Abstract
Hepatocellular carcinoma is the major cancer-related death worldwide, and the prognosis is grim. Therefore, there is an urgent need of detection at early stage, effective treatment options, and prevention of recurrence. Recently, liquid biopsies, such as circulating tumor cells, circulating tumor DNA, and microRNA, have accumulating evidence that is useful for these needs.
In this review, we summarize the current advance of liquid biopsies and discuss the perspective.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Hepatocellular carcinoma (HCC) is the second leading cause of cancer-related death, and its incidence has been increasing worldwide [1]. In Japan, HCC is the fifth most common cause of cancer-related death [2]. Due to advances in medical technology, mainly in the field of imaging, the rate of early detection of HCC has been increasing, and the emergence of molecularly targeted therapies has enabled HCC patients to receive systemic chemotherapy [3]. However, in many patients, the cancer is still detected in the intermediate or advanced stages, and these patients cannot receive curative treatments such as resection or radio frequency ablation. Even after curative treatments, the recurrence rate is high due to the existing liver damage [4].
For advanced cases, systemic chemotherapies have been the main treatment option. Currently, three molecularly targeted anticancer drugs—sorafenib [5], regorafenib [6], and lenvatinib [7]—are available for advanced HCC. However, due to the lack of a biomarker that can aid decision-making regarding which drug to use, these three drugs are generally used according to the physician’s choice in daily practice.
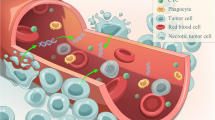
To help physicians choose the appropriate drug, liquid biopsies have already shown promising applications in clinical settings for some cancers, including colorectal cancer [8], breast cancer [9], and lung cancer [10]. These biopsies, which can be performed on blood or body fluids, provide genetic and epigenetic information and can be performed repeatedly with low invasiveness. Also, they provide real-time information about cancer status. In this paper, we review liquid biopsies, focusing particularly on blood-based circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), and microRNAs (miRNAs) from blood samples in HCC cases (Fig. 8.1). We also discuss current perspectives on these procedures.
1.1 Circulating Tumor Cells
The path to tumor metastasis mainly begins with tumor cells in systemic circulation [11]. CTCs originate from a primary organ and reach distant metastatic sites without being blocked by a barrier system; the idea for how this occurs was originally based on the “seed and soil” theory [12]. CTCs play an important role in diagnosis, because finding them helps in the early detection of cancer, and CTC analysis offers valuable information including DNA, RNA, and protein levels. This is because CTCs reflect the real status of the cancer, unlike tissue biopsies, which are collected before treatment.
Various techniques for liquid biopsies have been developed, mainly divided into either physical or biological methods. Recently, biological methods using antibody-based enrichment techniques have been developed. In particular, CellSearch, which can detect the epithelial cell adhesion molecule (EpCAM), was approved by the Food and Drug Administration in the USA (Fig. 8.2) [13]. This system consists of a CellPrep system, a CellSearch epithelial cell kit, and a CellSpotter analyzer. It enumerates CTCs of epithelial origin in whole blood using antibodies that bind to cytokeratins 8, 18, and 19, an antibody to CD45 conjugated to allophycocyanin, and the nuclear dye 4′,6-diamidino-2-phenylindole (DAPI). A meta-analysis with 998 HCC subjects including advanced cases showed a pooled sensitivity of CTC detection of 67% at a pooled specificity of 98% [14].
One limitation of early detection is poor sensitivity; with current techniques, it is difficult to detect a sufficient number of tumor cells with a high sensitivity and specificity, especially in early stages [15]. In addition, the epithelial-mesenchymal transition (EMT) is an important event that occurs when tumors shed into metastasis, and the EMT may disrupt epithelial cells and EpCAM expression [16]. In fact, only 35% of HCC-derived CTCs express EpCAM [17]. To overcome these challenges, new innovations are needed to detect higher numbers of CTCs and greater amounts of specific antibodies in addition to EpCAM.
1.2 Circulating Tumor DNA
Circulating tumor DNA (ctDNA) consists of extracellular nucleic acid fragments that flow into the plasma from tumor cells [18]. The mechanism of DNA shedding involves necrosis, apoptosis, and secretion by tumor cells. The concentration of ctDNA in a healthy individual is 1–10 ng/ml, and cancer patients have a higher amount [19]. Tumor-derived DNA fragments are shorter than non-tumor-derived DNA fragments, and after curative treatment, cell-free DNA (tumor- plus non-tumor-derived DNA) integrity increases and returns to normal levels [20]. This suggests that DNA integrity may be a potential predictive marker for the diagnosis, treatment, and monitoring of HCC. Compared to CTCs, the fraction of ctDNA can be more easily detected [21], and ctDNA shows the total heterogeneity of all the tumors and thus reflects the total tumor status.
Recent studies have focused not only on quantitative analysis but also on aberrations and methylation of DNA. The utility of analyzing ctDNA by next-generation sequencing (NGS) has been increasing. In addition, recent whole-genome sequencing showed the presence of somatic mutation of TERT promoter, the WNT/beta-catenin pathway, the PI3K-AKT-mTOR pathway, the MAPK-pathway in patients with HCC [22]. Ikeda et al. analyzed 32 ctDNA fragments in 26 HCC patients and detected 100% genetic alteration and serial ctDNA emergence of a new gene alteration [23]. Although the ctDNA demonstrated excellent sensitivity and specificity, the detection rate was strongly affected by the tumor burden and tumor type, namely, a rate of somatic mutation is 15–56% for operable cases; on the other hand, in advanced stages, the ctDNA detection rate rose to 74–88% [24]. Recently, Cai et al. analyzed 574 genes from the ctDNA of 4 HCC patients; 96.9% of these patients’ tissue mutations could also be detected in plasma samples [25]. The authors concluded that ctDNA could overcome the issue of tumor heterogeneity and track therapeutic responses in real time.
It is well known that methylation is a common epigenetic regulation mechanism and that the methylation of suppressor genes has been connected with carcinogenesis. Thanks to the development of NGS techniques, methylation can aid in the diagnosis of HCC [26].
Xu et al. have developed HCC-specific methylation marker panels comparing HCC tissue and peripheral blood mononuclear cell methylation as high as 1000 cases [27]. Compared to normal controls, this model achieved 85.7% sensitivity and 94.3% specificity. The main limitation of this method is the low detection rate in early stages, as mentioned above.
1.3 MicroRNAs
miRNAs are small, noncoding, interfering RNAs that are 21–30 nucleotides long; they play a crucial role in cellular processes and carcinogenesis [28]. Namely, the upregulation of miRNAs inhibits tumor suppressor genes, and the downregulation of miRNAs inhibits oncogenes. A total of over 2300 miRNAs have been identified. Recent studies have shown that miRNAs are associated with pathogenesis of cancers and not only genetic but also epigenetic modification of carcinogenesis [29, 30].
miRNAs have two ways of expanding into the bloodstream. One is secretion through cell death by apoptosis and necrosis, and the other is release in a package into a small membrane vehicle called an exosome. In the plasma, the miRNA binds to particular proteins such as Argonaute 2 and high-density lipoproteins and escapes RNase [31]. Cancer cells secrete exosomes with miRNAs to assist their invasion or metastasis.
Among miRNA groups, certain specific miRNAs are associated with HCC prognosis. In the liver, some miRNAs are associated with HBV and HCV infection and liver fibrosis. For example, the expression of miR-34 has been shown to be increased in patients with hepatic fibrosis, HCV, alcoholic disease, and NAFLDS, in addition to HCC [32].
Li et al. first demonstrated that miRNA expression profiles could be useful biomarkers for distinguishing between HCC with no HBV infection and HBV-positive HCC [33]. Zhou et al. identified using seven miRNAs and improved the diagnostic accuracy of HCC [34]. More recently, Lin et al. analyzed over 300 HCC and non-HCC patients and identified 7 different miRNAs (miR-29a, miR-29c, miR-133a, miR-143, miR-145, miR-192, and miR-505) with higher sensitivity than AFP and a similar specificity [35]. Based on these data, in China, this system has been approved for clinical use and is now widely available.
Regarding recurrence, miR122, miR26a, and miR29a may be predictive markers for recurrence [36]. If the quantity of these miRNAs with tumor markers can be monitored, effective treatment can be possible. The limitation of miRNAs is their diagnostic accuracy. The measurement methods used are quantitative RT-PCR, microarray, and NGS, and it is difficult to compare among these methods.
2 Perspective
Compared to their use in other malignancies such as lung cancer, breast cancer, and colon cancer, the advance of liquid biopsies in the field of HCC has been slow. One reason is the typical HCC problem of background liver damage. Most HCCs occur in the damaged liver due to HBV, HCV, and nonalcoholic steatohepatitis, and the difference in etiology may affect the results of mutation or the quantitative amount of CTCs, ctDNA, and miRNA. In addition, when comparing HCC data with controls, the controls depend on the background of the liver, and careful attention is needed to interpret the results.
Recently, a blood test called CancerSEEK, which uses protein biomarkers and somatic mutations for the early detection of HCC and seven other cancers, has shown promising results (Fig. 8.3) [37]. Surprisingly, the overall sensitivity and specificity are 98% and 99%, respectively, and the sensitivity for HCC is highest among stage I cancer cases. In addition, the plasma from all HCC cases has been shown to have at least one mutation in 16 genes including TERT, TP53, and PIK3CA. These results suggest that the detection rate of HCC, which is expected to have low sensitivity and specificity of detection due to background liver damage, can be made high by combining a protein marker with genetic alteration. However, although healthy individuals were chosen as the controls, the control panels were affected by inflammation and cirrhosis. The CancerSEEK method is also attractive in terms of its low cost. The research teams have tried to keep the cost below $500 per sample. If this is possible, the era of precision medicine, in terms of not only sensitivity and specificity but also low cost, will soon arrive.
For a decade, clinical trials have been conducted on new, effective molecularly targeted drugs. In addition to “all comer” clinical trials, some trials have focused on somatic mutations. If these mutations can be identified by CTCs or ctDNA, then suitable drugs against cancer can be found. For example, a copy number gain for FGF19 and FGF/FGF4 amplification is a predictive marker for sorafenib response [38, 39]. In non-small-cell lung cancer, the use of the EGFR mutation in liquid biopsies is now widely available. If such changes can be found before systemic chemotherapy, HCC patients are expected to achieve a good response.
Nowadays, clinical trials using immune checkpoint inhibitors are being widely conducted, including for HCC. In the USA, the PD-1 antibody nivolumab was approved for sorafenib-failed advanced HCC [40]. In non-small-cell lung cancer, PD-1 expression in over 50% of tumor cells was correlated with the improved efficacy of pembrolizumab [41]. Whether the amount of PD-1 expression is also a predictive marker in HCC is controversial; however, the amount of quantitative change in expression may be a predictive marker for efficacy in patients with HCC receiving immune checkpoint blockers.
In conclusion, we review liquid biopsies in patients with HCC. Liquid biopsies are useful as diagnostic, therapeutic, prognostic factors and will play a crucial role of the new era of precision medicine.
References
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–86.
Ikeda M, Morizane C, Ueno M, Okusaka T, Ishii H, Furuse J. Chemotherapy for hepatocellular carcinoma: current status and future perspectives. Jpn J Clin Oncol. 2018;48(2):103–14.
Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–14.
Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63(5):844–55.
Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359(4):378–90.
Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389(10064):56–66.
Kudo M, Finn RS, Qin S, Han KH, Ikeda K, Piscaglia F, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet. 2018;391(10126):1163–73.
Augestad KM, Merok MA, Ignatovic D. Tailored treatment of colorectal cancer: surgical, molecular, and genetic considerations. Clin Med Insights Oncol. 2017;11:1179554917690766.
Low SK, Zembutsu H, Nakamura Y. Breast cancer: the translation of big genomic data to cancer precision medicine. Cancer Sci. 2018;109(3):497–506.
O’Flaherty L, Wikman H, Pantel K. Biology and clinical significance of circulating tumor cell subpopulations in lung cancer. Transl Lung Cancer Res. 2017;6(4):431–43.
Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014;158(5):1110–22.
Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3(6):453–8.
Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10(20):6897–904.
Sun C, Liao W, Deng Z, Li E, Feng Q, Lei J, et al. The diagnostic value of assays for circulating tumor cells in hepatocellular carcinoma: a meta-analysis. Medicine. 2017;96(29):e7513.
Miller MC, Doyle GV, Terstappen LW. Significance of circulating tumor cells detected by the cellsearch system in patients with metastatic breast colorectal and prostate cancer. J Oncol. 2010;2010:617421.
Okajima W, Komatsu S, Ichikawa D, Miyamae M, Ohashi T, Imamura T, et al. Liquid biopsy in patients with hepatocellular carcinoma: circulating tumor cells and cell-free nucleic acids. World J Gastroenterol. 2017;23(31):5650–68.
Su YH, Kim AK, Jain S. Liquid biopsies for hepatocellular carcinoma. Transl Res. 2018;201:84–97.
Ng CKY, Di Costanzo GG, Terracciano LM, Piscuoglio S. Circulating cell-free DNA in hepatocellular carcinoma: current insights and outlook. Front Med. 2018;5:78.
Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32(6):579–86.
Jiang P, Chan CW, Chan KC, Cheng SH, Wong J, Wong VW, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A. 2015;112(11):E1317–25.
Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24.
Schulze K, Imbeaud S, Letouze E, Alexandrov LB, Calderaro J, Rebouissou S, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47(5):505–11.
Ikeda S, Lim JS, Kurzrock R. Analysis of tissue and circulating tumor DNA by next-generation sequencing of hepatocellular carcinoma: implications for targeted therapeutics. Mol Cancer Ther. 2018;17(5):1114–22.
Cabel L, Proudhon C, Buecher B, Pierga JY, Bidard FC. Circulating tumor DNA detection in hepatocellular carcinoma. Ann Oncol. 2018;29(5):1094–6.
Cai ZX, Chen G, Zeng YY, Dong XQ, Lin MJ, Huang XH, et al. Circulating tumor DNA profiling reveals clonal evolution and real-time disease progression in advanced hepatocellular carcinoma. Int J Cancer. 2017;141(5):977–85.
Yin CQ, Yuan CH, Qu Z, Guan Q, Chen H, Wang FB. Liquid biopsy of hepatocellular carcinoma: circulating tumor-derived biomarkers. Dis Markers. 2016;2016:1427849.
Xu RH, Wei W, Krawczyk M, Wang W, Luo H, Flagg K, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017;16(11):1155–61.
Morishita A, Masaki T. miRNA in hepatocellular carcinoma. Hepatol Res. 2015;45(2):128–41.
Ramassone A, Pagotto S, Veronese A, Visone R. Epigenetics and microRNAs in cancer. Int J Mol Sci. 2018;19(10). pii: E3139.
Wang L, Zhang X, Jia LT, Hu SJ, Zhao J, Yang JD, et al. c-Myc-mediated epigenetic silencing of MicroRNA-101 contributes to dysregulation of multiple pathways in hepatocellular carcinoma. Hepatology (Baltimore, Md). 2014;59(5):1850–63.
Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–8.
Mohamed AA, Ali-Eldin ZA, Elbedewy TA, El-Serafy M, Ali-Eldin FA, AbdelAziz H. MicroRNAs and clinical implications in hepatocellular carcinoma. World J Hepatol. 2017;9(23):1001–7.
Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, et al. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010;70(23):9798–807.
Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol. 2011;29(36):4781–8.
Lin XJ, Chong Y, Guo ZW, Xie C, Yang XJ, Zhang Q, et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: a multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. Lancet Oncol. 2015;16(7):804–15.
Canale M, Ulivi P, Foschi FG, Scarpi E, De Matteis S, Donati G, et al. Clinical and circulating biomarkers of survival and recurrence after radiofrequency ablation in patients with hepatocellular carcinoma. Crit Rev Oncol Hematol. 2018;129:44–53.
Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science (New York, NY). 2018;359(6378):926–30.
Kaibori M, Sakai K, Ishizaki M, Matsushima H, De Velasco MA, Matsui K, et al. Increased FGF19 copy number is frequently detected in hepatocellular carcinoma with a complete response after sorafenib treatment. Oncotarget. 2016;7(31):49091–8.
Arao T, Ueshima K, Matsumoto K, Nagai T, Kimura H, Hagiwara S, et al. FGF3/FGF4 amplification and multiple lung metastases in responders to sorafenib in hepatocellular carcinoma. Hepatology. 2013;57(4):1407–15.
El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389(10088):2492–502.
Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372(21):2018–28.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Suzuki, E., Chiba, T., Kato, N. (2019). Liquid Biopsy in Hepatocellular Carcinoma. In: Shimada, H. (eds) Biomarkers in Cancer Therapy. Springer, Singapore. https://doi.org/10.1007/978-981-13-7295-7_8
Download citation
DOI: https://doi.org/10.1007/978-981-13-7295-7_8
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-7294-0
Online ISBN: 978-981-13-7295-7
eBook Packages: MedicineMedicine (R0)