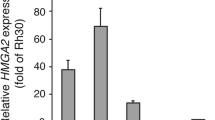
Abstract
Rhabdomyosarcoma (RMS) is an aggressive childhood mesenchymal tumor with two major molecular and histopathologic subtypes: fusion-positive (FP)RMS, characterized by the PAX3-FOXO1 fusion protein and largely of alveolar histology, and fusion-negative (FN)RMS, the majority of which exhibit embryonal tumor histology. Metastatic disease continues to be associated with poor overall survival despite intensive treatment strategies. Studies on RMS biology have provided some insight into autocrine as well as paracrine signaling pathways that contribute to invasion and metastatic propensity. Such pathways include those driven by the PAX3-FOXO1 fusion oncoprotein in FPRMS and signaling pathways such as IGF/RAS/MEK/ERK, PI3K/AKT/mTOR, cMET, FGFR4, and PDGFR in both FP and FNRMS. In addition, specific cytoskeletal proteins, G protein coupled receptors, Hedgehog, Notch, Wnt, Hippo, and p53 pathways play a role, as do specific microRNA. Paracrine factors, including secreted proteins and RMS-derived exosomes that carry cargo of protein and miRNA, have also recently emerged as potentially important players in RMS biology. This review summarizes the known factors contributing to RMS invasion and metastasis and their implications on identifying targets for treatment and a better understanding of metastatic RMS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Rhabdomyosarcoma clinical and molecular subtypes
Rhabdomyosarcoma (RMS) is the most common (~ 50%) soft tissue sarcoma in children and adolescents, with the head and neck region, genitourinary tract, and the extremities being the most common sites [1]. It is a malignant pediatric neoplasm characterized by early myogenic differentiation, implicating a skeletal myoblastic cell of origin [2]. Approximately 400 cases of RMS are diagnosed yearly within the USA with a slight male preponderance and an incidence rate of approximately 4.5 per million persons younger than 14 years of age [3]. While most RMS occurs sporadically, a minority is associated with specific genetic predisposing conditions, including Li-Fraumeni syndrome (germline TP53 mutation) [4], neurofibromatosis (NF1 gene mutation) [5], Noonan syndrome (mutations in PTPN11, KRAS, NRAS, SHOC2, RAF1, or SOS10) [6], and Beckwith Wiedemann syndrome (Chromosome 11p15 imprinting) [7], among others. Despite improvements in outcome over the past 4 decades due primarily to cooperative group clinical trials, patients with advanced stage and metastatic disease still fare poorly, with a 5-year overall survival of less than 30% [8].
The current World Health Organization (WHO) classification divides RMS into four major subtypes: embryonal (ERMS), alveolar (ARMS), spindle cell/sclerosing (SCRMS), and pleomorphic (PRMS), with ERMS and ARMS being the two most common RMS subtypes in children [9]. Figure 1 summarizes the major recurrent genetic abnormalities observed in ARMS and ERMS, respectively.
Rhabdomyosarcoma major histopathologic subtypes include embryonal (ERMS) and alveolar (ARMS) tumors, associated with distinct clinical and genomic characteristics. At the genomic level, the majority of ARMS cases harbor a chromosomal translocation involving primarily the PAX3 or PAX7 and the Forkhead box O1 (FOXO1) genes. Fusion-negative cases (FN) of histologic ARMS share clinical and genomic features of ERMS, such as loss of heterozygosity (LOH) at chromosome 11p15.5. ERMS is also characterized by specific recurrent chromosomal gains and losses, affecting cell cycle genes. In line with these findings, future classification of RMS will be based on fusion status, classifying rhabdomyosarcomas as either fusion positive (FPRMS) or fusion negative (FNRMS)
ERMS accounts for more than 60% of RMS cases in childhood [10, 11]. It has a relatively favorable prognosis, with 5-year survival rates approaching 70–80% with appropriate therapy. The presence of metastatic disease upon diagnosis is, however, associated with a worse outcome [12]. ERMS cells share morphological similarities to fetal muscle and are small round or spindle shaped with variable degrees of differentiation [11]. At the molecular level, ERMS is characterized by recurrent somatic mutations including loss of heterozygosity (LOH) at chromosome region 11p15.5, which harbors imprinted genes such as H19, insulin-like growth factor 2 (IGF2), and cyclin-dependent kinase inhibitor 1C (CDKN1C) [13,14,15]. Allelic loss at this region contributes to the loss of tumor suppressive functions of H19 and CDKN1C genes and the increased expression of oncogenes such as IGF2 and HRAS [16]. ERMS is also often associated with chromosomal gains (notably in chromosomes 2, 8, 12, and 13) or losses (notably in chromosomes 9 and 16) [17, 18]. Frequent mutations known to contribute to ERMS involve the proto-oncogenes encoding the RAS family of proteins [19, 20] and events leading to activation of the PI3K pathway [18, 21] and less common mutations in the tumor suppressor gene TP53 [22].
ARMS is less common than ERMS (20% of RMS cases), with histologic features of small round undifferentiated cells separated by fibrous septa. Up to 80% of ARMS are characterized by a chromosomal translocation involving the PAX3 or PAX7 gene, most commonly the t(2;13) translocation and a less common t(1;13) translocation [23]. Presence of the gene fusion in ARMS (ARMSp) is associated with worse prognosis and unfavorable outcome, whereas ARMS tumors that do not harbor the fusion gene (ARMSn) exhibit prognostic features and clinical outcomes similar to ERMS. Additionally, the aforementioned LOH at 11p15.5 and aneuploidy often associated with ERMS are also detected in ARMSn [24]. This has allowed a new molecular—rather than histopathologic—classification of RMS according to fusion status into fusion-positive (FPRMS) and fusion-negative (FNRMS) tumors [11]. Of note, some ARMS cases that lack the common PAX3/7-FOXO1 fusion gene may harbor alternative gene fusions [25]. Such genetic alterations include a translocation t(2;2)(q35;p23) which involves PAX3 and the gene coding for nuclear receptor coactivator NCOA1 or the translocation t(2;8)(q35;q13) generating PAX3-NCOA2 [26, 27]. Another rearrangement within the ARMS subtype results in the fusion protein PAX3-INO80D, with PAX-driven fusion gene-positive tumors having an expression profile consistent with the PAX3-FOXO1-positive ARMS cases or FPRMS [18]. Notably, an amplification of chromosome 12q13-q14 (with amplified CDK4 expression) has been associated with increased aggressiveness of FPRMS [28, 29].
Due to different primary sites of RMS, which may not even include skeletal muscle, there has been some debate regarding the cell of origin of RMS tumors. Some studies implicate mesenchymal stem cells, which gain activation of a myogenic transcription program, in addition to suggested cells of origin such as fetal myoblast cells or muscle side population or satellite cells, the latter being known to contribute to adult skeletal muscle regeneration [2, 30]. For instance, mutations in p53 [31], or p53 combined with Ptch1 gene silencing [32] in maturing myoblasts, contribute to ERMS development in mice. On the other hand, conditional expression of a mutant Smo allele showed endothelial progenitor cells to be responsible for RMS tumorigenesis [33], while another study showed that adipocytes with restricted Hedgehog (Hh) signaling lead to highly penetrant tumors with ERMS characteristics [34]. Together, these models demonstrate that specific genetic events in different cells of origin may lead to a common RMS histopathologic phenotype, activating a myogenic program that is interrupted by concomitant deregulation of other molecular pathways.
While localized RMS is associated with cure rates of up to 80% with current multimodality therapy, the outcome for patients who present with metastatic disease remains poor [1, 12]. Patients with metastatic disease, more than 3 sites of metastases, or either bone or bone marrow metastases, still fare extremely poorly with a 5-year survival rate of less than 10% [12]. Thus, identifying specific targeted approaches aimed at eliminating metastatic disease will be essential to improve outcomes for this subset of patients. Here, we summarize recent findings relating to the biology of metastasis in RMS and highlight areas of particular interest and promise in future therapeutic approaches.
2 PAX3-FOXO1 fusion oncogene as a driver of metastasis
The driver oncoprotein in FPRMS is due to a chromosomal translocation involving the fusion of the DNA binding domain of either PAX3 or PAX7 with the transactivation domain of FOXO1. Multiple studies have implicated the fusion oncoprotein in enhancing RMS metastasis, and this is clinically underlined by the fact that FPRMS is associated with higher rates of metastasis as well as increased propensity for bone and bone marrow metastases when compared to FNRMS [8, 35]. While both PAX3/7 and FOXO1 function as transcription factors, their resulting fusion oncoprotein has an altered function that augments transcription of target genes [36]. Several studies have attempted to characterize the downstream transcriptional pathways altered by PAX3-FOXO1. Expression studies in myoblasts have shown that PAX3-FOXO1 enhances invasion capacity and alters the transcription of around 70 mRNA and 27 miRNA targets [37]. Moreover, PAX-FOXO1 regulates the transcription of genes involved in cell proliferation and motility including c-Met, IGF-1, and CXCR4 [38,39,40]. Profiling of human tumor samples identified 171 differentially expressed genes in ARMS tumors compared to fetal skeletal muscle and 103 differentially expressed genes including RAC1, CCND1, and IGFBP2 between FP-ARMS and FN-ARMS [41]. In human tumor samples, various differentially regulated gene signatures have been characterized as possible markers to differentiate FPRMS from FNRMS cases [42, 43].
Downstream effectors regulated by PAX3-FOXO1 are thought to act by deregulating myogenic differentiation, enhancing cell proliferation, and promoting cell survival [36] (Fig. 2). Importantly, expression of PAX3-FOXO1 in mouse myoblasts, and also in ERMS cell lines, induces invasion, migration, and angiogenesis, further corroborating its suspected role in RMS cell metastasis [44, 45]. While the exact downstream effectors of PAX3-FOXO1 specifically contributing to distant metastases have yet to be characterized, they likely include paracrine factors and extracellular mediators in addition to the abovementioned pathways, some of which will be discussed in the ensuing sections of this review.
Contribution of the PAX3-FOXO1 fusion protein to RMS metastasis. Represented is the chromosomal translocation most commonly characterizing FPRMS, which involves the DNA binding domain of the PAX3 gene and the transactivation domain of FOXO1. The resulting PAX3-FOXO1 fusion oncoprotein has been implicated in FPRMS metastasis by regulating several signaling pathways involved in cellular invasion, proliferation, survival, motility, and differentiation, which in turn can promote RMS metastasis
3 Signaling pathways involved in RMS metastasis
Figure 3 depicts a summary of signaling molecules and pathways that have been implicated to date in RMS cell invasion and metastasis, which are discussed in detail in the sections below.
Signaling pathways involved in RMS metastasis. Dysregulation or amplification of signaling pathways involved in invasion and motility contribute to RMS metastasis in both FNRMS and FPRMS. From left to right: IL4R signaling and HGF binding to its c-MET receptor activate downstream JAK/STAT pathway and can stimulate MMP production. miR-874 inhibition of target protein GEFT is lost in RMS, leading to enhanced invasion and motility. IGF1 binding to its receptor can activate IGFBP2-mediated induction of MMP production and PI3K/Akt/mTOR signaling. miR-378a-3p reduces RMS metastasis by inhibiting IGF1R signaling but is downregulated in RMS. PDGFR signaling activates downstream MAPK, and PDGF binding to their receptor can also activate Ezrin which mediates RMS invasion through Rho-GTPase-mediated cytoskeletal remodeling. Hh signaling activates downstream NANOG which has been implicated in ERMS metastasis. Integrin α2β3 can activate NFκB signaling pathway, contributing to RMS growth and aggressiveness. miR-22 inhibits TACC1 and RABB5B which are involved in cell motility but is downregulated in RMS cells. GPCR surface expression is upregulated in RMS and can activate downstream signaling pathways including Hippo signaling as represented by activation of downstream YAP which then interacts with TEAD to regulate the expression of myogenic differentiation genes. Activated Notch receptor and its downstream targets Hes1 and Hey1 have been correlated with invasiveness of RMS cells. Abbreviations: IL4R interleukin-4 receptor, Jak Janus kinase, STAT signal transducer and activator of transcription, HGF hepatocyte growth factor, c-MET Met proto-oncogene receptor tyrosine kinase, IGF1R, insulin-like growth factor receptor 1, IGFPB2 insulin-like growth factor binding protein 2, MMP matrix metalloproteinase, IRS insulin receptor substrate, PI3K phosphoinositide-3 kinase, PDK1 phosphoinositide-dependent kinase-1, AKT serine/threonine kinase 1, mTOR mechanistic target of rapamycin, YB1 Y-box binding protein, PDGFR platelet-derived growth factor receptor, GRB2 growth factor receptor-bound protein 2, SOS son of sevenless, Ras guanosine-nucleotide-binding protein, Raf serine/threonine protein kinase, MEK mitogen-activated protein kinase kinase, ERK extracellular signal-regulated kinase. Ptch1 Patched 1, Smo smoothened homolog precursor, GLi glioma-associated oncogene, NANOG homobox protein, NFκB nuclear factor-kappa B, JNK c-Jun N-terminal kinase, GPCR G protein coupled receptor, RhoA Ras homolog family member A, MST mammalian sterile-twenty-like, YAP Yes-associated protein, TEAD transcriptional enhancer factor TEF, NICD Notch intracellular domain, GEFT Guanine nucleotide exchange factor T, TACC1 transforming acidic coiled-coil containing protein 1, RAB5B member of the RAS oncogene family
3.1 RTK signaling
The RAS/MEK/ERK and PI3K/AKT axes are commonly altered in RMS regardless of fusion status. Indeed, the most commonly occurring mutations in FNRMS involve the RAS and PIK3CA pathways. Common alterations in the RAS/MAPK and PI3K/AKT pathways exist in FNRMS and FPRMS [18]. While these pathways are necessary for RMS cell proliferation and survival, it is not clear whether they contribute directly to RMS invasion, angiogenesis, or metastatic ability. Notably, one study showed that caveolin-1 overexpression promotes migration and invasion of ERMS cells, which was reversed upon pharmacologic inhibition of ERK signaling, suggesting its essential contribution, at least to the caveolin-mediated invasive phenotype [46].
Importantly, the mTOR pathway is activated downstream of AKT, leading to the inactivation of eukaryotic initiation factor 4E (4E-BP1) and the activation of protein S6 kinase 1 (S6K1), which in turn enhances angiogenesis and HIF1α expression [47]. Several studies have shown that the mTOR pathway contributes to tumor invasion, and inhibition of mTOR signaling leads to reduced cellular migration, invasion, and angiogenesis in preclinical models of RMS [48, 49]. Indeed, combination therapies targeting both MEK/ERK and PI3K/mTOR pathways, or alternatively dual blockade of hedgehog (using Gli1/2 inhibitor) and PI3K/mTOR pathways, have been suggested as potentially synergistic in the treatment of RMS [50,51,52].
Another highly expressed receptor tyrosine kinase (RTK) within both RMS subtypes is the fibroblast growth factor receptor 4 (FGFR4) [53, 54]. Transcript levels of FGFR4 are increased in metastatic RMS tumors, and FGFR4 activating mutations at K535 and E550 lead to increased metastasis in a xenotransplanted human RMS model, while its knockdown reduces metastatic potential of RMS cells [55]. Concomitant with the fact that FGFR4 is a transcriptional target of PAX3 and PAX7, its expression is enhanced by the fusion oncoprotein leading to higher levels of expression in FPRMS [56]. Importantly, inhibitors of the PI3K/mTOR pathway may have efficacy in RMS tumors expressing mutant FGFR4, at least as demonstrated in preclinical models [57].
The insulin-like growth factors 1 (IGF1) and 2 (IGF2), along with their receptors (IGFR) and binding proteins (IGFBPs), have been associated with metastasis in various tumor types [58,59,60]. In patients with RMS, IGFBP2 plasma levels are elevated and correlate with metastatic stage, specifically in FNRMS. Gene silencing of IGFBP2 in vitro inhibited both RMS cell migration and invasion and decreased RMS lung metastases in a xenograft model [61]. IGF2 ligand overexpression is found in both FPRMS and FNRMS with loss of imprinting (LOI) leading to biallelic expression of IGF2 in FPRMS and LOH inducing an upregulation in FNRMS. In addition, IGF1R is upregulated in both subtypes [15, 42, 62]. Disappointingly, attempts at therapeutically targeting IGF1R signaling with small molecule inhibitors have been found to induce rapid resistance in part through the upregulation of alternative receptors such as platelet-derived growth factor receptor α (PDGFRα) [63].
c-MET is another RTK that is a physiological target of PAX3 and PAX7. In the case of FPRMS, knockdown of PAX3-FOXO1 via siRNA reduced MET mRNA and protein levels and decreased migration and invasion of Rh30 ARMS cells in vitro [64]. In mouse fibroblasts transduced with PAX3-FOXO1, Met expression was upregulated, leading to an anchorage-independence that required both MET and its ligand HGF/SF [65]. MET-mediated motility of ARMS cells was shown to be ERK2-dependent and ERK1- or mTOR-independent [66]. Although MET expression is upregulated by PAX3-FOXO1, its contribution to the metastatic behavior of RMS cells was evident in both ERMS and ARMS, and it is expressed in clinical tumors of both subtypes [67]. Inhibition of SPRY2, a protein found to interact with and stabilize MET, resulted in an inhibition of metastasis in both RMS subtypes via the subsequent inhibition of downstream ERK/MAPK signaling [68]. In vivo, ERMS cells expressing the constitutively active TPR-MET oncoprotein demonstrated an enhanced ability to migrate and metastasize, owing to an elevated production of VEGF and MMP-9 [69].
The anaplastic lymphoma kinase (ALK) is upregulated in RMS and to a larger extent in FPRMS [70, 71]. Its expression correlates with metastasis and poor prognosis, and in vitro siRNA silencing of ALK leads to decreased cellular invasion [72]. However, despite the effects seen in vitro, ALK inhibitors do not seem to affect ALK signaling in vivo [73], and clinical trials of ALK inhibition as monotherapy have failed to show significant responses in patients with RMS [74].
The platelet-derived growth factors PDGF-C and PDGF-D are also upregulated in RMS, as demonstrated through gene expression profiling of primary human tumor samples. Interestingly, elevated PDGF-D levels correlate with expression of genes involved in cell migration and wound healing, and stromal PDGFRβ, a PDGF-D receptor, is associated with ARMS tumors and positively correlates with metastasis [75]. Experimentally, inhibition of PDGF signaling leads to a decrease in rhabdosphere formation, stemness, and stromal cell infiltration, suggesting it plays a role in stromal remodeling [75].
Both FPRMS and FNRMS express high levels of the interleukin-4 receptor (IL4R), as well as high levels of its ligands IL4 and IL13. IL4R signaling activates downstream JAK/STAT, RAS/MAPK, and PI3K/AKT pathways and stimulates MMP production. Human ERMS cells treated with IL4 exhibit a decrease in expression of myogenic markers and an increase in migration ability. Furthermore, IL4R activation enhances propensity of metastasis into the lungs and lymph node involvement as demonstrated in an in vivo ARMS mouse model [76, 77].
3.2 Cytoskeletal proteins
Ezrin is a member of the ezrin/radixin/moesin (ERM) family of proteins, which regulate membrane-cytoskeleton interactions and are implicated in cytoskeletal remodeling through bidirectional activation with Rho GTPases [78]. Importantly, Ezrin is also a target of the c-Met RTK and is upregulated in highly metastatic murine RMS compared to non-metastatic cell lines, whereby the inhibition of Ezrin expression reduces metastasis [79]. Several mechanisms of action have been suggested to explain metastatic promotion by Ezrin, such as Ezrin-mediated activation of downstream MAPK and AKT signaling, and Ezrin-dependent HGC/c-MET associated metastasis [78]. Furthermore, the transcription factor Six-1, which controls migration during early myogenesis, leads to increased Ezrin expression and is associated with enhanced RMS metastasis. Importantly, both Ezrin and Six-1 are expressed in human RMS tissue, and expression correlates with advanced and metastatic stage of disease [79, 80].
Other cell surface proteins, such as VLA-2 (integrin α2β1), have been implicated in ERMS cell invasion. In the RD ERMS cell line, increased adhesion to collagen and laminin in vitro and enhanced formation of metastatic colonies in vivo were associated with VLA-2 expression, revealing the ability of surface integrins and adhesion molecules to favor RMS metastasis [81]. Mechanistically, Integrin α2β1 was found to respond to EGF stimulation which results in transient spreading of ERMS cells, through downstream activation of Rac1 intracellular signaling [82].
3.3 G protein and G protein-coupled receptors
The cannabinoid receptor-1 (CNR1), a G protein coupled receptor, is upregulated in FPRMS tumors and its expression is associated with increased metastatic potential and basement membrane invasion of FPRMS cells in vitro [42, 83]. In addition, using a murine model of metastatic FPRMS, pharmacological inhibition of CNR1 abrogated metastasis to the lungs, indicating it plays an important role in invasion and metastasis [83].
Rho/ROCK/ARHGAP25 signaling contributes to FPRMS cell invasion, as specific inhibition of ROCK2 limits invasion of ARMS cells. Since ROCK expression is similar in both ARMS and ERMS tissues, this differential effect was attributed to ROCK activation. Indeed, the upstream ROCK kinase inhibitor RhoE has low expression in FPRMS tumor tissue as compared to FNRMS tumors, and overexpression of RhoE reduces cellular invasion of FPRMS cells. Additionally, ARHGAP25, which inhibits Rac activity, is upregulated in FPRMS and its knockdown inhibits invasion. Thus, ROCK acts in FPRMS by upregulating ARHGAP25 and subsequently inhibiting Rac function, which subsequently promotes motility and cellular invasion [84].
Another G protein, Van Gogh-like 2 (VANGL2), is highly expressed in human RMS cells. It activates downstream signaling cascades involving Rac1 and RhoA proteins via protein-protein interactions and is a regulator of the non-canonical Wnt/PCP pathway. In RMS cells, VANGL2 is implicated in formation of rhabdomyospheres in vitro, and in xenograft growth and tumor-propagating cell maintenance in vivo, indicating a possible role for VANGL2/RhoA signaling in stemness in both FP and FNRMS [85].
3.4 Hedgehog signaling pathway
Hedgehog (Hh) signaling promotes tumor progression in a variety of cancer types [86, 87]. In RMS, Hh signaling is particularly associated with FNRMS. In fact, genes implicated in Hh signaling including Ptch1, Gli1, Gli3, and Myf5 are significantly upregulated, with a concomitant downregulation of MyoD1, in FNRMS compared to FPRMS [42, 88]. In turn, Hh activation increases ERMS self-renewal and invasive capacity, whereby Hh signaling inhibition reduces both phenotypes. Additionally, NANOG, which is also regulated by Hh signaling, has been demonstrated to act as a prognostic indicator particular to ERMS [89]. While SHH has low expression levels in ARMS cells, IHH and DHH genes exhibit high expression levels in all RMS cell lines which correlates with activation of Hh pathway pointing to a ligand-dependent activation of Hh signaling within RMS cells [90]. Indeed, heritable PTCH mutation as in Gorlin syndrome is associated with increased risk of RMS development [91]. Mice expressing low levels of normal PTCH develop tumors that resemble human RMS with differentially expressed genes including Secreted Phosphoprotein 1 (Spp1), matrix metalloproteinases (MMPs) such as Mmp-2 and Mmp-14, nuclear factor-kappa B (NF- κB), and the transcription factor p65 and its phosphorylated active isoform which have been linked to tumor cell invasion and angiogenesis [92].
3.5 Notch pathway
Activation of the Notch pathway involves the release of the Notch intracellular domain by gamma-secretase and subsequent transcriptional activation within the nucleus [93, 94]. Notch receptors are upregulated in both FPRMS and FNRMS; however, its downstream targets HES1 and HEY1 are significantly upregulated in FPRMS. Importantly, activation of the Notch pathway was correlated with degree of invasiveness and motility of RMS cell lines. Upon gamma-secretase inhibition, there was a significant decrease in RMS invasion and motility in vitro, indicating a possible role in mediating the invasive behavior of RMS cells [95].
Additionally, intracellular NOTCH1 (ICN1) was found to increase the number of tumor propagating cells in a zebrafish model of ERMS by upregulating the transcription repressor SNAIL1, causing a dedifferentiation of ERMS cells. This study implicates the NOTCH1/SNAIL1/MEF2C axis as a possible contributor to increased metastatic capacity of ERMS [96]. Of note, Notch inhibition leads to a downregulation of N-cadherin mRNA and protein levels. In turn, specific inhibition of N-cadherin through the use of antibodies reduces RMS invasion irrespective of histological/molecular subtype. Similarly, Notch-mediated regulation of α9-integrin expression regulates RMS adherence, motility, and invasion, indicating that both N-cadherin and α9-integrin may be implicated in RMS metastasis [97].
3.6 Wnt signaling pathway
Wnt signaling plays an important role in skeletal muscle differentiation, and studies suggest that canonical Wnt signaling is inactive in RMS [98, 99]. Silencing of the transcription factor c-fos promotes the development of RMS with highly invasive properties in mice harboring p53 deletion by inhibiting Wnt signaling [98]. Similarly, Wnt3a induces the differentiation of rhabdomyoblasts in ARMS, indicating a tumor suppressive role for Wnt/β-Catenin signaling in ARMS [100]. Additionally, activating canonical Wnt signaling by either GSK3 inhibition or Wnt3a-mediated stimulation suppresses ERMS growth mainly through inhibiting self-renewal capacity and depleting tumor propagating cells [101].
3.7 Hippo signaling pathway
The inhibition of MST/Hippo signaling in a genetically engineered FPRMS mouse model leads to enhanced invasion and proliferation of FPRMS tumors [102]. Yes-associated protein 1 (YAP1), a downstream effector of the Hippo pathway, has elevated expression levels in FNRMS. Interestingly, elevated YAP1 correlates with increased aggressive of FNRMS but not FPRMS. In mice, enhanced YAP1 expression in activated satellite cells contributes to ERMS induction and penetrance. YAP1 acts by interacting with TEAD1 and the subsequent regulation of MYOD1 transcriptional activity and myoblast differentiation [103].
3.8 The TP53 tumor suppressor protein
Mutation of TP53 leading to the loss of tumor suppressive function i has been described in FNRMS. Alternatively, loss of p53 activity in FNRMS may also be due to an increase in copy number of MDM2 gene [42]. However, the p53 pathway is also deregulated in FPRMS tumors, and TP53 mutation or loss of function is well established to be associated with poorer outcomes in RMS [104]. An ERMS zebrafish model harboring a loss-of-function mutation in tp53 (tp53del/del) revealed that while tp53 loss did not alter cancer cell stemness, tp53del/del ERMS exhibited more local invasiveness and metastatic propensity [105].
3.9 YB-1 transcription factor
Y-box binding protein 1 (YB-1) regulates transcription and mRNA expression and has been implicated in epithelial-mesenchymal transitions (EMT) which drives metastasis of cancer cells of epithelial origin [106, 107]. In sarcomas, YB-1 enhances tumor cell invasion by enhancing HIF1α protein expression. In fact, sarcomas including RMS show upregulated levels of YB-1, which correlate with HIF1α expression and with poorer patient outcomes. YB-1 expression leads to enhanced tumor cell migration, and inhibition of YB-1 decreases pulmonary metastasis in vivo [108].
3.10 Other proteins
In addition to the above, profiling gene expression experiments also identified upregulated genes in metastatic RMS tissue, showing high expression levels of two genes—FOXF1 and LMO4—which correlated with increased cellular migration. Decreasing their expression levels resulted in a 10-fold decrease in RMS invasiveness, underlining their implication in RMS invasive phenotype [109].
3.11 miRNA implicated in RMS invasion and metastasis
An observed downregulation of miR-874 expression in RMS tissues correlates with an increased expression of its downstream target protein GEFT. Experimentally, restoration of miR-874 expression inhibits migration and invasion of RMS cells. These cells also exhibit diminished wound-healing activity and reduced GEFT expression, whereas the restoration of GEFT reverses the miR-874-mediated inhibition of cellular invasion and motility [110].
Similarly, miR-22 is downregulated in RMS cells compared to normal muscle and its introduction into RMS cells inhibits adhesion-independent growth and migration in vitro as well as tumor dissemination in vivo. This is consistent with miR-22 function in targeting and inhibiting TACC1 and RAB5B expression, which are involved in RMS motility as well as ERBB3 expression [111]. In addition, downregulation of miR-378 family members is observed in RMS cell lines, and the ectopic expression of miR-378-3p in ARMS cells stimulates differentiation and reduces cell migration on one hand and cell proliferation by downregulating IGF1R/AKT signaling on the other. This indicates a tumor-suppressive function of these miRNAs in RMS and further implicates IGFR and AKT signaling in RMS invasion and metastasis [112].
4 Differentiation pathways and invasion potential in RMS
An interesting study utilizing a zebrafish model of RMS showed that mid-differentiated ERMS cells expressing myogenin were the first to intravasate the vasculature and exhibit enhanced migratory capacity. This is in contrast to myogenic factor 5 (Myf5)-expressing tumor cells, which are only able to invade vasculature after seeding by myogenin-positive cells [113]. While the relationship between level of differentiation and metastatic or invasive cellular ability is not yet well characterized at a mechanistic level, this observation points to the complex interplay between differentiation, cellular phenotype, and the acquisition of metastatic phenotype.
5 Paracrine signaling
Although much of what is known regarding RMS biology has focused on autocrine mechanisms of tumor progression, several studies have started to shed light on paracrine factors facilitating tumor invasion and distant metastatic spread. Effectors that have been investigated to date include secreted proteins and cargo-carrying exosomes that can act both locally and on distant sites. Figure 4 shows a schematic presentation of the different paracrine factors identified to date as possible mediators of RMS invasion, each of which is discussed in detail below.
Paracrine mediators of RMS invasion. Paracrine signaling pathways that contribute to RMS invasiveness and motility involve the release of proteins by RMS cells, including matrix metalloproteinases (MMPs) that degrade the extracellular matrix, interleukin-8 (IL-8) which enhances angiogenesis by acting on recipient endothelial cells, and tissue factor (TF) which stimulates platelet-derived microvesicle (PMVs) release. Microvesicles released by RMS cells include exosomes that can deliver their protein and miRNA cargo to both recipient rhabdomyosarcoma and stromal cells as well as distant organs via the bloodstream. RMS cells also express chemokine receptors such as CXCR4 and leukemia inhibitory factor receptor (LIFR) which respond to stimulation by SDF-1 and LIF respectively allowing their homing into distant organs. Sphingosine-1-phosphate (S1P) released by RMS cells subsequently enhances RMS invasion and metastasis through autocrine stimulation of RMS cells and paracrine stimulation of stromal cells
5.1 Secreted factors
Matrix metalloproteinases (MMPs) are proteolytic enzymes that degrade the extracellular matrix and contribute to cellular invasion, migration, and angiogenesis [114]. A higher level of MMP protein expression, namely MMP-1, MMP-2, and MMP-9, was detected in ARMS cell lines compared to ERMS, likely contributing to the metastatic propensity of ARMS cells. MMP expression may be regulated by the PAX3-FOXO1 fusion protein since the ectopic expression of the latter increases MMP production and RMS invasive capacity [45, 115].
Leukemia inhibitory factor (LIF) is a signaling cytokine that is secreted by bone marrow stromal cells, while its receptor, LIF-R, is expressed on both ARMS and ERMS cells, independent of PAX-FOXO1 expression. The LIF-LIFR access was found to mediate increased seeding of LIF-responding Rh30 ARMS cells to bone marrow, lymph nodes, and lung, while knockdown of LIF-R inhibited the phenotype, thus identifying the LIF-LIF-R axis as a possible regulator of RMS metastasis [116].
CXCR4, a chemokine receptor well known to be involved in tumor cell invasion, is also highly expressed in rhabdomyoblasts [40]. FPRMS cells express much higher levels compared to FNRMS, and ectopic expression of PAX3-FOXO1 enhances CXCR4 expression in the latter [40]. CXCR4 is known to respond to stimulation by SDF-1 chemokine which allows the homing of CXCR4+ cells into the bone marrow and lymph nodes [117]. Indeed, CXCR4–SDF-1 axis enhances FPRMS metastasis into the bone marrow, and the presence of SDF-1 promotes rhabdomyoblast migration and invasion, and also upregulates MMP expression [118]. On the other hand, in contrast to CXCR4 expression that is upregulated by the fusion protein in FPRMS cells, CXCR7 is expressed at higher levels within ERMS cell lines. CXCR7 responds to stimulation by both SDF1 and interferon-inducible T cell alpha chemoattractant (ITAC) chemokines leading to an activation of downstream signaling cascades involving the MAPK and AKT pathways. This in turn is associated with enhanced directed chemotaxis into the bone marrow and motility of RMS cells, but not tumor cell proliferation or survival [119].
Another secreted factor, IL-8, has been shown to enhance neovascularization in RMS tumors and contribute to its progression. IL-8 (CXCL8) expression increases under hypoxic conditions within RMS cells, which is dependent on AP-1 and NF-κB expression. IL-8 secreted by RMS cells acts through paracrine signaling on human umbilical vein endothelial cells (HUVECs), indicating an essential role in promoting angiogenesis under hypoxic conditions which is crucial for tumor metastasis [120].
The blood coagulation cascade and tissue factor (TF) release have been shown to enhance tumor metastasis and angiogenesis in a variety of cancer types [121,122,123]. Human RMS cells have been shown to express TF, which activates the coagulation cascade and thrombin formation. Thrombin, in turn, stimulates the release of circular membrane fragments called platelet-derived microvesicles (PMVs). PMVs enhance the metastasis of human RMS cells by stimulating AKT and MAPK p42/44 and transferring α2β3 integrin (CD41) to RMS cells, which enhances their adhesion to the endothelium. In fact, RMS cells covered with PMVs have an increased migratory potential into the bone marrow in vivo. TF-stimulated thrombin also acts at the level of blood vessels where it increases their permeability through the activation of complement proteins further enhancing tumor metastasis [124].
Another secreted protein, sphingosine-1-phosphate (S1P), regulates various processes implicated in cancer malignancy including tumor cell migration and metastasis. It is secreted into the extracellular fluid where it acts through autocrine/paracrine signaling by activating G protein coupled receptors and various downstream signaling pathways involved in angiogenesis, cytoskeletal remodeling, migration, and invasion [125]. S1P enhances the motility and invasiveness of RMS cells, and S1P inhibition reduces their metastatic ability in vivo [126]. Of note, S1P activity is suppressed by the endoplasmic reticulum (ER) membrane localized enzyme sphingosine-1-phosphate lyase (SGPL1), which acts to irreversibly cleave S1P. In some ARMS cells, SGPL1 has been shown to be overexpressed but mis-localized, due to a point mutation which interferes in its S1P degradation activity and thereby indirectly enhances metastatic potential through increased S1P activity [127].
5.2 Exosomes
Exosomes are extracellular nanovesicles that can transfer their cargo which includes lipids, proteins, and nucleic acids (coding and non-coding) from the parent cell responsible for their release to recipient cells [128]. Exosomes induce functional and behavioral changes in the target cell and are implicated in apoptosis, inflammation, and the release of angiogenic factors, cytokines, and growth factors that influence recipient cell activity [129]. In cancer, exosomes are released in higher quantities by cancer cells and have been demonstrated to participate in tumorigenesis by modulating the immune response, increasing invasive and migratory properties of recipient cells and establishing a metastatic niche by carrying and releasing oncogenic proteins and nucleic acids [130].
Expression array analysis of RMS cell-derived exosomes revealed that the miRNA content of these exosomes differed from their cellular content, with certain miRNAs being selectively enriched in the exosomes compared to the parent cell. Comparing miRNA content of exosomes from FPRMS and FNRMS cell lines, 31 miRNAs were found to be commonly deregulated, 10 of which are enriched in exosomes derived from either subtype [131]. Two commonly enriched exosomal miRNAs derived from all tested RMS cell lines, miR-1246 and miR-1268, are known to play a role in Wnt, Cadherin/Integrin, and p53/Ras signaling pathways, indicating that cargo delivery could regulate proliferation, angiogenesis and metastasis. Experimentally, RMS-derived exosomes from either subtype were found to increase the migration and invasion of normal recipient fibroblasts and endothelial cells both in vitro and in vivo in a dose-dependent manner, thus underlining their putative contribution to the metastatic process [131].
The difference in miRNA enriched within FNRMS exosomes compared to FPRMS exosomes suggests that paracrine signaling via exosomes may have different effects in the two subtypes, with specific pathways contributing to the aggressiveness of the fusion-positive subtype. In fact, certain miRNAs enriched in FPRMS-derived exosomes but not FNRMS-derived exosomes were represented in networks involving proteins such as CXCL8, YBX1, CCR2, and IGF1R that are implicated in tumor metastasis and stemness [131].
One of the miRNAs differentially expressed in exosomes, miR-486-5p, is a downstream target of the PAX3-FOXO1 fusion oncoprotein and indeed exhibits higher levels of expression in FPRMS-derived exosomes [132, 133]. Ectopic expression of the fusion oncoprotein PAX3-FOXO1 in C2C12 mouse myoblasts results in an increase in miR-486-5p expression level in both cells and exosomes, and treatment of recipient fibroblasts with FPRMS-derived exosomes increases cell migration and invasion in a miR-486-5p expression-dependent manner [133].
Proteomic profiling of RMS-derived exosomes revealed 81 common proteins between ERMS and ARMS exosomes, all of which have paracrine-signaling related functions involved in cancer growth and metastasis [134]. For instance, of the proteins common within the exosomes from both FPRMS and FNRMS cell lines, Annexin A2 (ANX A2) has been associated with metastatic behavior of different cancer types [134,135,136]. Additionally, common exosomal proteins within all tested RMS cell lines include those involved in integrin signaling pathway such as ACTB, CD147, FN1, and ITGB1, further suggesting a role in RMS metastasis [134]. Previous work in breast and pancreatic cell lines demonstrated that exosomal integrins determine sites of metastasis, for example, α6β4 and α6β1 integrins within exosomes were specifically associated with lung metastasis [137]. Whether exosome cargo of RMS tumors also contributes to metastatic organotropism has yet to be investigated. Of note, combined network analysis of the commonly enriched miRNAs and proteins within RMS-derived exosomes revealed a potential role for Vimentin signaling [134], which has been demonstrated to contribute to cancer metastasis in other tumor types [138, 139].
Notably, of the proteins found to be specific to ARMS exosomes, several were related to DNA replication and FAS signaling pathways, while those specific to ERMS exosomes were involved in TGF-beta, RAS, PI3K, IGF, and MAPK signaling pathways, further suggesting that different paracrine signaling mechanisms exist between the two subtypes [134]. How those proteins affect recipient stromal cells is an area of active current investigation, especially the effects on, and contribution of, recipient stromal and vascular cells in promoting tumor invasion, angiogenesis, distant seeding, and tumor growth.
6 Implications and future directions
Knowing that failure of current treatment of RMS is primarily due to either inadequacy of treatment of metastatic disease or to local invasion of tumor precluding effective local control, identification and targeting of pathways responsible for RMS invasion and metastasis are critical for therapeutic advancements. Recent clinical trials have evaluated a few of the identified targets that have been discussed in this review, with limited effects. For example, a small-molecule inhibitor of IGF1R revealed RMS sensitivity to this pathway inhibition; however, cells exhibited pathways of resistance including an EGFR-dependent mechanism of survival [140]. Promising immunotherapies are also being studied, such as antibody inhibition of CXCR4 which has been shown in preclinical in vitro studies to have some efficacy against migration and invasion of ARMS Rh30 cells. Activated and expanded natural killer (NKAE) cell therapy, another novel immunotherapy, completely suppressed ARMS implantation in vivo, and a combination of both of these treatments completely suppressed metastasis in mice [141].
Targeting fusion oncoprotein expression with methods such as siRNA-mediated gene silencing has also been proposed, where siRNA can be delivered via liposome-protamine-siRNA particles (LRP). While fusion oncoprotein expression was in fact reduced, more efficient methods of PAX3-FOXO1 targeting are needed [142]. Further work to verify the discussed pathways as mediators of metastasis will be essential for targeted therapies against metastatic disease. Studies combining inhibition of PI3K/AKT pathway with inhibitors of other proteins that promote RMS metastasis (e.g., IGF1R or mTOR) are ongoing, with promising preclinical results showing synergism [143, 144].
Efforts should also be made to detect biomarkers that are predictive of RMS metastasis and response to treatment; this would allow a more accurate prognosis of disease. One such promising biomarker may be miR-486-5p, where in a limited number of samples, its levels seemed to be higher in exosomes derived from FPRMS patient serum, deserving further validation in larger sample sizes [133]. Additionally, proteins that are commonly expressed within FPRMS- and FNRMS-derived exosomes and have been implicated in cancer metastasis, such as CD147, are interesting candidates for further evaluation as possible therapeutic targets [145,146,147].
References
Sultan, I., Qaddoumi, I., Yaser, S., Rodriguez-Galindo, C., & Ferrari, A. (2009). Comparing adult and pediatric rhabdomyosarcoma in the surveillance, epidemiology and end results program, 1973 to 2005: an analysis of 2,600 patients. Journal of Clinical Oncology. https://doi.org/10.1200/JCO.2008.19.7483.
Saab, R., Spunt, S. L., & Skapek, S. X. (2011). Myogenesis and rhabdomyosarcoma. In Current topics in developmental biology (Vol. 94, pp. 197–234). doi:https://doi.org/10.1016/B978-0-12-380916-2.00007-3.
Ries, L. A. G., Smith, M. A., Gurney, J. G., Linet, M., Tamra, T., Young, J. L., & Bunin, G. R. (1999). Cancer incidence and survival among children and adolescents: United States SEER Program 1975-1995. NIH Pub. No. 99-4649, 179 pp.
Mai, P. L., Best, A. F., Peters, J. A., DeCastro, R. M., Khincha, P. P., Loud, J. T., et al. (2016). Risks of first and subsequent cancers among TP53 mutation carriers in the National Cancer Institute Li-Fraumeni syndrome cohort. Cancer. https://doi.org/10.1002/cncr.30248.
Sung, L., Anderson, J. R., Arndt, C., Raney, R. B., Meyer, W. H., & Pappo, A. S. (2004). Neurofibromatosis in children with rhabdomyosarcoma: a report from the intergroup rhabdomyosarcoma study IV. The Journal of Pediatrics. https://doi.org/10.1016/j.jpeds.2004.02.026.
Jongmans, M. C. J., Van Der Burgt, I., Hoogerbrugge, P. M., Noordam, K., Yntema, H. G., Nillesen, W. M., et al. (2011). Cancer risk in patients with Noonan syndrome carrying a PTPN11 mutation. European Journal of Human Genetics, 19, 870–874. https://doi.org/10.1038/ejhg.2011.37.
Smith, A. C., Squire, J. A., Thorner, P., Zielenska, M., Shuman, C., Grant, R., Chitayat, D., Nishikawa, J. L., & Weksberg, R. (2001). Association of alveolar rhabdomyosarcoma with the Beckwith-Wiedemann syndrome. Pediatric and Developmental Pathology, 4, 550–558. https://doi.org/10.1007/s10024001-0110-6.
Breneman, J. C., Lyden, E., Pappo, A. S., Link, M. P., Anderson, J. R., Parham, D. M., et al. (2003). Prognostic factors and clinical outcomes in children and adolescents with metastatic rhabdomyosarcoma-a report from the intergroup rhabdomyosarcoma study IV. Journal of Clinical Oncology. https://doi.org/10.1200/JCO.2003.06.129.
Rudzinski, E. R., Anderson, J. R., Hawkins, D. S., Skapek, S. X., Parham, D. M., & Teot, L. A. (2015). The world health organization classification of skeletal muscle tumors in pediatric rhabdomyosarcoma a report from the children’s oncology group. Archives of Pathology & Laboratory Medicine. https://doi.org/10.5858/arpa.2014-0475-OA.
Ognjanovic, S., Linabery, A. M., Charbonneau, B., & Ross, J. A. (2009). Trends in childhood rhabdomyosarcoma incidence and survival in the United States, 1975-2005. Cancer. https://doi.org/10.1002/cncr.24465.
Parham, D. M., & Barr, F. G. (2013). Classification of Rhabdomyosarcoma and its molecular basis. Advances in Anatomic Pathology. https://doi.org/10.1097/PAP.0b013e3182a92d0d.
Oberlin, O., Rey, A., Lyden, E., Bisogno, G., Stevens, M. C. G., Meyer, W. H., et al. (2008). Prognostic factors in metastatic rhabdomyosarcomas: results of a pooled analysis from United States and European Cooperative Groups. Journal of Clinical Oncology. https://doi.org/10.1200/JCO.2007.14.7207.
Davicioni, E., Anderson, M. J., Finckenstein, F. G., Lynch, J. C., Qualman, S. J., Shimada, H., et al. (2009). Molecular classification of rhabdomyosarcoma - genotypic and phenotypic determinants of diagnosis: a report from the Children’s Oncology Group. The American Journal of Pathology. https://doi.org/10.2353/ajpath.2009.080631.
Scrable, H., Witte, D., Shimada, H., Seemayer, T., Wang-Wuu, S., Soukup, S., et al. (1989). Molecular differential pathology of rhabdomyosarcoma. Genes, Chromosomes & Cancer. https://doi.org/10.1002/gcc.2870010106.
Anderson, J., Gordon, A., McManus, A., Shipley, J., & Pritchard-Jones, K. (1999). Disruption of imprinted genes at chromosome region 11p15.5 in paediatric rhabdomyosarcoma. Neoplasia, 1(4), 340–348. https://doi.org/10.1038/sj.neo.7900052.
Anderson, J., Gordon, A., Pritchard-Jones, K., & Shipley, J. (1999). Genes, chromosomes, and rhabdomyosarcoma. Genes, Chromosomes & Cancer. https://doi.org/10.1002/(SICI)1098-2264(199912)26:4<275::AID-GCC1>3.0.CO;2-3.
Weber-Hall, S., Anderson, J., McManus, A., Abe, S., Nojima, T., Pinkerton, R., et al. (1996). Gains, losses, and amplification of genomic material in rhabdomyosarcoma analyzed by comparative genomic hybridization. Cancer Research, 56(14), 3220–3224.
Shern, J. F., Chen, L., Chmielecki, J., Wei, J. S., Patidar, R., Rosenberg, M., et al. (2014). Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discovery. https://doi.org/10.1158/2159-8290.CD-13-0639.
Stratton, M. R., Cooper, C. S., Gusterson, B. A., & Fisher, C. (1989). Detection of point mutations in N-ras and K-ras genes of human embryonal rhabdomyosarcomas using oligonucleotide probes and the polymerase chain reaction. Cancer Research, 49(22), 6324–6327.
Martinelli, S., McDowell, H. P., Delle Vigne, S., Kokai, G., Uccini, S., Tartaglia, M., & Dominici, C. (2009). RAS signaling dysregulation in human embryonal rhabdomyosarcoma. Genes, Chromosomes & Cancer. https://doi.org/10.1002/gcc.20702.
Petricoin, E. F., Espina, V., Araujo, R. P., Midura, B., Yeung, C., Wan, X., et al. (2007). Phosphoprotein pathway mapping: Akt/mammalian target of rapamycin activation is negatively associated with childhood rhabdomyosarcoma survival. Cancer Research. https://doi.org/10.1158/0008-5472.CAN-06-1344.
Mora, J., Dobrenis, A. M., Bussel, J. B., & Aledo, A. (2000). p53 mutation and MDM2 amplification frequency pediatric rhabdomyosarcoma tumors and cell lines. Medical and Pediatric Oncology. https://doi.org/10.1002/1096-911X(200008)35:2<96::AID-MPO2>3.0.CO;2-Z.
Olanich, M. E., & Barr, F. G. (2013). A call to ARMS: targeting the PAX3-FOXO1 gene in alveolar rhabdomyosarcoma. Expert Opinion on Therapeutic Targets. https://doi.org/10.1517/14728222.2013.772136.
Williamson, D., Missiaglia, E., De Reyniès, A., Pierron, G., Thuille, B., Palenzuela, G., et al. (2010). Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. Journal of Clinical Oncology. https://doi.org/10.1200/JCO.2009.26.3814.
Barr, F. G., Qualman, S. J., Macris, M. H., Melnyk, N., Lawlor, E. R., Strzelecki, D. M., Triche, T. J., Bridge, J. A., & Sorensen, P. H. B. (2002). Genetic heterogeneity in the alveolar rhabdomyosarcoma subset without typical gene fusions. Cancer Research, 62(16), 4704–4710.
Wachtel, M., Dettling, M., Koscielniak, E., Stegmaier, S., Treuner, J., Simon-Klingenstein, K., et al. (2004). Gene expression signatures identify rhabdomyosarcoma subtypes and detect a novel t(2;2)(q35;p23) translocation fusing PAX3 to NCOA1. Cancer Research. https://doi.org/10.1158/0008-5472.CAN-04-0844.
Sumegi, J., Streblow, R., Frayer, R. W., Cin, P. D., Rosenberg, A., Meloni-Ehrig, A., & Bridge, J. A. (2010). Recurrent t(2;2) and t(2;8) translocations in rhabdomyosarcoma without the canonical PAX-FOXO1 fuse PAX3 to members of the nuclear receptor transcriptional coactivator family. Genes, Chromosomes & Cancer. https://doi.org/10.1002/gcc.20731.
Park, S., Lee, J., Do, I. G., Jang, J., Rho, K., Ahn, S., Maruja, L., Kim, S. J., Kim, K. M., Mao, M., Oh, E., Kim, Y. J., Kim, J., & Choi, Y. L. (2014). Aberrant CDK4 amplification in refractory rhabdomyosarcoma as identified by genomic profiling. Scientific Reports, 4, 1–8. https://doi.org/10.1038/srep03623.
Barr, F. G., Duan, F., Smith, L. M., Gustafson, D., Pitts, M., Hammond, S., & Gastier-Foster, J. M. (2009). Genomic and clinical analyses of 2p24 and 12q13-q14 amplification in alveolar rhabdomyosarcoma: a report from the Children’s Oncology Group. Genes, Chromosomes & Cancer. https://doi.org/10.1002/gcc.20673.
Charytonowicz, E., Cordon-Cardo, C., Matushansky, I., & Ziman, M. (2009). Alveolar rhabdomyosarcoma: is the cell of origin a mesenchymal stem cell? Cancer Letters. https://doi.org/10.1016/j.canlet.2008.09.039.
Preussner, J., Zhong, J., Sreenivasan, K., Günther, S., Engleitner, T., Künne, C., et al. (2018). Oncogenic amplification of zygotic dux factors in regenerating p53-deficient muscle stem cells defines a molecular cancer subtype. Cell Stem Cell. https://doi.org/10.1016/j.stem.2018.10.011.
Rubin, B. P., Nishijo, K., Chen, H. I. H., Yi, X., Schuetze, D. P., Pal, R., et al. (2011). Evidence for an unanticipated relationship between undifferentiated pleomorphic sarcoma and embryonal rhabdomyosarcoma. Cancer Cell. https://doi.org/10.1016/j.ccr.2010.12.023.
Drummond, C. J., Hanna, J. A., Garcia, M. R., Devine, D. J., Heyrana, A. J., Finkelstein, D., et al. (2018). Hedgehog pathway drives fusion-negative rhabdomyosarcoma initiated from non-myogenic endothelial progenitors. Cancer Cell. https://doi.org/10.1016/j.ccell.2017.12.001.
Hatley, M. E., Tang, W., Garcia, M. R., Finkelstein, D., Millay, D. P., Liu, N., et al. (2012). A mouse model of rhabdomyosarcoma originating from the adipocyte lineage. Cancer Cell. https://doi.org/10.1016/j.ccr.2012.09.004.
Barr, F. G. (2001). Gene fusions involving PAX and FOX family members in alveolar rhabdomyosarcoma. Oncogene. https://doi.org/10.1038/sj.onc.1204599.
Linardic, C. M. (2008). PAX3-FOXO1 fusion gene in rhabdomyosarcoma. Cancer Letters. https://doi.org/10.1016/j.canlet.2008.03.035.
Loupe, J. M., Miller, P. J., Bonner, B. P., Maggi, E. C., Vijayaraghavan, J., Crabtree, J. S., & Hollenbach, A. D. (2016). Comparative transcriptomic analysis reveals the oncogenic fusion protein pax3-foxo1 globally alters mRNA and miRNA to enhance myoblast invasion. Oncogenesis., 5. https://doi.org/10.1038/oncsis.2016.53.
Epstein, J. A., Shapiro, D. N., Cheng, J., Lam, P. Y. P., & Maas, R. L. (1996). Pax3 modulates expression of the c-met receptor during limb muscle development. Proceedings of the National Academy of Sciences of the United States of America. https://doi.org/10.1073/pnas.93.9.4213.
Ayalon, D., Glaser, T., & Werner, H. (2001). Transcriptional regulation of IGF-I receptor gene expression by the PAX3-FKHR oncoprotein. Growth Hormone & IGF Research. https://doi.org/10.1054/ghir.2001.0244.
Tomescu, O., Xia, S. J., Strezlecki, D., Bennicelli, J. L., Ginsberg, J., Pawel, B., & Barr, F. G. (2004). Inducible short-term and stable long-term cell culture systems reveal that the PAX3-FKHR fusion oncoprotein regulates CXCR4, PAX3, and PAX7 expression. Laboratory Investigation, 84, 1060–1070. https://doi.org/10.1038/labinvest.3700125.
De Pittà, C., Tombolan, L., Albiero, G., Sartori, F., Romualdi, C., Jurman, G., et al. (2006). Gene expression profiling identifies potential relevant genes in alveolar rhabdomyosarcoma pathogenesis and discriminates PAX3-FKHR positive and negative tumors. International Journal of Cancer. https://doi.org/10.1002/ijc.21698.
Laé, M., Ahn, E. H., Mercado, G. E., Chuai, S., Edgar, M., Pawel, B. R., et al. (2007). Global gene expression profiling of PAX-FKHR fusion-positive alveolar and PAX-FKHR fusion-negative embryonal rhabdomyosarcomas. The Journal of Pathology. https://doi.org/10.1002/path.2170.
Davicioni, E., Finckenstein, F. G., Shahbazian, V., Buckley, J. D., Triche, T. J., & Anderson, M. J. (2006). Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Research. https://doi.org/10.1158/0008-5472.CAN-05-4578.
Anderson, J., Ramsay, A., Gould, S., & Pritchard-Jones, K. (2001). PAX3-FKHR induces morphological change and enhances cellular proliferation and invasion in rhabdomyosarcoma. The American Journal of Pathology. https://doi.org/10.1016/S0002-9440(10)61784-1.
Onisto, M., Slongo, M. L., Gregnanin, L., Gastaldi, T., Carli, M., & Rosolen, A. (2005). Expression and activity of vascular endothelial growth factor and metalloproteinases in alveolar and embryonal rhabdomyosarcoma cell lines. International Journal of Oncology.
Codenotti, S., Faggi, F., Ronca, R., Chiodelli, P., Grillo, E., Guescini, M., et al. (2019). Caveolin-1 enhances metastasis formation in a human model of embryonal rhabdomyosarcoma through Erk signaling cooperation. Cancer Letters. https://doi.org/10.1016/j.canlet.2019.02.013.
Martin, D. E., & Hall, M. N. (2005). The expanding TOR signaling network. Current Opinion in Cell Biology. https://doi.org/10.1016/j.ceb.2005.02.008.
Kaylani, S. Z., Xu, J., Srivastava, R. K., Kopelovich, L., Pressey, J. G., & Athar, M. (2013). Rapamycin targeting mTOR and hedgehog signaling pathways blocks human rhabdomyosarcoma growth in xenograft murine model. Biochemical and Biophysical Research Communications. https://doi.org/10.1016/j.bbrc.2013.05.001.
Wan, X., Shen, N., Mendoza, A., Khanna, C., & Helman, L. J. (2006). CCI-779 inhibits rhabdomyosarcoma xenograft growth by an antiangiogenic mechanism linked to the targeting of mTOR/Hif-1α/VEGF signaling. Neoplasia. https://doi.org/10.1593/neo.05820.
Guenther, M. K., Graab, U., & Fulda, S. (2013). Synthetic lethal interaction between PI3K/Akt/mTOR and Ras/MEK/ERK pathway inhibition in rhabdomyosarcoma. Cancer Letters. https://doi.org/10.1016/j.canlet.2013.05.010.
Renshaw, J., Taylor, K. R., Bishop, R., Valenti, M., De Haven Brandon, A., Gowan, S., et al. (2013). Dual blockade of the PI3K/AKT/mTOR (AZD8055) and RAS/MEK/ERK (AZD6244) pathways synergistically inhibits rhabdomyosarcoma cell growth in vitro and in vivo. Clinical Cancer Research. https://doi.org/10.1158/1078-0432.CCR-13-0850.
Graab, U., Hahn, H., & Fulda, S. (2015). Identification of a novel synthetic lethality of combined inhibition of hedgehog and PI3K signaling in rhabdomyosarcoma. Oncotarget. https://doi.org/10.18632/oncotarget.2726.
Baird, K., Davis, S., Antonescu, C. R., Harper, U. L., Walker, R. L., Chen, Y., et al. (2005). Gene expression profiling of human sarcomas: insights into sarcoma biology. Cancer Research. https://doi.org/10.1158/0008-5472.CAN-05-1699.
Khan, J., Wei, J. S., Ringnér, M., Saal, L. H., Ladanyi, M., Westermann, F., et al. (2001). Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nature Medicine. https://doi.org/10.1038/89044.
Taylor IV, J. G., Cheuk, A. T., Tsang, P. S., Chung, J. Y., Song, Y. K., Desai, K., et al. (2009). Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. The Journal of Clinical Investigation. https://doi.org/10.1172/JCI39703.
Crose, L. E. S., Etheridge, K. T., Chen, C., Belyea, B., Talbot, L. J., Bentley, R. C., & Linardic, C. M. (2012). FGFR4 blockade exerts distinct antitumorigenic effects in human embryonal versus alveolar rhabdomyosarcoma. Clinical Cancer Research. https://doi.org/10.1158/1078-0432.CCR-10-3063.
McKinnon, T., Venier, R., Yohe, M., Sindiri, S., Gryder, B. E., Shern, J. F., Kabaroff, L., Dickson, B., Schleicher, K., Chouinard-Pelletier, G., Menezes, S., Gupta, A., Zhang, X., Guha, R., Ferrer, M., Thomas, C. J., Wei, Y., Davani, D., Guidos, C. J., Khan, J., & Gladdy, R. A. (2018). Functional screening of FGFR4-driven tumorigenesis identifies PI3K/mTOR inhibition as a therapeutic strategy in rhabdomyosarcoma. Oncogene., 37, 2630–2644. https://doi.org/10.1038/s41388-017-0122-y.
Shariat, S. F., Lamb, D. J., Kattan, M. W., Nguyen, C., Kim, J., Beck, J., et al. (2002). Association of preoperative plasma levels of insulin-like growth factor I and insulin-like growth factor binding proteins-2 and -3 with prostate cancer invasion, progression, and metastasis. Journal of Clinical Oncology. https://doi.org/10.1200/JCO.20.3.833.
Dunlap, S. M., Celestino, J., Wang, H., Jiang, R., Holland, E. C., Fuller, G. N., & Zhang, W. (2007). Insulin-like growth factor binding protein 2 promotes glioma development and progression. Proceedings of the National Academy of Sciences of the United States of America. https://doi.org/10.1073/pnas.0703145104.
Jones, J. I., & Clemmons, D. R. (1995). Insulin-like growth factors and their binding proteins: biological actions. Endocrine Reviews. https://doi.org/10.1210/edrv-16-1-3.
Tombolan, L., Orso, F., Guzzardo, V., Casara, S., Zin, A., Bonora, M., et al. (2011). High IGFBP2 expression correlates with tumor severity in pediatric rhabdomyosarcoma. The American Journal of Pathology. https://doi.org/10.1016/j.ajpath.2011.07.018.
Makawita, S., Ho, M., Durbin, A. D., Thorner, P. S., Malkin, D., & Somers, G. R. (2009). Expression of insulin-like growth factor pathway proteins in rhabdomyosarcoma: IGF-2 expression is associated with translocation-negative tumors. Pediatric and Developmental Pathology. https://doi.org/10.2350/08-05-0477.1.
Huang, F., Hurlburt, W., Greer, A., Reeves, K. A., Hillerman, S., Chang, H., et al. (2010). Differential mechanisms of acquired resistance to insulin-like growth factor-I receptor antibody therapy or to a small-molecule inhibitor, BMS-754807, in a human rhabdomyosarcoma model. Cancer Research. https://doi.org/10.1158/0008-5472.CAN-10-0391.
Kikuchi, K., Tsuchiya, K., Otabe, O., Gotoh, T., Tamura, S., Katsumi, Y., et al. (2008). Effects of PAX3-FKHR on malignant phenotypes in alveolar rhabdomyosarcoma. Biochemical and Biophysical Research Communications. https://doi.org/10.1016/j.bbrc.2007.11.017.
Taulli, R., Scuoppo, C., Bersani, F., Accornero, P., Forni, P. E., Miretti, S., et al. (2006). Validation of Met as a therapeutic target in alveolar and embryonal rhabdomyosarcoma. Cancer Research. https://doi.org/10.1158/0008-5472.CAN-05-4292.
Otabe, O., Kikuchi, K., Tsuchiya, K., Katsumi, Y., Yagyu, S., Miyachi, M., et al. (2017). MET/ERK2 pathway regulates the motility of human alveolar rhabdomyosarcoma cells. Oncology Reports. https://doi.org/10.3892/or.2016.5213.
Ferracini, R., Olivero, M., Di Renzo, M. F., Martano, M., De Giovanni, C., Nanni, P., et al. (1996). Retrogenic expression of the MET proto-oncogene correlates with the invasive phenotype of human rhabdomyosarcomas. Oncogene.
Saini, M., Verma, A., & Mathew, S. J. (2018). SPRY2 is a novel MET interactor that regulates metastatic potential and differentiation in rhabdomyosarcoma. Cell Death & Disease, 9, 1–15. https://doi.org/10.1038/s41419-018-0261-2.
Skrzypek, K., Kusienicka, A., Szewczyk, B., Adamus, T., Lukasiewicz, E., Miekus, K., & Majka, M. (2015). Constitutive activation of MET signaling impairs myogenic differentiation of rhabdomyosarcoma and promotes its development and progression. Oncotarget. https://doi.org/10.18632/oncotarget.5145.
Pillay, K., Govender, D., & Chetty, R. (2002). ALK protein expression in rhabdomyosarcomas. Histopathology. https://doi.org/10.1046/j.1365-2559.2002.01534.x.
Van Gaal, J. C., Flucke, U. E., Roeffen, M. H. S., De Bont, E. S. J. M., Sleijfer, S., Mavinkurve-Groothuis, A. M. C., et al. (2012). Anaplastic lymphoma kinase aberrations in rhabdomyosarcoma: clinical and prognostic implications. Journal of Clinical Oncology. https://doi.org/10.1200/JCO.2011.37.8588.
Gasparini, P., Casanova, M., Villa, R., Collini, P., Alaggio, R., Zin, A., et al. (2016). Anaplastic lymphoma kinase aberrations correlate with metastatic features in pediatric rhabdomyosarcoma. Oncotarget. https://doi.org/10.18632/oncotarget.10368.
van Erp, A. E. M., Hillebrandt-Roeffen, M. H. S., van Houdt, L., Fleuren, E. D. G., van der Graaf, W. T. A., & Versleijen-Jonkers, Y. M. H. (2017). Targeting anaplastic lymphoma kinase (ALK) in rhabdomyosarcoma (RMS) with the second-generation ALK inhibitor ceritinib. Targeted Oncology, 12, 815–826. https://doi.org/10.1007/s11523-017-0528-z.
Schöffski, P., Wozniak, A., Leahy, M. G., Aamdal, S., Rutkowski, P., Bauer, S., et al. (2018). The tyrosine kinase inhibitor crizotinib does not have clinically meaningful activity in heavily pre-treated patients with advanced alveolar rhabdomyosarcoma with FOXO rearrangement: European Organisation for Research and Treatment of Cancer phase 2 trial. European Journal of Cancer. https://doi.org/10.1016/j.ejca.2018.02.011.
Ehnman, M., Missiaglia, E., Folestad, E., Selfe, J., Strell, C., Thway, K., et al. (2013). Distinct effects of ligand-induced PDGFRα and PDGFRβ signaling in the human rhabdomyosarcoma tumor cell and stroma cell compartments. Cancer Research. https://doi.org/10.1158/0008-5472.CAN-12-1646.
Hosoyama, T., Aslam, M. I., Abraham, J., Prajapati, S. I., Nishijo, K., Michalek, J. E., et al. (2011). IL-4R drives dedifferentiation, mitogenesis, and metastasis in rhabdomyosarcoma. Clinical Cancer Research. https://doi.org/10.1158/1078-0432.CCR-10-3445.
Nanni, P., Nicoletti, G., Palladini, A., Astolfi, A., Rinella, P., Croci, S., et al. (2009). Opposing control of rhabdomyosarcoma growth and differentiation by myogenin and interleukin 4. Molecular Cancer Therapeutics. https://doi.org/10.1158/1535-7163.MCT-08-0678.
Curto, M., & McClatchey, A. I. (2004). Ezrin... a metastatic detERMinant? Cancer Cell. https://doi.org/10.1016/S1535-6108(04)00031-5.
Yu, Y., Khan, J., Khanna, C., Helman, L., Meltzer, P. S., & Merlino, G. (2004). Expression profiling identifies the cytoskeletal organizer ezrin and the developmental homeoprotein Six-1 as key metastatic regulators. Nature Medicine. https://doi.org/10.1038/nm966.
Yu, Y., Davicioni, E., Triche, T. J., & Merlino, G. (2006). The homeoprotein Six1 transcriptionally activates multiple protumorigenic genes but requires ezrin to promote metastasis. Cancer Research. https://doi.org/10.1158/0008-5472.CAN-05-2360.
Chan, B. M. C., Matsuura, N., Takada, Y., Zetter, B. R., & Hemler, M. E. (1991). In vitro and in vivo consequences of VLA-2 expression on rhabdomyosarcoma cells. Science. https://doi.org/10.1126/science.2011740.
Leabu, M., Uniyal, S., Xie, J., Xu, Y. Q., Vladau, C., Morris, V. L., & Chan, B. M. C. (2005). Integrin α2β1 modulates EGF stimulation of Rho GTPase-dependent morphological changes in adherent human rhabdomyosarcoma RD cells. Journal of Cellular Physiology. https://doi.org/10.1002/jcp.20163.
Marshall, A. D., Lagutina, I., & Grosveld, G. C. (2011). PAX3-FOXO1 induces cannabinoid receptor 1 to enhance cell invasion and metastasis. Cancer Research. https://doi.org/10.1158/0008-5472.CAN-11-0924.
Thuault, S., Comunale, F., Hasna, J., Fortier, M., Planchon, D., Elarouci, N., et al. (2016). The RhoE/ROCK/ARHGAP25 signaling pathway controls cell invasion by inhibition of Rac activity. Molecular Biology of the Cell. https://doi.org/10.1091/mbc.E16-01-0041.
Hayes, M. N., McCarthy, K., Jin, A., Oliveira, M. L., Iyer, S., Garcia, S. P., et al. (2018). Vangl2/RhoA signaling pathway regulates stem cell self-renewal programs and growth in rhabdomyosarcoma. Cell Stem Cell. https://doi.org/10.1016/j.stem.2018.02.002.
Karhadkar, S. S., Bova, G. S., Abdallah, N., Dhara, S., Gardner, D., Maitra, A., et al. (2004). Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature. https://doi.org/10.1038/nature02962.
Scales, S. J., & de Sauvage, F. J. (2009). Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends in Pharmacological Sciences. https://doi.org/10.1016/j.tips.2009.03.007.
Zibat, A., Missiaglia, E., Rosenberger, A., Pritchard-Jones, K., Shipley, J., Hahn, H., & Fulda, S. (2010). Activation of the hedgehog pathway confers a poor prognosis in embryonal and fusion gene-negative alveolar rhabdomyosarcoma. Oncogene. https://doi.org/10.1038/onc.2010.368.
Satheesha, S., Manzella, G., Bovay, A., Casanova, E. A., Bode, P. K., Belle, R., Feuchtgruber, S., Jaaks, P., Dogan, N., Koscielniak, E., & Schäfer, B. W. (2016). Targeting hedgehog signaling reduces self-renewal in embryonal rhabdomyosarcoma. Oncogene., 35, 2020–2030. https://doi.org/10.1038/onc.2015.267.
Almazán-Moga, A., Zarzosa, P., Molist, C., Velasco, P., Pyczek, J., Simon-Keller, K., Giralt, I., Vidal, I., Navarro, N., Segura, M. F., Soriano, A., Navarro, S., Tirado, O. M., Ferreres, J. C., Santamaria, A., Rota, R., Hahn, H., Sánchez de Toledo, J., Roma, J., & Gallego, S. (2017). Ligand-dependent hedgehog pathway activation in rhabdomyosarcoma: the oncogenic role of the ligands. British Journal of Cancer, 117, 1314–1325. https://doi.org/10.1038/bjc.2017.305.
Bresler, S. C., Padwa, B. L., & Granter, S. R. (2016). Nevoid basal cell carcinoma syndrome (Gorlin syndrome). Head and Neck Pathology. https://doi.org/10.1007/s12105-016-0706-9.
De Bortoli, M., Castellino, R. C., Skapura, D. G., Shen, J. J., Su, J. M., Russell, H. V., et al. (2007). Patched haploinsufficient mouse rhabdomyosarcoma overexpress secreted phosphoprotein 1 and matrix metalloproteinases. European Journal of Cancer. https://doi.org/10.1016/j.ejca.2007.02.008.
Kopan, R. (2002). Notch: a membrane-bound transcription factor. Journal of Cell Science.
Artavanis-Tsakonas, S., Rand, M. D., & Lake, R. J. (1999). Notch signaling: cell fate control and signal integration in development. Science. https://doi.org/10.1126/science.284.5415.770.
Roma, J., Masià, A., Reventós, J., De Toledo, J. S., & Gallego, S. (2011). Notch pathway inhibition significantly reduces rhabdomyosarcoma invasiveness and mobility in vitro. Clinical Cancer Research. https://doi.org/10.1158/1078-0432.CCR-10-0166.
Ignatius, M. S., Hayes, M. N., Lobbardi, R., Chen, E. Y., McCarthy, K. M., Sreenivas, P., & Langenau, D. M. (2017). The NOTCH1/SNAIL1/MEF2C pathway regulates growth and self-renewal in embryonal rhabdomyosarcoma. Cell Reports. https://doi.org/10.1016/j.celrep.2017.05.061.
Masià, A., Almazán-Moga, A., Velasco, P., Reventós, J., Torán, N., Sánchez De Toledo, J., et al. (2012). Notch-mediated induction of N-cadherin and α9-integrin confers higher invasive phenotype on rhabdomyosarcoma cells. British Journal of Cancer, 107, 1374–1383. https://doi.org/10.1038/bjc.2012.411.
Singh, S., Vinson, C., Gurley, C. M., Nolen, G. T., Beggs, M. L., Nagarajan, R., et al. (2010). Impaired Wnt signaling in embryonal rhabdomyosarcoma cells from p53/c-fos double mutant mice. The American Journal of Pathology. https://doi.org/10.2353/ajpath.2010.091195.
Soglio, D. B. D., Rougemont, A. L., Absi, R., Giroux, L. M., Sanchez, R., Barrette, S., & Fournet, J. C. (2009). Beta-catenin mutation does not seem to have an effect on the tumorigenesis of pediatric rhabdomyosarcomas. Pediatric and Developmental Pathology. https://doi.org/10.2350/08-11-0553.1.
Annavarapu, S. R., Cialfi, S., Dominici, C., Kokai, G. K., Uccini, S., Ceccarelli, S., et al. (2013). Characterization of Wnt/β-catenin signaling in rhabdomyosarcoma. Laboratory Investigation, 93, 1090–1099. https://doi.org/10.1038/labinvest.2013.97.
Chen, E. Y., DeRan, M. T., Ignatius, M. S., Brooke Grandinetti, K., Clagg, R., McCarthy, K. M., et al. (2014). Glycogen synthase kinase 3 inhibitors induce the canonical WNT/β-catenin pathway to suppress growth and self-renewal in embryonal rhabdomyosarcoma. Proceedings of the National Academy of Sciences of the United States of America. https://doi.org/10.1073/pnas.1317731111.
Oristian, K. M., Crose, L. E. S., Kuprasertkul, N., Bentley, R. C., Lin, Y. T., Williams, N., & Linardic, C. M. (2018). Loss of MST/Hippo signaling in a genetically engineered mouse model of fusion-positive rhabdomyosarcoma accelerates tumorigenesis. Cancer Research. https://doi.org/10.1158/0008-5472.CAN-17-3912.
Tremblay, A. M., Missiaglia, E., Galli, G. G., Hettmer, S., Urcia, R., Carrara, M., et al. (2014). The Hippo transducer YAP1 transforms activated satellite cells and is a potent effector of embryonal rhabdomyosarcoma formation. Cancer Cell. https://doi.org/10.1016/j.ccr.2014.05.029.
Seki, M., Nishimura, R., Yoshida, K., Shimamura, T., Shiraishi, Y., Sato, Y., et al. (2015). Integrated genetic and epigenetic analysis defines novel molecular subgroups in rhabdomyosarcoma. Nature Communications. https://doi.org/10.1038/ncomms8557.
Ignatius, M. S., Hayes, M. N., Moore, F. E., Tang, Q., Garcia, S. P., Blackburn, P. R., et al. (2018). Tp53 deficiency causes a wide tumor spectrum and increases embryonal rhabdomyosarcoma metastasis in zebrafish. eLife. https://doi.org/10.7554/eLife.37202.
Evdokimova, V., Ovchinnikov, L. P., & Sorensen, P. H. B. (2006). Y-box binding protein 1: providing a new angle on translational regulation. Cell Cycle. https://doi.org/10.4161/cc.5.11.2784.
Evdokimova, V., Tognon, C., Ng, T., Ruzanov, P., Melnyk, N., Fink, D., et al. (2009). Translational activation of Snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell. https://doi.org/10.1016/j.ccr.2009.03.017.
El-Naggar, A. M., Veinotte, C. J., Cheng, H., Grunewald, T. G. P., Negri, G. L., Somasekharan, S. P., et al. (2015). Translational activation of HIF1α by YB-1 promotes sarcoma metastasis. Cancer Cell. https://doi.org/10.1016/j.ccell.2015.04.003.
Armeanu-Ebinger, S., Bonin, M., Häbig, K., Poremba, C., Koscielniak, E., Godzinski, J., & Seitz, G. (2011). Differential expression of invasion promoting genes in childhood rhabdomyosarcoma. International Journal of Oncology. https://doi.org/10.3892/ijo.2011.921.
Shang, H., Liu, Y., Li, Z., Liu, Q., Cui, W., Zhang, L., Pang, Y., Liu, C., & Li, F. (2019). MicroRNA-874 functions as a tumor suppressor in rhabdomyosarcoma by directly targeting GEFT. American Journal of Cancer Research, 9(4), 668–681.
Bersani, F., Lingua, M. F., Morena, D., Foglizzo, V., Miretti, S., Lanzetti, L., et al. (2016). Deep sequencing reveals a novel miR-22 regulatory network with therapeutic potential in rhabdomyosarcoma. Cancer Research. https://doi.org/10.1158/0008-5472.CAN-16-0709.
Megiorni, F., Cialfi, S., McDowell, H. P., Felsani, A., Camero, S., Guffanti, A., Pizer, B., Clerico, A., de Grazia, A., Pizzuti, A., Moles, A., & Dominici, C. (2014). Deep sequencing the microRNA profile in rhabdomyosarcoma reveals down-regulation of miR-378 family members. BMC Cancer, 14, 1–17. https://doi.org/10.1186/1471-2407-14-880.
Ignatius, M. S., Chen, E., Elpek, N. M., Fuller, A. Z., Tenente, I. M., Clagg, R., et al. (2012). In vivo imaging of tumor-propagating cells, regional tumor heterogeneity, and dynamic cell movements in embryonal rhabdomyosarcoma. Cancer Cell. https://doi.org/10.1016/j.ccr.2012.03.043.
Kessenbrock, K., Plaks, V., & Werb, Z. (2010). Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. https://doi.org/10.1016/j.cell.2010.03.015.
Diomedi-Camassei, F., Boldrini, R., Ravà, L., Donfrancesco, A., Boglino, C., Messina, E., & Callea, F. (2004). Different pattern of matrix metalloproteinases expression in alveolar versus embryonal rhabdomyosarcoma. Journal of Pediatric Surgery. https://doi.org/10.1016/j.jpedsurg.2004.07.014.
Wysoczynski, M., Miekus, K., Jankowski, K., Wanzeck, J., Bertolone, S., Janowska-Wieczorek, A., et al. (2007). Leukemia inhibitory factor: a newly identified metastatic factor in rhabdomyosarcomas. Cancer Research. https://doi.org/10.1158/0008-5472.CAN-06-1021.
Teicher, B. A., & Fricker, S. P. (2010). CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clinical Cancer Research. https://doi.org/10.1158/1078-0432.CCR-09-2329.
Libura, J., Drukala, J., Majka, M., Tomescu, O., Navenot, J. M., Kucia, M., et al. (2002). CXCR4-SDF-1 signaling is active in rhabdomyosarcoma cells and regulates locomotion, chemotaxis, and adhesion. Blood. https://doi.org/10.1182/blood-2002-01-0031.
Grymula, K., Tarnowski, M., Wysoczynski, M., Drukala, J., Barr, F. G., Ratajczak, J., et al. (2010). Overlapping and distinct role of CXCR7-SDF-1/ITAC and CXCR4-SDF-1 axes in regulating metastatic behavior of human rhabdomyosarcomas. International Journal of Cancer. https://doi.org/10.1002/ijc.25245.
Wysoczynski, M., Shin, D. M., Kucia, M., & Ratajczak, M. Z. (2010). Selective upregulation of interleukin-8 by human rhabdomyosarcomas in response to hypoxia: therapeutic implications. International Journal of Cancer. https://doi.org/10.1002/ijc.24732.
Lima, L. G., & Monteiro, R. Q. (2013). Activation of blood coagulation in cancer: implications for tumour progression. Bioscience Reports. https://doi.org/10.1042/BSR20130057.
Hu, L., Lee, M., Campbell, W., Perez-Soler, R., & Karpatkin, S. (2004). Role of endogenous thrombin in tumor implantation, seeding, and spontaneous metastasis. Blood. https://doi.org/10.1182/blood-2004-03-1047.
Poon, R. T. P., Lau, C. P. Y., Ho, J. W. Y., Yu, W. C., Fan, S. T., & Wong, J. (2003). Tissue factor expression correlates with tumor angiogenesis and invasiveness in human hepatocellular carcinoma. Clinical Cancer Research.
Wysoczynski, M., Liu, R., Kucia, M., Drukala, J., & Ratajczak, M. Z. (2010). Thrombin regulates the metastatic potential of human rhabdomyosarcoma cells: distinct role of PAR1 and PAR3 signaling. Molecular Cancer Research. https://doi.org/10.1158/1541-7786.MCR-10-0019.
Nakajima, M., Nagahashi, M., Rashid, O. M., Takabe, K., & Wakai, T. (2017). The role of sphingosine-1-phosphate in the tumor microenvironment and its clinical implications. Tumor Biology. https://doi.org/10.1177/1010428317699133.
Schneider, G., Bryndza, E., Abdel-Latif, A., Ratajczak, J., Maj, M., Tarnowski, M., et al. (2013). Bioactive lipids S1P and C1P are prometastatic factors in human rhabdomyosarcoma, and their tissue levels increase in response to radio/chemotherapy. Molecular Cancer Research. https://doi.org/10.1158/1541-7786.MCR-12-0600.
Adamus, A., Engel, N., & Seitz, G. (2019). SGPL1321 mutation: one main trigger for invasiveness of pediatric alveolar rhabdomyosarcoma. Cancer Gene Therapy, 1–14. https://doi.org/10.1038/s41417-019-0132-8.
Théry, C., Zitvogel, L., & Amigorena, S. (2002). Exosomes: composition, biogenesis and function. Nature Reviews. Immunology. https://doi.org/10.1038/nri855.
Valadi, H., Ekström, K., Bossios, A., Sjöstrand, M., Lee, J. J., & Lötvall, J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology. https://doi.org/10.1038/ncb1596.
Tai, Y. L., Chen, K. C., Hsieh, J. T., & Shen, T. L. (2018). Exosomes in cancer development and clinical applications. Cancer Science. https://doi.org/10.1111/cas.13697.
Ghayad, S. E., Rammal, G., Ghamloush, F., Basma, H., Nasr, R., Diab-Assaf, M., Chelala, C., & Saab, R. (2016). Exosomes derived from embryonal and alveolar rhabdomyosarcoma carry differential miRNA cargo and promote invasion of recipient fibroblasts. Scientific Reports, 6, 1–15. https://doi.org/10.1038/srep37088.
Hanna, J. A., Garcia, M. R., Lardennois, A., Leavey, P. J., Maglic, D., Fagnan, A., Go, J. C., Roach, J., Wang, Y. D., Finkelstein, D., & Hatley, M. E. (2018). PAX3-FOXO1 drives miR-486-5p and represses miR-221 contributing to pathogenesis of alveolar rhabdomyosarcoma. Oncogene., 37, 1991–2007. https://doi.org/10.1038/s41388-017-0081-3.
Ghamloush, F., Ghayad, S. E., Rammal, G., Fahs, A., Ayoub, A. J., Merabi, Z., Harajly, M., Zalzali, H., & Saab, R. (2019). The PAX3-FOXO1 oncogene alters exosome miRNA content and leads to paracrine effects mediated by exosomal miR-486. Scientific Reports, 9, 1–12. https://doi.org/10.1038/s41598-019-50592-4.
Rammal, G., Fahs, A., Kobeissy, F., Mechref, Y., Zhao, J., Zhu, R., et al. (2019). Proteomic profiling of rhabdomyosarcoma-derived exosomes yield insights into their functional role in paracrine signaling. Journal of Proteome Research. https://doi.org/10.1021/acs.jproteome.9b00157.
Li, S., Zou, H., Shao, Y. Y., Mei, Y., Cheng, Y., Hu, D. L., et al. (2017). Pseudogenes of annexin A2, novel prognosis biomarkers for diffuse gliomas. Oncotarget. https://doi.org/10.18632/oncotarget.22197.
Murphy, A. G., Foley, K., Rucki, A. A., Xia, T., Jaffee, E. M., Huang, C. Y., & Zheng, L. (2017). Stromal Annexin A2 expression is predictive of decreased survival in pancreatic cancer. Oncotarget. https://doi.org/10.18632/oncotarget.22433.
Hoshino, A., Costa-Silva, B., Shen, T. L., Rodrigues, G., Hashimoto, A., Tesic Mark, M., & Lyden, D. (2015). Tumour exosome integrins determine organotropic metastasis. Nature. https://doi.org/10.1038/nature15756.
Richardson, A. M., Havel, L. S., Koyen, A. E., Konen, J. M., Shupe, J., Wiles, W. G., et al. (2018). Vimentin is required for lung adenocarcinoma metastasis via heterotypic tumor cell–cancer-associated fibroblast interactions during collective invasion. Clinical Cancer Research. https://doi.org/10.1158/1078-0432.CCR-17-1776.
Satelli, A., & Li, S. (2011). Vimentin in cancer and its potential as a molecular target for cancer therapy. Cellular and Molecular Life Sciences. https://doi.org/10.1007/s00018-011-0735-1.
Huang, F., Greer, A., Hurlburt, W., Han, X., Hafezi, R., Wittenberg, G. M., et al. (2009). The mechanisms of differential sensitivity to an insulin-like growth factor-1 receptor inhibitor (BMS-536924) and rationale for combining with EGFR/HER2 inhibitors. Cancer Research. https://doi.org/10.1158/0008-5472.CAN-08-0835.
Vela, M. (2018). Anti CXCR4 antibody combined with activated and expanded natural killer cells for sarcoma immunotherapy. Journal of Clinical Oncology. https://doi.org/10.1200/jco.2018.36.15_suppl.11541.
Rengaswamy, V., Zimmer, D., Süss, R., & Rössler, J. (2016). RGD liposome-protamine-siRNA (LPR) nanoparticles targeting PAX3-FOXO1 for alveolar rhabdomyosarcoma therapy. Journal of Controlled Release. https://doi.org/10.1016/j.jconrel.2016.05.063.
Anderson, J. L., Park, A., Akiyama, R., Tap, W. D., Denny, C. T., & Federman, N. (2015). Evaluation of in vitro activity of the class i PI3K inhibitor buparlisib (BKM120) in pediatric bone and soft tissue sarcomas. PLoS One. https://doi.org/10.1371/journal.pone.0133610.
Van Erp, A. E. M., Versleijen-Jonkers, Y. M. H., Van Der Graaf, W. T. A., & Fleuren, E. D. G. (2018). Targeted therapy-based combination treatment in rhabdomyosarcoma. Molecular Cancer Therapeutics. https://doi.org/10.1158/1535-7163.MCT-17-1131.
Xin, X., Zeng, X., Gu, H., Li, M., Tan, H., Jin, Z., Hua, T., Shi, R., & Wang, H. (2016). CD147/EMMPRIN overexpression and prognosis in cancer: a systematic review and meta-analysis. Scientific Reports, 6, 1–12. https://doi.org/10.1038/srep32804.
Dana, P., Kariya, R., Vaeteewoottacharn, K., Sawanyawisuth, K., Seubwai, W., Matsuda, K., et al. (2017). Upregulation of CD147 promotes metastasis of cholangiocarcinoma by modulating the epithelial-to-mesenchymal transitional process. Oncology Research. https://doi.org/10.3727/096504016X14813899000565.
Caudron, A., Battistella, M., Feugeas, J. P., Pagès, C., Basset-Seguin, N., Sadoux, A., et al. (2015). EMMPRIN/CD147 as a novel independent prognostic biomarker in melanoma. Journal of Clinical Oncology. https://doi.org/10.1200/jco.2015.33.15_suppl.e20027.
Acknowledgments
All figures were created using BioRender.com
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ramadan, F., Fahs, A., Ghayad, S.E. et al. Signaling pathways in Rhabdomyosarcoma invasion and metastasis. Cancer Metastasis Rev 39, 287–301 (2020). https://doi.org/10.1007/s10555-020-09860-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10555-020-09860-3