dis. 2015;6(7):e1808–e1808.
Huang LX, Hu CY, Jing L, Wang MC, Xu M, Wang J, Wang Y, Nan KJ, Wang SH. MicroRNA-219-5p inhibits epithelial-mesenchymal transition and metastasis of colorectal cancer by targeting lymphoid enhancer-binding factor 1. Cancer Sci. 2017;108:1985–95.
Zhang Z, Liu X, Feng B, Liu N, Wu Q, Han Y, Nie Y, Wu K, Shi Y, Fan D. STIM1, a direct target of microrna-185, promotes tumor metastasis and is associated with poor prognosis in colorectal cancer. Oncogene. 2014;34(37):4808–20.
He Z, Yu L, Luo S, Li M, Li J, Li Q, Sun Y, Wang C. Mir-296 inhibits the metastasis and epithelial-mesenchymal transition of colorectal cancer by targeting S100A4. BMC Cancer. 2017. https://doi.org/10.1186/s12885-017-3121-z.
Bentley K, Chakravartula S. The temporal basis of angiogenesis. Philos Trans Royal Soc B: Biol Sci. 2017;372(1720):20150522.
Betz C, Lenard A, Belting H, Affolter M. Cell behaviors and dynamics during angiogenesis. Development. 2016;143(13):2249–60.
Li J, Sun X, Wang Z, Chen L, Li D, Zhou J, Liu M. Regulation of vascular endothelial cell polarization and migration by Hsp70/Hsp90-organizing protein. PLoS ONE. 2012;7(4):e36389.
Goradel N, Mohammadi N, Haghi-Aminjan H, Farhood B, Negahdari B, Sahebkar A. Regulation of tumor angiogenesis by micrornas: state of the art. J Cell Physiol. 2018;234(2):1099–110.
Wang Y, Wang L, Chen C, Chu X. New insights into the regulatory role of microrna in tumor angiogenesis and clinical implications. Molecular Cancer. 2018. https://doi.org/10.1186/s12943-018-0766-4.
Leone P, Buonavoglia A, Fasano R, Solimando A, De Re V, Cicco S, Vacca A, Racanelli V. Insights into the regulation of tumor angiogenesis by micro-Rnas. J Clin Med. 2019;8(12):2030.
Soheilifar M, Grusch M, Neghab H, Amini R, Maadi H, Saidijam M, Wang Z. Angioregulatory micrornas in colorectal cancer. Cancers. 2019;12(1):71.
Zhou J. Microrna regulation network in colorectal cancer metastasis. World J Biol Chem. 2014;5(3):301.
Chen X, Zeng K, Xu M, Liu X, Hu X, Xu T, He B, Pan Y, Sun H, Wang S. P53-induced Mir-1249 inhibits tumor growth, metastasis, and angiogenesis by targeting VEGFA And HMGA2. Cell Death Dis. 2019. https://doi.org/10.1038/s41419-018-1188-3.
Zhou Q, Zhu Y, Wei X, Zhou J, Chang L, Sui H, Han Y, Piao D, Sha R, Bai Y. Mir-590-5P inhibits colorectal cancer angiogenesis and metastasis by regulating nuclear factor 90/vascular endothelial growth factor a axis. Cell Death Dis. 2016;7(10):e2413–e2413.
Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J, Zhou K, Liu X, Ren X, Wang F, Hu J, Zhu X, Yang W, Liao W, Li G, Ding Y, Liang L. Cancer-derived exosomal Mir-25–3P promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. 2018. https://doi.org/10.1038/s41467-018-07810-w.
Wang S, Zhang Z, Gao Q. Transfer of microrna-25 by colorectal cancer cell-derived extracellular vesicles facilitates colorectal cancer development and metastasis. Molecular Ther - Nucleic Acids. 2021;23:552–64.
Muhammad S. Micrornas in colorectal cancer: role in metastasis and clinical perspectives. World J Gastroenterol. 2014;20(45):17011.
Baumjohann D, Ansel K. Microrna-mediated regulation of T helper cell differentiation and plasticity. Nat Rev Immunol. 2013;13(9):666–78.
Zhu K, Pan Q, Zhang X, Kong L, Fan J, Dai Z, Wang L, Yang X, Hu J, Wan J, Zhao Y, Tao Z, Chai Z, Zeng H, Tang Z, Sun H, Zhou J. Mir-146A enhances angiogenic activity of endothelial cells in hepatocellular carcinoma by promoting PDGFRA expression. Carcinogenesis. 2013;34(9):2071–9.
Verghese E, Drury R, Green C, Holliday D, Lu X, Nash C, Speirs V, Thorne J, Thygesen H, Zougman A, Hull M, Hanby A, Hughes T. Mir-26B is down-regulated in carcinoma-associated fibroblasts from ER-positive breast cancers leading to enhanced cell migration and invasion. J Pathol. 2013;231(3):388–99.
Mitra A, Zillhardt M, Hua Y, Tiwari P, Murmann A, Peter M, Lengyel E. Micrornas reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer Discov. 2012;2(12):1100–8.
Lu L, Thai T, Calado D, Chaudhry A, Kubo M, Tanaka K, Loeb G, Lee H, Yoshimura A, Rajewsky K, Rudensky A. Foxp3-dependent Microrna155 confers competitive fitness to regulatory T cells by targeting SOCS1 Protein. Immunity. 2009;30(1):80–91.
Zhou X, Huang X, Liang S, Tang S, Wu S, Huang T, Mo Z, Wang Q. Identifying Mirna and gene modules of colon cancer associated with pathological stage by weighted Gene Co-expression network analysis. Onco Targets Ther. 2018;11:2815–30.
Makondi P, Wei P, Huang C, Chang Y. Development of novel predictive mirna/target gene pathways for colorectal cancer distance metastasis to the liver using a bioinformatic approach. PLoS ONE. 2019;14(2):e0211968.
Cai X, Yin Y, Li N, Zhu D, Zhang J, Zhang C, Zen K. Re-POLARIZATION OF TUMOR-ASSOCIATED MACROPHAGES TO PRO-INFLAMMATORY M1 macrophages by microrna-155. J Mol Cell Biol. 2012;4(5):341–3.
Lin X, Wang S, Sun M, Zhang C, Wei C, Yang C, Dou R, Liu Q, Xiong B. Mir-195–5P/NOTCH2-mediated EMT modulates IL-4 secretion in colorectal cancer to affect M2-Like TAM polarization. Journal of Hematology & Oncology. 2019. https://doi.org/10.1186/s13045-019-0810-x8.
Wei C, Yang C, Wang S, Shi D, Zhang C, Lin X, Liu Q, Dou R, Xiong B. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Molecular Cancer. 2019. https://doi.org/10.1186/s12943-019-0976-4.
Zhou M, Chen J, Zhou L, Chen W, Ding G, Cao L. pancreatic Cancer Derived Exosomes Regulate The Expression Of TLR4 in Dendritic Cells Via Mir-203. Cell Immunol. 2014;292(1–2):65–9.
Huang L, Wang X, Wen C, Yang X, Song M, Chen J, Wang C, Zhang B, Wang L, Iwamoto A, Wang J, Liu H. Hsa-Mir-19A is associated with lymph metastasis and mediates The TNF-α induced epithelial-to-mesenchymal transition in colorectal cancer. Scientific Reports. 2015. https://doi.org/10.1038/srep13350.
Shen Z, Zhou R, Liu C, Wang Y, Zhan W, Shao Z, Liu J, Zhang F, Xu L, Zhou X, Qi L, Bo F, Ding Y, Zhao L. Microrna-105 is involved in TNF-Α-related tumor microenvironment enhanced colorectal cancer progression. Cell Death Dis. 2017;8(12):3213.
Huang X, Zhu X, Yu Y, Zhu W, Jin L, Zhang X, Li S, Zou P, Xie C, Cui R. Dissecting Mirna signature in colorectal cancer progression and metastasis. Cancer Lett. 2021;501:66–82.
Polakis P. Wnt signaling in cancer. Cold Spring Harb Perspect Biol. 2012;4(5):a008052–a008052.
Novellasdemunt L, Antas P, Li V. Targeting Wnt signaling in colorectal cancer. A review in the theme: cell signaling: proteins, pathways and mechanisms. Am J Physiol-Cell Physiol. 2015;309(8):511–21.
Valeri N, Braconi C, Gasparini P, Murgia C, Lampis A, Paulus-Hock V, Hart J, Ueno L, Grivennikov S, Lovat F, Paone A, Cascione L, Sumani K, Veronese A, Fabbri M, Carasi S, Alder H, Lanza G, Gafa R, Moyer M, Ridgway R, Cordero J, Nuovo G, Frankel W, Rugge M, Fassan M, Groden J, Vogt P, Karin M, Sansom O, Croce C. Microrna-135B promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell. 2014;25(4):469–83.
Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink J, Bolijn A, Meijer G, Agami R. Regulation of the adenomatous polyposis coli gene by the Mir-135 family in colorectal cancer. Can Res. 2008;68(14):5795–802.
Prossomariti A, Piazzi G, D’Angelo L, Miccoli S, Turchetti D, Alquati C, Montagna C, Bazzoli F, Ricciardiello L. Mir-155 is downregulated in familial adenomatous polyposis and modulates WNT signaling by targeting AXIN1 And TCF4. Mol Cancer Res. 2018;16(12):1965–76.
Ma Y, Zhang P, Wang F, Zhang H, Yang Y, Shi C, Xia Y, Peng J, Liu W, Yang Z, Qin H. Elevated oncofoetal Mir-17–5P expression regulates colorectal cancer progression by repressing its target gene P130. Na Commun. 2012. https://doi.org/10.1038/ncomms2276.
Li T, Lai Q, Wang S, Cai J, Xiao Z, Deng D, He L, Jiao H, Ye Y, Liang L, Ding Y, Liao W. Microrna-224 sustains Wnt/Β-catenin signaling and promotes aggressive phenotype of colorectal cancer. J Exp Clin Cancer Res. 2016. https://doi.org/10.1186/s13046-016-0287-1.
Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S, Wang Y, Wang T, Hou Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal Lncrna H19. Theranostics. 2018;8(14):3932–48.
Li Y, Lauriola M, Kim D, Francesconi M, D’Uva G, Shibata D, Malafa M, Yeatman T, Coppola D, Solmi R, Cheng J. Adenomatous polyposis coli (APC) regulates Mir17-92 cluster through Β-catenin pathway in colorectal cancer. Oncogene. 2016;35(35):4558–68.
Tili E, Michaille J, Croce C. Micrornas play a central role in molecular dysfunctions linking inflammation with cancer. Immunol Rev. 2013;253(1):167–84.
Thomas H. Mir-100 and Mir-125B induce cetuximab resistance In CRC. Nat Rev Gastroenterol Hepatol. 2017;14(12):691–691.
Lu Y, Zhao X, Liu Q, Li C, Graves-Deal R, Cao Z, Singh B, Franklin J, Wang J, Hu H, Wei T, Yang M, Yeatman T, Lee E, Saito-Diaz K, Hinger S, Patton J, Chung C, Emmrich S, Klusmann J, Fan D, Coffey R. Lncrna MIR100HG-derived Mir-100 And Mir-125B mediate cetuximab resistance via Wnt/Β-catenin signaling. Nat Med. 2017;23(11):1331–41.
Zhang N, Li X, Wu C, Dong Y, Cai M, Mok M, Wang H, Chen J, Ng S, Chen M, Sung J, Yu J. Microrna-7 is a novel inhibitor Of YY1 contributing to colorectal tumorigenesis. Oncogene. 2012;32(42):5078–88.
Liu Y, Liu R, Yang F, Cheng R, Chen X, Cui S, Gu Y, Sun W, You C, Liu Z, Sun F, Wang Y, Fu Z, Ye C, Zhang C, Li J, Chen X. Mir-19A promotes colorectal cancer proliferation and migration by targeting TIA1. Molecular Cancer. 2017. https://doi.org/10.1186/s12943-017-0625-8.
Falzone L, Scola L, Zanghì A, Biondi A, Di Cataldo A, Libra M, Candido S. Integrated analysis of colorectal cancer microrna datasets: identification of micrornas associated with tumor development. Aging. 2018;10(5):1000–14.
Fu J, Tang W, Du P, Wang G, Chen W, Li J, Zhu Y, Gao J, Cui L. Identifying microrna-mrna regulatory network in colorectal cancer by a combination of expression profile and bioinformatics analysis. BMC Syst Biol. 2012. https://doi.org/10.1186/1752-0509-6-68.
Phesse T, Flanagan D, Vincan E. Frizzled7: a promising Achilles’ heel for targeting the wnt receptor complex to treat cancer. Cancers. 2016;8(5):50.
Tang Q, Zou Z, Zou C, Zhang Q, Huang R, Guan X, Li Q, Han Z, Wang D, Wei H, Gao X, Wang X. Microrna-93 suppress colorectal cancer development via wnt/β-catenin pathway downregulating. Tumor Biol. 2014;36(3):1701–10.
Fasihi A, M Soltani B, Atashi A, Nasiri S. Introduction of hsa-miR-103a and hsa-miR-1827 and hsa-miR-137 as new regulators of Wnt signaling pathway and their relation to colorectal carcinoma. J Cell Biochem. 2018;119:5104–17.
Hwang W, Yang M. Numb is involved in the non-random segregation of subcellular vesicles in colorectal cancer stem cells. Cell Cycle. 2016;15(20):2697–703.
Liu B, Zhou W, Jiang H, Xiang Z, Wang L. Mir-1303 promotes the proliferation, migration and invasion of prostate cancer cells through regulating The Wnt/Β-catenin pathway by targeting DKK3. Exp Therapeutic Med. 2019. https://doi.org/10.3892/etm.2019.8120.
Dong-xu W, Jia L, Su-juan Z. Microrna-185 is a novel tumor suppressor by negatively modulating the Wnt/Β-catenin pathway in human colorectal cancer. Indian J Cancer. 2015;52(7):182.
Zhang Y, Guo L, Li Y, Feng G, Teng F, Li W, Zhou Q. Microrna-494 promotes cancer progression and targets adenomatous polyposis coli in colorectal cancer. Molecular Cancer. 2018. https://doi.org/10.1186/s12943-017-0753-1.
Cao J, Yan X, Liu T, Han X, Yu J, Liu S, Wang L. Microrna-552 promotes tumor cell proliferation and migration by directly targeting DACH1 Via The Wnt/Β-catenin signaling pathway in colorectal cancer. Oncol Lett. 2017;14(3):3795–802.
Ji S, Ye G, Zhang J, Wang L, Wang T, Wang Z, Zhang T, Wang G, Guo Z, Luo Y, et al. miR-574-5p negatively regulates Qki6/7 to impact beta-catenin/Wnt signalling and the development of colorectal cancer. Gut. 2013;62:716–26.
Campbell P, Groehler A, Lee K, Ouellette M, Khazak V, Der C. K-Ras promotes growth transformation and invasion of immortalized human pancreatic cells by raf and phosphatidylinositol 3-kinase signaling. Can Res. 2007;67(5):2098–106.
Lena M. New strategies for colorectal cancer screening. World J Gastroenterol. 2013;19(12):1855.
Bahrami A, Hassanian S, ShahidSales S, Farjami Z, Hasanzadeh M, Anvari K, Aledavood A, Maftouh M, Ferns G, Khazaei M, Avan A. Targeting RAS signaling pathway as a potential therapeutic target in the treatment of colorectal cancer. J Cell Physiol. 2017;233(3):2058–66.
Dong C, Davis R, Flavell R. MAP kinases in the immune response. Annu Rev Immunol. 2002;20(1):55–72.
Hommes D. Mitogen activated protein (MAP) kinase signal transduction pathways and novel anti-inflammatory targets. Gut. 2003;52(1):144–51.
Sun Y, Liu W, Liu T, Feng X, Yang N, Zhou H. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduction. 2015;35(6):600–4.
Taupin D, Podolsky D. Mitogen-activated protein kinase activation regulates intestinal epithelial differentiation. Gastroenterology. 1999;116(5):1072–80.
Troppmair J, Bruder J, Munoz H, Lloyd P, Kyriakis J, Banerjee P, Avruch J, Rapp U. Mitogen-activated protein kinase/extracellular signal-regulated protein kinase activation by oncogenes, serum, And 12-O-tetradecanoylphorbol-13-acetate requires raf and is necessary for transformation. J Biol Chem. 1994;269(9):7030–5.
Fang J, Richardson B. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6(5):322–7.
Arrington A, Heinrich E, Lee W, Duldulao M, Patel S, Sanchez J, Garcia-Aguilar J, Kim J. Prognostic and predictive roles Of KRAS mutation in colorectal cancer. Int J Mol Sci. 2012;13(12):12153–68.
Nishimura J, Handa R, Yamamoto H, Tanaka F, Shibata K, Mimori K, Takemasa I, Mizushima T, Ikeda M, Sekimoto M, Ishii H, Doki Y, Mori M. Microrna-181A is associated with poor prognosis of colorectal cancer. Oncol Rep. 2012;28(6):2221–6.
Sun H, Chen X, Ji H, Wu T, Lu H, Zhang Y, Li H, Li Y. Mir-494 is an independent prognostic factor and promotes cell migration and invasion in colorectal cancer by directly targeting PTEN. Int J Oncol. 2014;45(6):2486–94.
Wu W, Yang J, Feng X, Wang H, Ye S, Yang P, Tan W, Wei G, Zhou Y. Microrna-32 (Mir-32) regulates phosphatase and tensin homologue (PTEN) expression and promotes growth, migration, and invasion in colorectal carcinoma cells. Molecular Cancer. 2013. https://doi.org/10.1186/1476-4598-12-30.
Fonseca A, Ramalhete S, Mestre A, Pires Neves R, Marreiros A, Castelo-Branco P, Roberto V. Identification of colorectal cancer associated biomarkers: an integrated analysis of mirna expression. Aging. 2021;13(18):21991–2029.
Lu Y, Yang H, Yuan L, et al. Overexpression of miR-335 confers cell proliferation and tumour growth to colorectal carcinoma cells. Mol Cell Biochem. 2016;412:235–45.
Kent O, Mendell J, Rottapel R. Transcriptional regulation of Mir-31 by oncogenic KRAS mediates metastatic phenotypes by repressing RASA1. Mol Cancer Res. 2016;14(3):267–77.
Sun D, Yu F, Ma Y, Zhao R, Chen X, Zhu J, Zhang C, Chen J, Zhang J. Microrna-31 Activates the RAS pathway and functions as an oncogenic microrna in human colorectal cancer by repressing RAS P21 gtpase activating protein 1 (RASA1). J Biol Chem. 2013;288(13):9508–18.
Sun X, Lin F, Sun W, Zhu W, Fang D, Luo L, Li S, Zhang W, Jiang L. Exosome-transmitted mirna-335-5P promotes colorectal cancer invasion and metastasis by facilitating EMT via targeting RASA1. Molecular Therapy - Nucleic Acids. 2021;24:164–74.
You Q, Li H, Liu Y, Xu Y, Miao S, Yao G, Xue Y, Geng J, Jin X, Meng H. Microrna-650 targets inhibitor of growth 4 to promote colorectal cancer progression via mitogen activated protein kinase signaling. Oncol Lett. 2018. https://doi.org/10.3892/ol.2018.8910.
Rasmussen M, Lyskjær I, Jersie-Christensen R, Tarpgaard L, Primdal-Bengtson B, Nielsen M, Pedersen J, Hansen T, Hansen F, Olsen J, Pfeiffer P, Ørntoft T, Andersen C. Mir-625–3P regulates oxaliplatin resistance by targeting MAP2K6-P38 signalling in human colorectal adenocarcinoma cells. Nature Communications. 2016. https://doi.org/10.1038/ncomms12436.
Hata T, Mokutani Y, Takahashi H, Inoue A, Munakata K, Nagata K, Haraguchi N, Nishimura J, Hata T, Matsuda C, Murata K, Mizushima T, Doki Y, Mori M, Yamamoto H. Identification of microrna-487B as a negative regulator of liver metastasis by regulation Of KRAS in colorectal cancer. Int J Oncol. 2016;50(2):487–96.
Michael MZ, O’Connor SM, Pellekaan NGV, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Canc Res. 2003;1:882–91.
Wei W, Nian X, Wang S, Jiao H, Wang Y, Xiao Z, Yang R, Ding Y, Ye Y, Liao W. Mir-422A inhibits cell proliferation in colorectal cancer by targeting AKT1 and MAPK1. Cancer Cell Intern. 2017. https://doi.org/10.1186/s12935-017-0461-3.
Koveitypour Z, Panahi F, Vakilian M, Peymani M, Seyed Forootan F, Nasr Esfahani M, Ghaedi K. Signaling pathways involved in colorectal cancer progression. Cell Biosci. 2019. https://doi.org/10.1186/s13578-019-0361-4.
Niu Y, Zhang J, Tong Y, Li J, Liu B. Mir-145-5P restrained cell growth, invasion, migration and tumorigenesis via modulating RHBDD1 in colorectal cancer via The EGFR-associated signaling pathway. Int J Biochem Cell Biol. 2019;117:105641.
Ye S, Song W, Xu XG, Zhao XY, Yang L. IGF2BP2 promotes colorectal cancer cell proliferation and survival through interfering with RAF-1 degradation by miR-195. FEBS Lett. 2016;590:1641–50.
Zhu H, Dougherty U, Robinson V, Mustafi R, Pekow J, Kupfer S, Li Y, Hart J, Goss K, Fichera A, Joseph L, Bissonnette M. EGFR signals downregulate tumor suppressors Mir-143 And Mir-145 in western diet-promoted murine colon cancer: role Of G1 regulators. Mol Cancer Res. 2011;9(7):960–75.
Kent O, Fox-Talbot K, Halushka M. RREB1 repressed Mir-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene. 2012;32(20):2576–85.
Fan C, Lin B, Huang Z, Cui D, Zhu M, Ma Z, Zhang Y, Liu F, Liu Y. Microrna-873 inhibits colorectal cancer metastasis by targeting ELK1 and STRN4. Oncotarget. 2018;10(41):4192–204.
Xu L, Zhang Y, Wang H, Zhang G, Ding Y, Zhao L. Tumor suppressor Mir-1 restrains epithelial-mesenchymal transition and metastasis of colorectal carcinoma via the MAPK And PI3K/AKT pathway. J Trans Med. 2014. https://doi.org/10.1186/s12967-014-0244-8.
Gong B, Liu WW, Nie WJ, et al. MiR-21/RASA1 axis affects malignancy of colon cancer cells via RAS pathways. World J Gastroenterol. 2015;21:1488–97.
Falzone L, Salomone S, Libra M. Evolution of cancer pharmacological treatments at the turn of the third millennium. Front Pharmacol. 2018. https://doi.org/10.3389/fphar.2018.01300.
Liao WT, Ye YP, Zhang NJ, et al. MicroRNA-30b functions as a tumour suppressor in human colorectal cancer by targeting KRAS, PIK3CD and BCL2. J Pathol. 2014;232:415–27.
Zhou Y, Feng X, Liu Y, Ye S, Wang H, Tan W, Tian T, Qiu Y, Luo H. Down-regulation of Mir-126 is associated with colorectal cancer cells proliferation, migration and invasion by targeting IRS-1 Via The AKT And ERK1/2 signaling pathways. PLoS ONE. 2013;8(11):e81203.
Liu Y, Zhou Y, Feng X, An P, Quan X, Wang H, Ye S, Yu C, He Y, Luo H. Microrna-126 functions as a tumor suppressor in colorectal cancer cells by targeting Cxcr4 via the Akt And Erk1/2 signaling pathways. Int J Oncol. 2013;44(1):203–10.
Wang H, An H, Wang B, Liao Q, Li W, Jin X, Cui S, Zhang Y, Ding Y, Zhao L. Mir-133A represses tumour growth and metastasis in colorectal cancer by targeting LIM And SH3 protein 1 and inhibiting the MAPK pathway. Eur J Cancer. 2013;49(18):3924–35.
Kent O, McCall M, Cornish T, Halushka M. Lessons from Mir-143/145: the importance of cell-type localization of mirnas. Nucleic Acids Res. 2014;42(12):7528–38.
Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, Cai X, Wang K, Wang G, Ba Y, Zhu L, Wang J, Yang R, Zhang Y, Ren Z, Zen K, Zhang J, Zhang C. Role Of Mir-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28(10):1385–92.
Fei B, Wang X, Fang X. Microrna-143 replenishment re-sensitizes colorectal cancer cells harboring mutant, but not wild-type. KRAS To Paclitaxel Treatment Tumor Biol. 2015;37(5):5829–35.
Yin Y, Yan Z, Lu N, Xu Q, He J, Qian X, Yu J, Guan X, Jiang B, Liu L. Downregulation of Mir-145 associated with cancer progression and VEGF transcriptional activation by targeting N-RAS and IRS1. Biochimica et Biophysica Acta (BBA)–Gene Regulatory Mechanisms. 2013;1829(2):239–47.
Wang Z, Zhang X, Yang Z, Du H, Wu Z, Gong J, Yan J, Zheng Q. Mir-145 regulates PAK4 via the MAPK pathway and exhibits an antitumor effect in human colon cells. Biochem Biophys Res Commun. 2012;427(3):444–9.
Shen K, Liang Q, Xu K, Cui D, Jiang L, Yin P, Lu Y, Li Q, Liu J. Mir-139 inhibits invasion and metastasis of colorectal cancer by targeting the type i insulin-like growth factor receptor. Biochem Pharmacol. 2012;84(3):320–30.
Ribeiro-Neto F, Urbani J, Lemee N, Lou L, Altschuler D. On The mitogenic properties of rap1b: camp-induced G <Sub>1</Sub> /S entry requires activated and phosphorylated rap1b. Proc Natl Acad Sci. 2002;99(8):5418–23.
Wang B, Shen Z, Gao Z, Zhao G, Wang C, Yang Y, Zhang J, Yan Y, Shen C, Jiang K, Ye Y, Wang S. Mir-194, commonly repressed in colorectal cancer, suppresses tumor growth by regulating the MAP4K4/C-Jun/MDM2 signaling pathway. Cell Cycle. 2015;14(7):1046–58.
Zhang N, Lu C, Chen L. Mir-217 regulates tumor growth and apoptosis by targeting the MAPK signaling pathway in colorectal cancer. Oncol Lett. 2016;12(6):4589–97.
Sun D, Wang C, Long S, Ma Y, Guo Y, Huang Z, Chen X, Zhang C, Chen J, Zhang J. C/EBP-Β-activated microrna-223 promotes tumour growth through targeting RASA1 in human colorectal cancer. Br J Cancer. 2015;112(9):1491–500.
Jiang M, Zhou L, Xu N, An Q. Hydroxysafflor yellow a inhibited lipopolysaccharide-induced non-small cell lung cancer cell proliferation, migration, and invasion by suppressing the PI3K/AKT/Mtor And ERK/MAPK signaling pathways. Thoracic Cancer. 2019;10(6):1319–33.
Zhao D, Sui Y, Zheng X. Mir-331-3P inhibits proliferation and promotes apoptosis by targeting HER2 through the PI3K/Akt And ERK1/2 pathways in colorectal cancer. Oncol Rep. 2015;35(2):1075–82.
Liu X, Wang Y, Zhao J. Microrna-337 inhibits colorectal cancer progression by directly targeting KRAS and suppressing the AKT and ERK pathways. Oncol Rep. 2017;38(5):3187–96.
Weng W, Leung W, Pang Y, Hsu H. Lauric acid can improve the sensitization of cetuximab in KRAS/BRAF mutated colorectal cancer cells by retrievable microrna-378 expression. Oncol Rep. 2015;35(1):107–16.
Chai J, Wang S, Han D, Dong W, Xie C, Guo H. Microrna-455 inhibits proliferation and invasion of colorectal cancer by targeting RAF proto-oncogene serine/threonine-protein kinase. Tumor Biol. 2014;36(2):1313–21.
Fan C, Lin Y, Mao Y, Huang Z, Liu A, Ma H, Yu D, Maitikabili A, Xiao H, Zhang C, Liu F, Luo Q, Ouyang G. Microrna-543 suppresses colorectal cancer growth and metastasis by targeting KRAS, MTA1 and HMGA2. Oncotarget. 2016;7(16):21825–39.
Fang Y, Sun B, Li Z, Chen Z, Xiang J. Mir-622 inhibited colorectal cancer occurrence and metastasis by suppressing K-ras. Mol Carcinog. 2015;55(9):1369–77.
Zhao J, Cao J, Zhou L, Du Y, Zhang X, Yang B, Gao Y, Wang Y, Ma N, Yang W. Mir-1260B inhibitor enhances the chemosensitivity of colorectal cancer cells to fluorouracil by targeting PDCD4/IGF1. Oncol Lett. 2018. https://doi.org/10.3892/ol.2018.9307.
Akamine T, Morodomi Y, Harada Y, et al. miR-3148 is a novel onco-microRNA that potentiates tumor growth in vivo. Antic- ancer Res. 2018;38:5693–701.
Jung B, Staudacher J, Beauchamp D. Transforming growth factor Β superfamily signaling in development of colorectal cancer. Gastroenterology. 2017;152(1):36–52.
Xu Y, Pasche B. TGF-Β signaling alterations and susceptibility to colorectal cancer. Hum Mol Genet. 2007;16(R1):R14–20.
Vychytilova-Faltejskova P, Slaby O. Pathophysiology roles and translational opportunities of mirnas in colorectal cancer. MicroRNA Human Malig. 2022. https://doi.org/10.1016/B978-0-12-822287-4.00007-4.
Xiao J, Lv D, Zhou J, Bei Y, Chen T, Hu M, Zhou Q, Fu S, Huang Q. Therapeutic inhibition of Mir-4260 suppresses colorectal cancer via targeting MCC and SMAD4. Theranostics. 2017;7(7):1901–13.
Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13(2):97–110.
Chen L, Gibbons D, Goswami S, Cortez M, Ahn Y, Byers L, Zhang X, Yi X, Dwyer D, Lin W, Diao L, Wang J, Roybal J, Patel M, Ungewiss C, Peng D, Antonia S, Mediavilla-Varela M, Robertson G, Jones S, Suraokar M, Welsh J, Erez B, Wistuba I, Chen L, Peng D, Wang S, Ullrich S, Heymach J, Kurie J, Qin F. Metastasis is regulated via microrna-200/ZEB1 axis control of tumour Cell PD-L1 expression and intratumoral immunosuppression. Nat Commun. 2014. https://doi.org/10.1038/ncomms6241.
Pecot C, Rupaimoole R, Yang D, Akbani R, Ivan C, Lu C, Wu S, Han H, Shah M, Rodriguez-Aguayo C, Bottsford-Miller J, Liu Y, Kim S, Unruh A, Gonzalez-Villasana V, Huang L, Zand B, Moreno-Smith M, Mangala L, Taylor M, Dalton H, Sehgal V, Wen Y, Kang Y, Baggerly K, Lee J, Ram P, Ravoori M, Kundra V, Zhang X, Ali-Fehmi R, Gonzalez-Angulo A, Massion P, Calin G, Lopez-Berestein G, Zhang W, Sood A. Tumour angiogenesis regulation by the Mir-200 family. Nat Commun. 2013. https://doi.org/10.1038/ncomms3427.
Toiyama Y, Hur K, Tanaka K, Inoue Y, Kusunoki M, Boland C, Goel A. Serum Mir-200C is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann Surg. 2014;259(4):735–43.
Hur K, Toiyama Y, Takahashi M, Balaguer F, Nagasaka T, Koike J, Hemmi H, Koi M, Boland C, Goel A. Microrna-200C modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2012;62(9):1315–26.
Rigoutsos I, Lee SK, Nam SY, Anfossi S, Pasculli B, Pichler M, Jing Y, Rodriguez-Aguayo C, Telonis AG, Rossi S, Ivan C, Ivkovic TC, Fabris L, Clark PM, Ling H, Shimizu M, Redis RS, Shah MY, Zhang X, Okugawa Y, Jung EJ, Tsirigos A, Huang L, Ferdin J, Gafà R, Spizzo R, Nicoloso MS, Paranjape AN, Shariati M, Tiron A, Yeh JJ, Teruel-Montoya R, Xiao L, Melo SA, Menter D, Jiang Z-Q, Flores ER, Negrini M, Goel A, Bar-Eli M, Mani SA, Liu CG, Lopez-Berestein G, Berindan-Neagoe I, Esteller M, Kopetz S, Lanza G, Calin GA. N-BLR, a primate-specific non-coding transcript leads to colorectal cancer invasion and migration. Genome Biol. 2017. https://doi.org/10.1172/JCI65508.
Zhang GJ, Xiao HX, Tian HP, Liu ZL, Xia SS, Zhou T. Upregulation of microRNA-155 promotes the migration and invasion of colorectal cancer cells through the regulation of claudin-1 expression. Int J Mol Med. 2013;31(6):1375–80.
Hahn S, Jackstadt R, Siemens H, Hünten S, Hermeking H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 2013;32(23):3079–95.
Kopan R, Ilagan M. The canonical notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–33.
Meurette O, Mehlen P. Notch signaling in the tumor microenvironment. Cancer Cell. 2018;34(4):536–48.
Maillard I, Pear W. Notch and cancer: best to avoid the ups and downs. Cancer Cell. 2003;3(3):203–5.
Ntziachristos P, Lim J, Sage J, Aifantis I. From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell. 2014;25(3):318–34.
Ramadan F, Fahs A, Ghayad S, Saab R. Signaling pathways in rhabdomyosarcoma invasion and metastasis. Cancer Metastasis Rev. 2020;39(1):287–301.
Huang B, Yang H, Cheng X, Wang D, Fu S, Shen W, Zhang Q, Zhang L, Xue Z, Li Y, Da Y, Yang Q, Li Z, Liu L, Qiao L, Kong Y, Yao Z, Zhao P, Li M, Zhang R. Trf/Mir-1280 suppresses stem cell-like cells and metastasis in colorectal cancer. Can Res. 2017;77(12):3194–206.
Niu L, Yang W, Duan L, Wang X, Li Y, Xu C, Liu C, Zhang Y, Zhou W, Liu J, Zhao Q, Hong L, Fan D. Biological implications and clinical potential of metastasis-related mirna in colorectal cancer. Molecular Therapy - Nucleic Acids. 2021;23:42–54.
Zhang X, Ai F, Li X, Tian L, Wang X, Shen S, Liu F. Microrna-34A suppresses colorectal cancer metastasis by regulating notch signaling. Oncol Lett. 2017;14(2):2325–33.
O’Brien J, Hayder H, Zayed Y, Peng C. Overview of microrna biogenesis, mechanisms of actions, and circulation. Front Endocrinol. 2018. https://doi.org/10.3389/fendo.2018.00402.
Oliveto S, Mancino M, Manfrini N, Biffo S. Role of micrornas in translation regulation and cancer. World J Biol Chem. 2017;8(1):45.
Si W, Shen J, Zheng H, Fan W. The role and mechanisms of action of micrornas in cancer drug resistance. Clin Epigenetics. 2019. https://doi.org/10.1186/s13148-018-0587-8.
Stark V, Facey C, Viswanathan V, Boman B. The role of mirnas, mirna clusters, and isomirs in development of cancer stem cell populations in colorectal cancer. Int J Mol Sci. 2021;22(3):1424.
Balzeau J, Menezes M, Cao S, Hagan J. The LIN28/Let-7 pathway in cancer. Front Genetics. 2017. https://doi.org/10.3389/fgene.2017.00031.
Chandra Boinpelly V, Verma R, Srivastav S, Srivastava R, Shankar S. Α-Mangostin-encapsulated PLGA nanoparticles inhibit colorectal cancer growth by inhibiting notch pathway. J Cell Mol Med. 2020;24(19):11343–54.
La X, Zhang L, Yang Y, Li H, Song G, Li Z. Tumor-secreted GRP78 facilitates the migration of macrophages into tumors by promoting cytoskeleton remodeling. Cell Signal. 2019;60:1–16.
Laganà A, Russo F, Sismeiro C, Giugno R, Pulvirenti A, Ferro A. Variability in the incidence of mirnas and genes in fragile sites and the role of repeats and cpg islands in the distribution of genetic material. PLoS ONE. 2010;5(6):e11166.
Lamy P, Andersen C, Dyrskjøt L, Tørring N, Ørntoft T, Wiuf C. Are micrornas located in genomic regions associated with cancer? Br J Cancer. 2006;95(10):1415–8.
Wynendaele J, Böhnke A, Leucci E, Nielsen SJ, Lambertz I, Hammer S, Sbrzesny N, Kubitza D, Wolf A, Gradhand E, Balschun K, Braicu I, Sehouli J, Darb-Esfahani S, Denkert C, Thomssen C, Hauptmann S, Lund A, Marine JC, Bartel F. An illegitimate microRNA target site within the 3’ UTR of MDM4 affects ovarian cancer progression and chemosensitivity. Can Res. 2010;70(23):9641–9.
Mishra P, Mishra P, Banerjee D, Bertino J. Mirsnps or mir-polymorphisms, new players in microrna mediated regulation of the cell: introducing microrna pharmacogenomics. Cell Cycle. 2008;7(7):853–8.
Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, Muller RU, Straka E, Su L, Burki EA, Crowell RE, Patel R, Kulkarni T, Homer R, Zelterman D, Kidd KK, Zhu Y, Christiani DC, Belinsky SA, Slack FJ, Weidhaas JB. A SNP in a let-7 microRNA complementary site in the KRAS 3’ untranslated region increases non-small cell lung cancer risk. Can Res. 2008;68(20):8535–40.
Brueckner B, Stresemann C, Kuner R, Mund C, Musch T, Meister M, Sültmann H, Lyko F. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Can Res. 2007;67(4):1419–23.
Lehmann U, Hasemeier B, Christgen M, Muller M, Romermann D, Langer F, et al. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J Pathol. 2008;214(1):17–24.
Nadal E, Chen G, Gallegos M, Lin L, Ferrer-Torres D, Truini A, Wang Z, Lin J, Reddy R, Llatjos R, Escobar I, Moya J, Chang A, Cardenal F, Capellà G, Beer D. Epigenetic inactivation of microrna-34B/C predicts poor disease-free survival in early-stage lung adenocarcinoma. Clin Cancer Res. 2013;19(24):6842–52.
Saito Y, Liang G, Egger G, Friedman J, Chuang J, Coetzee G, Jones P. Specific activation of microrna-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9(6):435–43.
Han L, Witmer P, Casey E, Valle D, Sukumar S. DNA methylation regulates microrna expression. Cancer Biol Ther. 2007;6(8):1290–4.
Lujambio A, Calin G, Villanueva A, Ropero S, Sánchez-Céspedes M, Blanco D, Montuenga L, Rossi S, Nicoloso M, Faller W, Gallagher W, Eccles S, Croce C, Esteller M. A microrna DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci. 2008;105(36):13556–61.
Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J, Prosper F, Garcia-Foncillas J. Epigenetic regulation of microrna expression in colorectal cancer. Int J Cancer. 2009;125(11):2737–43.
Reddy K. Microrna (Mirna) in cancer. Cancer Cell Intern. 2015;15(1):1–6.
Wiesen J, Tomasi T. Dicer is regulated by cellular stresses and interferons. Mol Immunol. 2009;46(6):1222–8.
Iliou M, da Silva-Diz V, Carmona F, Ramalho-Carvalho J, Heyn H, Villanueva A, Muñoz P, Esteller M. Impaired DICER1 function promotes stemness and metastasis in colon cancer. Oncogene. 2013;33(30):4003–15.
Vaksman O, Hetland T, Trope’ C, Reich R, Davidson B. Argonaute, dicer, and drosha are up-regulated along tumor progression in serous ovarian carcinoma. Hum Pathol. 2012;43(11):2062–9.
Shigeyasu K, Okugawa Y, Toden S, Boland C, Goel A. Exportin-5 functions as an oncogene and a potential therapeutic target in colorectal cancer. Clin Cancer Res. 2016;23(5):1312–22.
Pothoulakis C, Torre-Rojas M, Duran-Padilla M, Gevorkian J, Zoras O, Chrysos E, Chalkiadakis G, Baritaki S. CRHR2/Ucn2 signaling is a novel regulator of Mir-7/YY1/Fas circuitry contributing to reversal of colorectal cancer cell resistance to fas-mediated apoptosis. Int J Cancer. 2017;142(2):334–46.
Humphreys K, McKinnon R, Michael M. Mir-18A inhibits CDC42 and plays a tumour suppressor role in colorectal cancer cells. PLoS ONE. 2014;9(11):e112288.
Cruz-Gil S, Sanchez-Martinez R, Gomez de Cedron M, Martin-Hernandez R, Vargas T, Molina S, Herranz J, Davalos A, Reglero G, Ramirez de Molina A. Targeting the lipid metabolic axis ACSL/SCD in colorectal cancer progression by therapeutic mirnas: mir-19B-1 role. J Lipid Res. 2018;59(1):14–24.
Lu D, Tang L, Zhuang Y, Zhao P. Mir-17–3P regulates the proliferation and survival of colon cancer cells by targeting Par4. Mol Med Rep. 2017. https://doi.org/10.3892/mmr.2017.7863.
Chen B, Liu Y, Jin X, Lu W, Liu J, Xia Z, Yuan Q, Zhao X, Xu N, Liang S. Microrna-26A regulates glucose metabolism by direct targeting PDHX in colorectal cancer cells. BMC Cancer. 2014;14(1):1–10.
Jin H, Shi X, Zhao Y, Peng M, Kong Y, Qin D, Lv X. Microrna-30A mediates cell migration and invasion by targeting metadherin in colorectal cancer. Technol Cancer Res Treat. 2018;17:153303381875810.
Qin Y, Huo Z, Song X, Chen X, Tian X, Wang X. Mir-106A regulates cell proliferation and apoptosis of colon cancer cells through targeting the PTEN/PI3K/AKT signaling pathway. Oncol Lett. 2017. https://doi.org/10.3892/ol.2017.7715.
Liu J, Chen Z, Xiang J, Gu X. Microrna-155 acts as a tumor suppressor in colorectal cancer by targeting CTHRC1 In�vitro. Oncol Lett. 2018;15(4):5561–8.
Li J, Xia L, Zhou Z, Zuo Z, Xu C, Song H, Cai J. Mir-186-5P upregulation inhibits proliferation, metastasis and epithelial-to-mesenchymal transition of colorectal cancer cell by targeting ZEB1. Arch Biochem Biophys. 2018;640:53–60.
Zhao J, Zhang P, Wang X. YBX1 promotes tumor progression via the PI3K/AKT signaling pathway in laryngeal squamous cell carcinoma. Trans Cancer Res. 2021;10(11):4859–69.
Gulei D, Magdo L, Jurj A, Raduly L, Cojocneanu-Petric R, Moldovan A, Moldovan C, Florea A, Pasca S, Pop L, Moisoiu V, Budisan L, Pop-Bica C, Ciocan C, Buiga R, Muresan M, Stiufiuc R, Ionescu C, Berindan-Neagoe I. The silent healer: Mir-205–5P Up-regulation inhibits epithelial to mesenchymal transition in colon cancer cells by indirectly up-regulating E-cadherin expression. Cell Death & Disease. 2018;9(2):66.
Chen S, Wang Y, Su Y, Zhang L, Zhang M, Li X, Wang J, Zhang X. Mir-205–5P/PTK7 axis is involved in the proliferation, migration and invasion of colorectal cancer cells. Mol Med Rep. 2018;17(5):6253–60.
Hu J, He G, Lan X, Zeng Z, Guan J, Ding Y, Qian X, Liao W, Ding Y, Liang L. Inhibition Of ATG12-mediated autophagy by Mir-214 enhances radiosensitivity in colorectal cancer. Oncogenesis. 2018;7(2):16.
Wang D, Li Y, Zhang C, Li X, Yu J. Mir-216A-3P inhibits colorectal cancer cell proliferation through direct targeting COX-2 and ALOX5. J Cell Biochem. 2017;119(2):1755–66.
Liao D, Li T, Ye C, Zeng L, Li H, Pu X, Ding C, He Z, Huang G. Mir-221 inhibits autophagy and targets TP53INP1 in colorectal cancer cells. Exp Ther Med. 2017. https://doi.org/10.3892/etm.2017.5522.
Qu R, Hao S, Jin X, Shi G, Yu Q, Tong X, Guo D. Microrna-374B reduces the proliferation and invasion of colon cancer cells by regulation Of LRH-1/Wnt signaling. Gene. 2018;642:354–61.
Yan F, Tu Z, Duan L, Wang D, Lin F. Microrna-383 suppresses cell proliferation and invasion in colorectal cancer by directly targeting paired box 6. Mol Med Rep. 2018. https://doi.org/10.3892/mmr.2018.8682.
He S, Wang G, Ni J, Zhuang J, Zhuang S, Wang G, Ye Y, Xia W. Microrna-511 inhibits cellular proliferation and invasion in colorectal cancer by directly targeting hepatoma-derived growth factor. Oncol Res Featur Preclin Clin Cancer Ther. 2018;26(9):1355–63.
Li K, Fang Y, Liao J, Duan J, Feng L, Luo X, Liang Z. Upregulation of Mir-598 promotes cell proliferation and cell cycle progression in human colorectal carcinoma by suppressing INPP5E expression. Mol Med Rep. 2017;17:2991–7.
Shen J, Li M. Microrna-744 inhibits cellular proliferation and invasion of colorectal cancer by directly targeting oncogene notch1. Oncol Res Featur Preclin Clin Cancer Ther. 2018;26(9):1401–9.
Li J, Xu J, Yan X, Jin K, Li W, Zhang R. Suppression of capn4 by microrna-1271 impedes the proliferation and invasion of colorectal cancer cells. Biomed Pharmacother. 2018;99:162–8.
Li M, Qian X, Zhu M, Li A, Fang M, Zhu Y, Zhang J. Mir-1273G-3P promotes proliferation, migration and invasion of lovo cells via cannabinoid receptor 1 through activation Of ERBB4/PIK3R3/Mtor/S6K2 signaling pathway. Mol Med Rep. 2018;17:4619–26.
Bakhsh T, Alhazmi S, Alburae N, Farsi A, Alzahrani F, Choudhry H, Bahieldin A. Exosomal mirnas as a promising source of biomarkers in colorectal cancer progression. Int J Mol Sci. 2022;23(9):4855.
Nedaeinia R, Manian M, Jazayeri M, Ranjbar M, Salehi R, Sharifi M, Mohaghegh F, Goli M, Jahednia S, Avan A, Ghayour-Mobarhan M. Circulating exosomes and exosomal micrornas as biomarkers in gastrointestinal cancer. Cancer Gene Ther. 2016;24(2):48–56.
Ding L, Lan Z, Xiong X, Ao H, Feng Y, Gu H, Yu M, Cui Q. The dual role of micrornas in colorectal cancer progression. Int J Mol Sci. 2018;19(9):2791.
Vijayan M, Reddy P. Peripheral biomarkers of stroke: focus on circulatory micrornas. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2016;1862(10):1984–93.
Igaz I, Szőnyi M, Varga P, Topa L. Potential relevance of micrornas in the diagnostics of inflammatory bowel diseases. Orv Hetil. 2014;155(13):487–91.
Galamb O, Barták B, Kalmár A, Nagy Z, Szigeti K, Tulassay Z, Igaz P, Molnár B. Diagnostic and prognostic potential of tissue and circulating long non-coding rnas in colorectal tumors. World J Gastroenterol. 2019;25(34):5026–48.
Ahmed F, Ahmed F, Ahmed F, Gouda M, Gouda M, Ahmed N, Ahmed N, Hussein L. Quantification of micrornas by absolute dpcr for the diagnostic screening of colon cancer. J Colon Rectal Cancer. 2019;1(3):10–37.
He H, Wang N, Yi X, Tang C, Wang D. Low-level serum Mir-24-2 is associated with the progression of colorectal cancer. Cancer Biomark. 2018;21(2):261–7.
Chen X, Ma N, Zhou Z, Wang Z, Hu Q, Luo J, Mei X, Yang Z, Zhang L, Wang X, Feng Y, Yu X, Ma J, Guo X. Estrogen receptor mediates the radiosensitivity of triple-negative breast cancer cells. Med Sci Monit. 2017;23:2674–83.
Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka H, Boland C, Goel A. Circulating microrna-203 predicts prognosis and metastasis in human colorectal cancer. Gut. 2015;66(4):654–65.
Ng L, Wan T, Man J, Chow A, Iyer D, Chen G, Yau T, Lo O, Foo D, Poon J, Leung W, Pang R, Law W. Identification of serum Mir-139-3P as a non-invasive biomarker for colorectal cancer. Oncotarget. 2017;8(16):27393–400.
Li N, Ma J, Guarnera M, Fang H, Cai L, Jiang F. Digital PCR quantification of mirnas in sputum for diagnosis of lung cancer. J Cancer Res Clin Oncol. 2013;140(1):145–50.
Wang P, Jing F, Li G, Wu Z, Cheng Z, Zhang J, Zhang H, Jia C, Jin Q, Mao H, Zhao J. Absolute quantification of lung cancer related microrna by droplet digital PCR. Biosens Bioelectron. 2015;74:836–42.
Campomenosi P, Gini E, Noonan D, Poli A, D’Antona P, Rotolo N, Dominioni L, Imperatori A. A comparison between quantitative PCR and droplet digital PCR technologies for circulating microrna quantification in human lung cancer. BMC Biotechnology. 2016. https://doi.org/10.1186/s12896-016-0292-7.
Conte D, Verri C, Borzi C, Suatoni P, Pastorino U, Sozzi G, Fortunato O. Novel method to detect micrornas using chip-based quantstudio 3D digital PCR. BMC Genomics. 2015. https://doi.org/10.1186/s12864-015-2097-9.
Kjaer-Frifeldt S, Hansen T, Nielsen B, Joergensen S, Lindebjerg J, Soerensen F, dePont Christensen R, Jakobsen A. The prognostic importance Of Mir-21 in stage II colon cancer: a population-based study. Br J Cancer. 2012;107(7):1169–74.
Schee K, Boye K, Abrahamsen T, Fodstad Ø, Flatmark K. Clinical Relevance Of Microrna Mir-21, Mir-31, Mir-92A, Mir-101, Mir-106A And Mir-145 In Colorectal Cancer. BMC Cancer. 2012. https://doi.org/10.1186/1471-2407-12-505.
Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland C, Goel A. Serum Mir-21 as a diagnostic and prognostic biomarker in colorectal cancer. J National Cancer Inst. 2013;105(12):849–59.
Strubberg A, Madison B. Micrornas in the etiology of colorectal cancer: pathways and clinical implications. Dis Model Mech. 2017;10(3):197–214.
Oue N, Anami K, Schetter A, Moehler M, Okayama H, Khan M, Bowman E, Mueller A, Schad A, Shimomura M, Hinoi T, Aoyagi K, Sasaki H, Okajima M, Ohdan H, Galle P, Yasui W, Harris C. High Mir-21 expression from FFPE tissues is associated with poor survival and response to adjuvant chemotherapy in colon cancer. Int J Cancer. 2013;134(8):1926–34.
Rapado-González Ó, Álvarez-Castro A, López-López R, Iglesias-Canle J, Suárez-Cunqueiro M, Muinelo-Romay L. Circulating micrornas as promising biomarkers in colorectal cancer. Cancers. 2019;11(7):898.
Mody K, Bekaii-Saab T. Clinical trials and progress in metastatic colon cancer. Surg Oncol Clin. 2018;27(2):349–65.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tariq, L., Arafah, A., Sehar, N. et al. Novel insights on perils and promises of miRNA in understanding colon cancer metastasis and progression. Med Oncol 40, 282 (2023). https://doi.org/10.1007/s12032-023-02099-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-023-02099-2