Abstract
In this study, horseradish peroxidase was extracted, purified, and immobilized on a calcium alginate-starch hybrid support by covalent bonding and entrapment. The immobilized HRP was used for the biodegradation of phenol red dye. A 3.74-fold purification was observed after precipitation with ammonium sulfate and dialysis. An immobilization yield of 88.33%, efficiency of 56.89%, and activity recovery of 50.26% were found. The optimum pH and temperature values for immobilized and free HRP were 5.0 and 50 °C and 6.5 and 60 °C, respectively. The immobilized HRP showed better thermal stability than its free form, resulting in a considerable increase in half-life time (t1/2) and deactivation energy (Ed). The immobilized HRP maintained 93.71% of its initial activity after 45 days of storage at 4 °C. Regarding the biodegradation of phenol red, immobilized HRP resulted in 63.57% degradation after 90 min. After 10 cycles of reuse, the immobilized HRP was able to maintain 43.06% of its initial biodegradative capacity and 42.36% of its enzymatic activity. At the end of 15 application cycles, a biodegradation rate of 8.34% was observed. In conclusion, the results demonstrate that the immobilized HRP is a promising option for use as an industrial biocatalyst in various biotechnological applications.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The use of natural biopolymers, such as collagen, cellulose, keratin, carrageenan, chitin, chitosan, and alginate has become increasingly important for enzyme immobilization, as immobilized enzymes have received prominent attention in biotechnological applications compared to free enzymes [1,2,3,4]. The immobilization of enzymes offers numerous operational and processing advantages, including enhanced stability, reusability, hyperactivity, selectivity, specificity, and resistance to inhibitors and harsh chemicals, as well as improved purity and yield of the final product [5,6,7,8].
The augmented enzymatic stability after immobilization can be primarily attributed to various factors. These include the prevention of dissociation of subunits in multisubunit enzymes, prevention of aggregation, prevention of autolysis or proteolysis by proteases, and the rigid structure imparted to the enzyme through multipoint covalent bonding. Moreover, immobilization can create a favorable microenvironment for enzymatic catalysis [9,10,11,12,13]. However, enzyme immobilization can lead to a reduction in enzymatic activity due to partial blocking of active sites during the immobilization process, increased mass transfer limitations between enzyme and substrate, and conformational changes. Therefore, the quest for simple and efficient immobilization methods that minimize these drawbacks is of paramount importance [4, 9].
Immobilized enzymes have a wide range of applications in different industrial bioprocesses, including biomedical, pharmaceutical, biotechnological, and bioengineering fields [2, 14, 15]. In recent years, numerous immobilization techniques and various solid supports have been successfully developed and applied in different industrial areas [5, 6, 16].
Among the diverse supports applicable to enzyme immobilization, the combination of biopolymers to form a hybrid support represents an interesting alternative, as it allows the combination of different characteristics in the same support (such as greater tolerance to different pHs, thermal resistance, and mechanical stability) [17,18,19]. It also allows the use of different immobilization techniques, such as covalent bonding, adsorption, entrapment, or encapsulation [2, 20]. In this context, the combination of sodium alginate with potato starch to form Ca alginate-starch beads has emerged as an attractive option. This combination results in a non-toxic, abundantly available, physiologically inert, cost-effective, and biocompatible support. Both starch and alginate possess these desirable properties, and the preparation of this hybrid support is straightforward [21]. Furthermore, alginate contributes with its gelling properties in the presence of divalent ions, facilitating the formation of alginate granules. On the other hand, the addition of starch primarily contributes to reducing enzymatic leaching, improving mechanical strength, and enhancing enzyme adhesion to the support in comparison to starch-free alginate beads [21,22,23,24,25,26].
Among the strategies employed for enzyme immobilization via covalent bonding, glutaraldehyde is commonly used as an activating agent due to its high versatility [27, 28]. Glutaraldehyde reacts with amino groups in proteins or hydroxyls in polymers, connecting the biopolymeric chains through inter- or intramolecular interactions [29], and can provide the immobilized enzyme with lower enzymatic leaching and greater thermal stability and durability, as well as mechanical improvements [30, 31].
Enzymes serve as biocatalysts, aiding in intricate chemical reactions within favorable experimental and environmental settings [32]. Horseradish peroxidase (HRP, EC 1.11.1.7), an oxidoreductase enzyme found in the roots of Armoracia rusticana, is widely used as an effective catalyst for the removal of phenols. Owing to its abundant availability and ease of extraction and purification, HRP has been extensively studied as an efficient and environmentally friendly biocatalyst for use in various industrial applications [3]. In the presence of hydrogen peroxide, HRP has the capacity to catalyze the generation of organic radicals, subsequently initiating the polymerization process of phenolic compounds [33,34,35]. However, similar to other enzymes, free HRP exhibits low catalytic stability under harsh processing conditions and is difficult to recover and isolate from the substrate and product. Therefore, immobilizing HRP to allow for its reuse and improved stability has proven to be a desirable task [3, 31,32,33].
Immobilized HRP can be employed in several areas, but significant effort has been dedicated to the degradation of environmentally harmful pollutants such as dyes. The presence of dyes in water bodies presents a risk to aquatic life due to reduced dissolved oxygen levels in the water, caused by the blocking of sunlight in the water system and resistance to photochemical reactions [36, 37]. The textile industry is the main source of dye-containing wastewater, as 10–25% of the applied dye is lost during different processes, and 2–20% is discharged into various environmental components. Similarly, food and pharmaceutical industries also have elevated levels of dyes in their wastewater [38]. Phenol red is one of the most toxic dyes linked to aquatic pollution and is widely used in pharmaceutical industries, coal processing, petroleum refineries, and dye and chemical industries. Given its high toxicity and carcinogenic effects, its removal from aqueous matrices is essential [39]. Considering that conventional methods of wastewater treatment suffer from drawbacks, such as high operational costs, generation of toxic by-products and sludge, and the need for additional treatments [40], enzymatic biodegradation has proven to be a potential alternative, as it has the advantages of environmental compatibility and reaction specificity [41, 42].
Previous studies have successfully demonstrated the biodegradative potential of immobilized HRP against various pollutants [3, 43,44,45,46]. However, no reports were found on its extraction and immobilization on a Ca alginate-starch hybrid support. Therefore, the present study aimed to immobilize HRP extracted from horseradish roots on a Ca alginate-starch hybrid support using glutaraldehyde as a cross-linking agent. This study evaluated the thermodynamic and kinetic properties, thermal and operational stability, and storage stability of free and immobilized HRP. Finally, the practical use of free and immobilized HRP for the degradation of phenol red dye was investigated in a batch reactor.
Materials and Methods
Chemical/Reagents
Food-grade sodium alginate (lot CHILE0I02511, purity > 99%, < 74 μm) was obtained from GastronomyLab (Brasília, Distrito Federal, Brazil). Soluble potato starch p.a. ((C6H10O5)n, < 75 μm, lot 23262, purity > 99%) was commercially obtained from Cinética (Itapevi, São Paulo, Brazil). Glutaraldehyde (lot STBJ8154, 50 wt. % in H2O, quality level: 200), sodium dodecyl sulfate (lot 129KO187V, purity > 99%), β-mercaptoethanol (lot STBG7404, purity > 99%), glycine (lot SLCK6757, purity > 98.5%), Tris (lot BCCG1869, purity > 99%), acrylamide (lot BCCC0307, purity > 99%), N,N,N′,N′-tetramethylethylenediamine (lot BCCC2915, purity > 99%), bovine serum albumin (lyophilized powder, A7906, lot SLBP1160V, 66 kDa, purity > 98%, quality level: 300), and horseradish peroxidase (lyophilized powder, 77332, 150 U/mg, lot BCBV2718, 40 kDa, quality level: 100) (used as HRP standard for electrophoresis) were obtained from Sigma-Aldrich (Burlington, MA, USA). Calcium chloride (lot DCBC4342, purity > 99%), dibasic sodium phosphate anhydrous (lot DCB00207, purity > 99%), and methanol (lot DCBC5308V, purity > 99%) were obtained from Vetec (Duque de Caxias, Rio de Janeiro, Brazil). Guaiacol (lot 53368, purity > 99%), Coomassie brilliant blue R-250 (lot 51452, purity > 98%), Coomassie brilliant blue G-250 (lot 41962, purity > 98%), and acetic acid (lot 60756, purity > 99%) were obtained from Neon Reagentes Analíticos (Suzano, São Paulo, Brazil). Potassium ferricyanide (lot 01946, purity > 99%) was obtained from Química Moderna (Barueri, São Paulo, Brazil). 4-Aminoantipyrine (lot 07101616, purity > 97%), bromophenol blue (lot 03112013, purity > 99%), and phosphoric acid (lot 15010026, 85 wt.% in H2O, purity > 99%) were purchased from Nuclear (Diadema, São Paulo, Brazil). Phenol p.a (lot 211897, purity > 99%), phenol red (lot 86285, quality level: 200), ammonium persulfate (lot 39794, purity > 98%), hydrogen peroxide (lot 138087, 30%, quality level: 100), sodium acetate (lot 216068, purity > 99%), sodium borate (lot 189408, purity > 98%), and ammonium sulfate (lot 249751, purity > 99%) were purchased from Synth (Diadema, São Paulo, Brazil). Boric acid (lot Q0011, purity > 99%) was purchased from Qhemis (Jundiaí, São Paulo, Brazil). All other chemical reagents used in this study were of analytical grade and used as received according to the supplier’s instructions. Horseradish roots (Armoracia rusticana) were commercially acquired in the city of Venâncio Aires, Rio Grande do Sul, Brazil.
HRP Extraction and Purification
The extraction of peroxidase from Armoracia rusticana roots was performed according to a methodology adapted from Muenchen et al. [47] and Fernandes et al. [48]. The mature horseradish roots, previously washed, peeled, and diced, were homogenized in a blender (Li-2.0-N, Skymsen, 50–60 Hz, 800 W, Brusque, Santa Catarina, Brazil) with 0.1 M sodium phosphate buffer solution (pH 7.0) at the ratio of 40 g of vegetable to 200 mL of buffer solution, for 3 min. The mixture was then kept under magnetic stirring for 15 min (22 ± 2 °C) and the crude extract was then filtered through four layers of gauze. The filtrate was centrifuged (CR21GIII, Hitachi, Tokyo, Japan) at 6000 rpm for 15 min at 4 °C.
After centrifugation, enzymatic precipitation was performed according to the methodology proposed by Zeraik et al. [49]. First, ammonium sulfate (NH4)2SO4 was slowly added to the supernatant under constant magnetic stirring at 4 °C until 40% saturation was reached. The solution was stored for 24 h under static conditions at 4 °C, followed by centrifugation at 10,000 rpm and discarding of the precipitate. Next, (NH4)2SO4 was slowly added to the supernatant until it reached 85% saturation, and the solution was kept under constant magnetic stirring at 4 °C. It was kept for 24 h under static conditions (4 °C) and centrifuged again at 10,000 rpm. The supernatant was discarded and the precipitate containing peroxidase was resuspended in a small amount of ultrapure water (Milli-Q, Millipore, Burlington, MA, USA) with conductivity equivalent to 18.2 MΩ/cm. Dialysis was then performed to remove ions and other salts present in the enzyme solution. For this, the enzymatic solution was transferred to a dialysis bag (3.5 kDa, Thermo Fisher Scientific, São Paulo, São Paulo, Brazil) and dialyzed against ultrapure water at 4 °C under constant magnetic stirring. The external solution was exchanged with the dialysis membrane every hour for a total time of 24 h. Finally, the enzyme solution was stored in Falcon-type polypropylene flasks under constant refrigeration (4 °C) and protected from light.
The enzyme purification factor (PF) was calculated by means of the ratio between the specific enzymatic activity of the enzyme solution after purification/dialysis (SEA2) and that in the crude extract (SEA1), according to Eq. 1.
where the specific enzymatic activity of the dialysate and the crude extract correspond, respectively, to the ratio between the enzymatic activity (U/mL) and the protein content (mg/mL) of the enzymatic solution after the dialysis process and between the enzymatic activity (U/mL) and the protein content (mg/mL) of the enzymatic solution after centrifugation at 6000 rpm.
The purification yield was obtained as the ratio between the total enzymatic activity after purification and the total enzymatic activity of the crude extract according to Eq. 2.
Polyacrylamide Gel Electrophoresis (SDS-PAGE)
To confirm the success of the purification and determine the molecular weight of the extracted HRP, we performed a polyacrylamide gel electrophoresis (PAGE) assay with sodium dodecyl sulfate (SDS), following the methodology described by Köktepe et al. [50]. The assay was conducted using a Mini Protean III electrophoresis cell (Bio-Rad, St. Louis, MO, USA). Sample solutions (16 μL) were homogenized with 4 μL of sample buffer solution (0.1% bromophenol blue, 0.5 M β-mercaptoethanol, 10% SDS, 2% glycine, and 0.5 M Tris–HCl) and heated in a water bath (MA159, Marconi, Piracicaba, São Paulo, Brazil) at 100 °C for 5 min. A 12% resolving gel (combining 2000 μL of 30% acrylamide, 1250 μL of 1.5 M Tris buffer, 50 μL of 10% SDS, 1700 μL of ultrapure water, 10 μL of tetramethylethylenediamine (TEMED) and 15 μL of 25% ammonium persulfate (APS)) and a 4.5% stacking gel (containing 330 μL of 30% acrylamide, 312 μL of 1 M Tris buffer, 25 μL of 10% SDS, 1830 μL of ultrapure water, 10 μL of TEMED and 10 μL of APS) were prepared. The gel was run at a constant current of 80 V for 3 h, followed by 110 V until the tracer dye reached the bottom of the plate. Proteins were stained using the Coomassie brilliant blue solution staining technique, which involved immersing the gel into a container containing the staining solution (0.1% Coomassie brilliant blue R-250, 25% methanol, 1% acetic acid, and 74% ultrapure water) and keeping it under constant stirring (60 rpm) on an orbital shaker table (AGK300D, MontLab, Campinas, São Paulo, Brazil) at room temperature (22 ± 3 °C) for 2 h. The gel was then washed with a bleaching solution (1% acetic acid, 6.25% methanol, and 92.75% ultrapure water) under constant stirring on an orbital shaker table (60 rpm) at room temperature (22 ± 3 °C). The molecular weight was estimated by comparison with molecular weight markers (14.4 to 116 kDa, Thermo Fisher Scientific, São Paulo, São Paulo, Brazil).
Determination of HRP Enzymatic Activity and Protein Quantification
To determine the enzymatic activity of free and immobilized HRP, guaiacol was used as the substrate, and the reaction was evaluated spectrophotometrically [51]. For free HRP, 100 μL of 0.1 M guaiacol, 100 μL of 0.01 M hydrogen peroxide, 20 μL of enzyme solution, and 2780 μL of 0.1 M sodium phosphate buffer (pH 6.0) were added into a 3-mL quartz cell. The reaction was conducted at room temperature (22 ± 3 °C). For immobilized HRP, the same volumes of hydrogen peroxide and guaiacol were used, followed by appropriate amounts of immobilized HRP and phosphate buffer for a total volume of 3 mL. The absorbance (λ = 470 nm) was read in an Ultraviolet–Visible/UV–Vis molecular absorption spectrophotometer (Genesys 10S, Thermo Scientific, Waltham, MA, USA) after 1 min of reaction at 25 °C. For the blank, the enzyme solution was replaced by 0.1 M sodium phosphate buffer (pH 6.0). Enzymatic activity was calculated from the absorbances obtained according to Eq. 3, where one unit of enzyme activity (U) of HRP corresponds to the amount of enzyme required to catalyze the conversion of 1 μmol of guaiacol (ε = 26,600 L/(mol.cm)) per minute.
where Δabsorbance is the difference between final absorbance (t = 1 min) and initial absorbance (at t = 0), Vtotal (mL) corresponds to the total volume of solution present in the cuvette, ε corresponds to the molar absorptivity coefficient of guaiacol (26,600 L/(mol.cm)), and Venzyme (mL) corresponds to the volume of enzyme solution added to the cuvette.
The total protein content was determined according to the Bradford method [52] using bovine serum albumin (BSA) as a standard. In this assay, the color of the complex formed by the reaction of Coomassie brilliant blue dye (G-250) with proteins, in the presence of phosphoric acid, is determined spectrophotometrically at 595 nm.
HRP Immobilization
HRP was immobilized on Ca alginate-starch beads through entrapment in a calcium alginate matrix and covalent bond formation using glutaraldehyde as a cross-linking agent, according to the methodology adapted from Bilal et al. [43]. The HRP enzyme solution obtained after dialysis (55.34 U/mL and protein content of 12 mg/mL) was added to a 20 mL solution containing predefined concentrations of sodium alginate (4.0% w/v) and potato starch (1.0% w/v) to obtain a final concentration of 50 mg of protein per g of support (corresponding to 9.22 U/mL of gel). The mixture was left under gentle magnetic stirring for approximately 5 min at room temperature (22 ± 3 °C). Then, 400 μL of glutaraldehyde solution (1% v/v) was added and mixed by magnetic stirring. The obtained gel was extruded dropwise into a CaCl2 solution (200 mM) using a syringe (needle diameter of 1 mm) under gentle magnetic stirring at 4 °C (employing an ice bath for temperature maintenance). The immobilized HRP beads were separated using a sieve, washed with ultrapure water, and immersed in a glutaraldehyde solution (0.02% v/v) at 4 °C without stirring for further hardening. Afterward, the beads were washed extensively until no proteins were visible in the washing solutions by UV–Vis spectrophotometer (λ = 280 nm) and stored in polypropylene Falcon tubes at 4 °C protected from light. Ca alginate-starch beads without added enzyme were used as the control, and the same procedure was followed as described above.
The average bead size and spherical equivalent diameter were determined by employing the methodology described by Urrea et al. [31]. The average bead size (VB, mL) was determined as the ratio of the volume of the alginate/HRP solution used (VA, mL) to the total number of beads obtained (NB). The equivalent spherical diameter (DB, mm) was then calculated using Eq. 4.
where 10 (mm/cm) corresponds to the conversion factor.
Evaluation of the Biocatalytic Performance of Immobilized HRP
The biocatalytic performance of immobilized HRP was evaluated according to the methodology described by Sheldon and Van Pelt [53]. Immobilization yield, immobilization efficiency, and recovery of immobilized activity were evaluated. Equation 5 was used to determine the percentage immobilization yield (IY).
The immobilization efficiency (IE) was calculated according to Eq. 6, corresponding to the amount of enzyme that remained active and accessible/functional after immobilization.
Another parameter that gives an idea of the success of total immobilization is activity recovery (AR) [53], defined as the multiplication of the immobilization yield (IY, %) by the immobilization efficiency (IE, %) divided by 100. Equation 7 was used to calculate the AR.
Characterization of Free and Immobilized HRP
Optimal pH and Temperature Conditions
To determine the optimal pH for free and immobilized HRP, enzymatic activity was evaluated over a pH range of 4.0 to 9.0 at 25 °C, according to the methods of Bilal et al. [54] and Zeyadi and Almulaiky [55]. To adjust the pH, acetate (100 mM, pH 4.0 to 5.5), phosphate (100 mM, pH 6.0 to 7.5), and borate (100 mM, pH 8.0 to 9.0) buffers were used. To evaluate the effect of temperature variation on the enzymatic activity of free and immobilized HRP, we subjected the enzymes to temperatures ranging from 20 to 85 °C (pH 6.0) [55]. Thus, the mixtures of guaiacol, hydrogen peroxide, and phosphate buffer (in the volumes and concentrations described in the “Determination of HRP Enzymatic Activity and Protein Quantification” section) were placed in a laboratory water bath and heated to the desired temperature. The enzyme was then added, and the enzymatic activity was measured after 1 min of reaction, as per “Determination of HRP Enzymatic Activity and Protein Quantification” section. The enzymatic activity of both assays (pH and temperature) was evaluated in static conditions. The enzymatic activity of free and immobilized HRP under optimal pH or temperature conditions was considered 100% activity, and the other activities of the same parameter were calculated as relative percentages.
Kinetic Parameters
The determination of the kinetic parameters of free and immobilized HRP was performed by varying the concentration of guaiacol (1 to 10 mM) and, subsequently, of hydrogen peroxide (0.2 to 2.0 mM). The enzyme activity assay was performed following the methodology described in the “Determination of HRP Enzymatic Activity and Protein Quantification” section. The Michaelis–Menten constant (KM) and the maximum reaction speed (Vmax) were determined using the Michaelis–Menten model and Lineweaver–Burk linearization, according to Eq. 8.
where Vi (mM/min) is the catalytic reaction rate, KM (mM) is the Michaelis–Menten constant, Vmax (mM/min) is the maximum reaction velocity, and [S] is the substrate concentration (mM). The turnover number (kcat) (1/min) for free and immobilized HRP was calculated using Eq. 9.
where Vmax (mM/min) is the maximum reaction velocity and [ET] corresponds to the total concentration (mM) of enzymes in the reaction medium.
The catalytic specificity constant (1/(min.mM)) for the free and immobilized HRP was calculated as the ratio between the turnover number (kcat) (1/min) and the Michaelis–Menten constant (KM) (mM), according to Eq. 10.
Thermostability and Thermodynamic Parameters
To evaluate the thermostability of free and immobilized HRP, the enzymes (free or immobilized) were incubated in phosphate buffer solution (pH 6.0, 0.1 M) at different temperatures (65, 68, 70, and 72 °C). A laboratory water bath was used to maintain the temperature, and the residual enzyme activity was checked as described in the “Determination of HRP Enzymatic Activity and Protein Quantification” section every 5 min for a total period of 30 min of incubation. Dedicated enzyme and buffer mixtures were used for each collection time at each evaluated temperature. The entire procedure was performed in triplicate. To calculate the residual enzyme activity, the initial activity was taken as 100%, and the others were calculated as relative activities using Eq. 11.
where R corresponds to residual enzyme activity, UF to activity after incubation at the temperature evaluated, and UI to initial activity.
The thermodynamic parameters were calculated according to the deactivation model described by Zeyadi and Almulaiky [55] and Rigo et al. [56]. The deactivation energy (Ed) was determined using nonlinear regression and considering Arrhenius law, according to Eq. 12.
where kd (1/min) is the deactivation constant, A (1/min) is the pre-exponential factor, Ed (J/mol) is the deactivation energy, R (8.314 J/(mol.K)) is the universal gas constant, and T (K) is the temperature.
The enzyme half-life (t1/2) corresponds to the time required for the enzyme to lose half of its initial activity, that is, the time required for the initial enzymatic activity to drop to 50% [57]. Equation 13 was used to determine the half-life.
where t1/2 (min) is the half-life of the enzyme and kd (1/min) is the kinetic constant for deactivation.
The stabilization factor (SF) at a specific temperature can be calculated by dividing the half-life of immobilized HRP by the half-life of free HRP [58]. To calculate the Gibbs free energy (ΔG) of thermal inactivation at different temperatures from the first-order velocity constant of the inactivation process, Eq. 14 was employed.
where R (8.314 J/(mol.K)) is the universal gas constant, T (K) is the temperature, kd (1/min) is the kinetic deactivation constant, h (6.626 × 10–34 J.s) is Planck’s constant, and k (1.381 × 10−23 J/K) is Boltzmann’s constant.
From the inactivation energy, the enthalpy (ΔH) was calculated according to Eq. 15.
where Ed (J/mol) is the inactivation energy, R (8.314 J/(mol.K)) is the universal gas constant, and T (K) is the temperature.
From the Gibbs free energy, the entropy (ΔS) was calculated according to Eq. 16.
The decimal reduction time or D-value corresponds to the exposure time of an enzyme to a temperature that preserves 10% of the enzyme activity [55]. For the determination of the D-value, Eq. 17 was used.
where kd (1/min) is the kinetic constant for deactivation.
Storage Time
The free HRP solution and the dried beads of immobilized extracted HRP were stored in closed polypropylene Falcon tubes, protected from light, at 4 °C (under refrigeration) and 25 °C (room temperature) for 3 months. We evaluated the residual enzymatic activity over time, following the procedure outlined in the “Determination of HRP Enzymatic Activity and Protein Quantification” section, to verify the enzymatic stability during the storage period. The initial enzymatic activity (first day) was considered 100% activity, and subsequent activities were calculated as a percentage relative to the initial activity.
Structural Characterization of Supports and Derivatives
The structural and morphological characterization of the hybrid support, the extracted HRP, and the immobilized HRP was performed by means of Fourier transform infrared spectroscopy with attenuated total reflectance (FT-IR-ATR) and scanning electron microscopy with energy dispersive spectroscopy (SEM/EDS) assays. The FT-IR-ATR assay was performed at the University of Santa Cruz do Sul (UNISC) and the SEM/EDS assay was performed at the Science and Technology Park (TECNOVATES) of the University of Vale do Taquari. Before conducting the FT-IR-ATR and SEM/EDS assays, the samples were dehydrated according to methodologies adapted from Cunha et al. [59] and Taketa et al. [60].
For the FT-IR-ATR assay, the samples were positioned on a universal UATR accessory (registration number L1250050, PerkinElmer, Waltham, MA, USA) with a horizontal base and single beam reflection on a diamond crystal, equipped with a circular zinc selenide photocatalyst (2 mm diameter) positioned in an FT-IR/FT-NIR Spectrum 400 spectrometer equipment (PerkinElmer, Waltham, MA, USA). Spectra were obtained using a 6333 nm He/Ne laser, wavelength of 4000–400 cm−1, 4 scans, resolution of 4 cm−1, and DTGS detector, according to the methodology adapted from Alver and Metin [61] and Souza and Corbellini [62].
The morphologies of the hybrid support and immobilized HRP were verified by SEM (Carl Zeiss, LS-10, Oberkochen, Baden-Württemberg, Germany) with an accelerating voltage of 10 kV. The dehydrated beads were fixed with carbon tape on a stub and covered with a thin gold layer. The elemental distribution was analyzed by energy dispersive spectroscopy (EDS) [63].
Biodegradation of Phenol Red Dye
Phenol Quantification
The phenolic content present in the phenol red dye solution was quantified using the colorimetric method, employing potassium ferricyanide and 4-aminoantipyrine, according to the methodology adapted from Zhang and Cai [64] and Vineh et al. [65]. To construct the calibration curve, 600 μL of borate buffer (0.1 M, pH 9.0), 600 μL of potassium ferricyanide solution (4.0% w/v), 600 μL of 4-aminoantipyrine (4-AAP) solution (0.01% w/v), and 50 μL of aqueous phenolic solution at concentrations of 10, 25, 50, 100, 200, 300, 400, and 500 mg/L were employed. After 10 min of reaction, the absorbance was read in a UV–Vis spectrophotometer at 510 nm. For the blank, 650 μL borate buffer (0.1 M, pH 9.0), 600 μL 4.0% potassium ferricyanide, and 600 μL 0.01% 4-AAP were employed. Using the absorbance values obtained, a calibration curve was drawn, which was subsequently used to determine the phenolic concentration in the phenol red dye.
Biodegradation of Phenol Red Dye by Free and Immobilized HRP
Biodegradation of phenol red dye was performed in dedicated flasks containing phenol red dye solution at a concentration of 100 mg/L and pH 6.0, similar to what has been previously reported by Yaseen and Scholz [66] in crude textile effluent, hydrogen peroxide (concentration in solution of 0.1 M), and adequate amounts of free or immobilized extracted enzyme to obtain 0.0015 U per mL of dye solution. The experiments were conducted at 22 ± 3 °C under constant magnetic stirring in a batch system. The biodegradation of the dye was monitored for a total period of 90 min, and samples were collected over time (1, 5, 10, 20, 30, 40, 60, and 90 min). In addition, different biodegradation tests (with and without free enzyme and/or on alginate beads, as well as in the presence and absence of hydrogen peroxide) were performed to confirm that the biodegradation in fact occurred due to enzymatic catalytic activity.
The biodegradation of phenol red dye was evaluated using the colorimetric method for phenol quantification (the “Phenol Quantification” section), verifying the phenolic concentration remaining in the solution after the designated time periods. All experiments were performed in triplicate and the results were expressed as mean values. The biodegradation rate was calculated for each condition based on Eq. 18 [65].
where C0 and Cf are the concentration of phenol (mg/L) before and after treatment, respectively.
Reusability of Immobilized HRP
The reusability of immobilized HRP was evaluated through the biodegradation of phenol red dye (100 mg/L, pH 6.0) with hydrogen peroxide (0.1 M) and immobilized HRP (0.0015 U/mL) in a batch reactor with continuous magnetic stirring at room temperature (22 ± 3 °C). At the end of each cycle, the immobilized HRP was separated, washed with ultrapure water, and reused for the biodegradation of a new aliquot of dye solution for a total of 15 cycles. At the end of each cycle, an aliquot of the supernatant was collected for the determination of the phenolic concentration, according to the procedure described in “Phenol Quantification” section. The initial biodegradation obtained in the first cycle was considered 100%, and subsequent biodegradation rates were calculated as relative percentages. Throughout the reuse cycles, the residual enzymatic activity was evaluated following the procedure described in the “Determination of HRP Enzymatic Activity and Protein Quantification” section, with the enzymatic activity after the first cycle corresponding to 100% and the following activities calculated as a relative percentage. All experiments and analyses were performed in triplicate. Also, after 15 cycles of reuse, a quantity of the immobilized HRP beads was collected, washed, dried, and analyzed using SEM and EDS, as previously described in the “Structural Characterization of Supports and Derivatives” section.
Statistical Analysis
All experiments were performed in triplicate. The results obtained were submitted to analysis of variance (ANOVA) and Tukey’s test with a 95% confidence level (p < 0.05) using the software PAST version 4.03.
Results and Discussion
Extraction and Purification of HRP
HRP was extracted and purified from horseradish roots. The three purification steps for the crude HRP enzyme solution are shown in Table 1. The crude HRP enzyme solution showed a specific enzyme activity of 1.23 U/mg. The HRP was precipitated with ammonium sulfate to a maximum of 85% saturation, resulting in a specific enzyme activity of 3.37 U/mg, representing a 2.73-fold purification. After dialysis, the specific activity obtained was 4.61 U/mg, resulting in a 3.74-fold purification with a yield of 37.74% ± 0.77. This yield was found suitable for the intended application.
Guo et al. [67] performed the extraction and purification of lignin peroxidase from the fungus Pichia methanolica and obtained a purification factor of 2.02 after dialysis, a value lower than that observed in the present study. Joel et al. [68] obtained a purification factor of cabbage peroxidase of 3.88 after precipitation with (NH4)2SO4, with a yield of 56.5%. After dialysis, the purification factor was 19.32, with a yield of 43.03%, which was higher than that obtained for the extracted HRP in the present study. Additionally, Guo et al. [67] and Joel et al. [68] observed a significant improvement in the purification factor of the crude extract after using a Sephadex chromatographic column following the dialysis process. This indicates that greater purification could be obtained for the HRP extracted in this work if chromatographic techniques were employed after dialysis. However, chromatographic techniques directly impact the cost of obtaining HRP, making it more expensive [69].
HRP Characterization
The SDS-PAGE assay can be used to obtain information about molecular weights, protein combinations, and degree of purity [55]. In Fig. 1, the results obtained for the SDS-PAGE assay are presented. Both the standard HRP (columns 1 and 2) and the enzyme solution obtained after dialysis (columns 3 and 4) showed the presence of protein bands between 25 and 66.2 kDa, with the main bands observed at approximately 27 kDa, 44 kDa, and 65 kDa. The bands observed between 25 and 60 kDa can be attributed to different peroxidases present in the roots, resulting from differences in amino acid sequences or the degree of glycosylation [50]. In columns 1, 2, 3, and 4, the presence of the band often attributed to HRP around 44 kDa is observed [70]. The protein bands present around 65 kDa are commonly attributed to horseradish myrosinase [71, 72] and could be observed with higher intensity in the standard HRP solution.
Columns 5 and 6, containing the crude extract, showed the presence of several low-intensity bands, indicating the presence of proteins with different molecular weights. After the purification and dialysis process (columns 3 and 4), the number of observed bands decreased compared to the crude extract (columns 5 and 6), as well as the concentration of some proteins, resulting in a higher intensity of staining on the gel. This confirms that precipitation with (NH4)2SO4 followed by dialysis was able to promote an adequate purification of the extracted HRP enzyme solution.
Immobilization of HRP on Ca-Alginate/Starch Hybrid Support
The extracted HRP was covalently immobilized by entrapment on a Ca alginate-starch hybrid support using glutaraldehyde as a cross-linking agent to improve thermostability, catalytic characteristics when compared to free enzyme, and to allow enzyme reuse. The addition of 50 mg of protein/g support (9.22 U/mL) resulted in an immobilization yield of 88.33% ± 0.98, an efficiency of 56.89% ± 2.20, and an activity recovery of 50.26% ± 2.25. The immobilization yield obtained in the present study was slightly higher than the immobilization yield obtained by Bilal et al. [43] of 86.27% for HRP that had been extracted and immobilized on Ca alginate beads cross-linked with glutaraldehyde. In another study, Bilal and Asgher [73] obtained an immobilization yield of 89.3% for the immobilization of manganese peroxidase on Ca alginate beads. Among the main difficulties reported for HRP immobilization is enzyme leakage, which directly impacts the immobilization yield. Therefore, although the addition of starch to the support composition indicates an improvement in immobilization yield, and the cross-linking performed with glutaraldehyde can help to further minimize enzyme leakage, these strategies cannot completely eliminate it [31].
The efficiency value obtained in the present study was higher than that observed by Melo et al. [74] for the immobilization of HRP on chitosan–polyethylene glycol nanoparticles (51.7%), indicating that the Ca alginate-starch hybrid support better preserved the catalytic activity of HRP, possibly due to the better enzyme stabilization and intermolecular crosslinking caused by the addition of glutaraldehyde [29]. Activity recovery is directly influenced by the values obtained for the immobilization yield and efficiency. Also, the efficiency and activity recovery are influenced by the pore size of the gel, so beads with small pores impose a restriction on the diffusion of the substrate to the active site of the enzyme, resulting in a decrease in the values of these parameters [44].
The spherical equivalent diameter (DB) of the alginate beads obtained from 20 repetitions of 1 mL extrusion of the Ca alginate-starch gel was also evaluated. The number of beads obtained per mL of extruded solution ranged from 40 to 43. The DB obtained for the granules was 3.59 ± 0.04 mm, similar to those obtained by Shen et al. [75] in the production of Ca alginate beads for phosphorus adsorption, and by Urrea et al. [31] in the immobilization of HRP on Ca alginate beads. The drip extrusion technique, used in the present study, is typically employed to produce beads with a diameter between 2 and 5 mm [76]. One of the main parameters influencing the size of alginate beads is the diameter of the extrusion needle used [77], so larger gauge needles result in larger diameter beads.
Characterization of Free and Immobilized HRP
Effects of Different pH and Temperature Values on Enzyme Activity
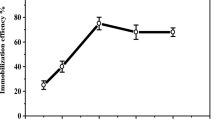
The pH and temperature parameters influence the activity of various enzymes and, consequently, their application in various industrial segments [78]. In this study, the influence of these parameters on the activity of free and immobilized HRP was studied by incubating the enzyme at pH values ranging from 4.0 to 9.0 and temperatures between 20 and 85 °C, and the enzyme activity was evaluated using a UV/Vis spectrophotometer. The results obtained for the enzymatic activity of free and immobilized HRP are shown in Fig. 2A. Free HRP showed maximum catalytic activity at pH 6.5, whereas immobilized HRP showed maximum catalytic activity at pH 5.0. The shift in optimum pH caused by enzyme immobilization can be explained by ionic interactions between the support matrix and H+ or OH− ions of the buffer solution, which can influence the enzyme microenvironment and, consequently, the optimum pH [43]. Furthermore, the specific method of immobilization employed and the structure and load of the matrix can contribute to the observed optimum pH shift [35]. Structural changes in HRP caused by immobilization can also influence its optimal pH [79, 80]. Free HRP demonstrated considerable stability at pH 5.5 to 7.0, showing 96.44% relative activity at pH 5.5, and 98.70% at pH 7.0. However, when exposed to pH values below 5.5, free HRP showed a considerable decrease in its catalytic activity, with only 68.04% activity remaining at pH 4.0. This profile was also observed with greater intensity at pH values above 7.0, resulting in a residual enzyme activity of 35.28% at pH 9.0.
Effect of pH (A) (temperature 25 °C, H2O2 concentration 0.3 mM in solution, guaiacol concentration 3.3 mM in solution) and temperature (B) (at pH 6.0, H2O2 concentration 0.3 mM in solution, guaiacol concentration 3.3 mM in solution) on free and immobilized horseradish peroxidase activity: (black diamond (♦)) free horseradish peroxidase and (white square (□)) immobilized horseradish peroxidase. Each point corresponds to the mean ± standard deviation of three independent experiments performed in triplicate. Note: A free horseradish peroxidase, where 100% relative activity (pH 6.5) is equivalent to 1.16 U/mL, and immobilized horseradish peroxidase, where 100% relative activity (pH 5.0) is equivalent to 0.66 U/mL; B free horseradish peroxidase, where 100% relative activity (at 60 °C) is equivalent to 4.165 U/mL, and immobilized horseradish peroxidase, where 100% relative activity (at 50 °C) is equivalent to 0.875 U/mL
In contrast, immobilized HRP was stable at pH values from 5.0 to 6.0 and, like free HRP, also showed a loss of catalytic activity as the pH values moved away from this stability range. However, the decrease observed for immobilized HRP was less than that for free HRP at acidic pH, with residual enzymatic activity at pH 4.0, corresponding to 82.68%. In alkaline pH, immobilized HRP was more sensitive than free HRP for values from 6.5 to 8.0, and more resistant at pH values of 8.5 and 9.0, showing a catalytic activity of 67.79% and 61.44%, respectively. The higher resistance to the decrease in catalytic activity observed for immobilized HRP compared to free HRP, especially at acidic (4.0 and 4.5) and alkaline (8.5 and 8.0) pH values, can be attributed to the presence of electrostatic interactions between the support and the biocatalyst, causing a lower impact on the biologically active conformation and consequently, better resistance to pH variations [3, 45, 79]. Furthermore, multipoint covalent attachment can restrict protein flexibility, which may protect the immobilized enzyme from denaturation at high acidic and alkaline pH levels, typically resulting in an expanded pH profile for the immobilized enzyme [81].
Similar to the findings of the present study, other authors observed a shift in the optimum pH for immobilized HRP employing the most varied supports [33, 82, 83]. Additionally, the broad catalytic activity exhibited by immobilized HRP in different pH ranges is an interesting factor regarding the treatment of compounds in aqueous matrices, which generally show pH levels close to neutrality, but can suffer variations to more acidic or alkaline ranges [66].
The influence of temperature on the catalytic activity of free and immobilized HRP is shown in Fig. 2B. Free HRP showed an increase in catalytic activity up to its maximum at 60 °C, after which enzymatic activity declined, resulting in only 0.54% activity at 85 °C. The increase in enzyme catalytic activity up to 60 °C can be explained by the increase in the enzyme reaction rate. However, temperatures higher than 60 °C led to denaturation of the protein structure and consequent loss of catalytic activity [84]. Immobilized HRP showed an increase in catalytic activity until 50 °C (optimal temperature) was reached. For temperatures above 50 °C, the immobilized HRP was shown to be thermally stable, preserving 90.15% of the catalytic activity at 85 °C. Immobilized HRP was found to be more thermally stable than free HRP at most of the temperatures evaluated. The enhanced thermal resistance of immobilized HRP can be attributed to the protection conferred by the presence of covalent bonds between the support and the enzyme, in addition to the entrapment of HRP, reducing conformational changes in the enzyme structure at high temperatures, preventing denaturation [32, 34]. The results obtained in the present work regarding temperature are consistent with previous studies involving HRP, which have demonstrated that enzyme immobilization confers greater thermal stability to the enzyme [32, 82, 85, 86].
Kinetic Parameters of Free and Immobilized HRP
The immobilization process affects the distribution of the substrate in the active sites of the enzyme [32], impacting the observed kinetic parameters. In this study, the values of kM, Vmax, kcat, and kcat/kM of free and immobilized HRP were evaluated for guaiacol and hydrogen peroxide, and the results are shown in Table 2 (for more details, see Fig. S1). After immobilization, there was an increase in the values obtained for kM and a decrease in Vmax, kcat, and kcat/kM when immobilized HRP was compared to free HRP for both guaiacol and hydrogen peroxide. When comparing guaiacol and hydrogen peroxide for the same enzymatic form (free or immobilized), higher values were obtained in all parameters for hydrogen peroxide, indicating a higher enzymatic affinity for guaiacol (due to higher kM values). In addition, higher Vmax, kcat, and kcat/kM values observed for hydrogen peroxide compared to guaiacol indicate that the hydrolysis of this molecule occurs faster than the consumption of guaiacol for product formation (tetraguaiacol). The increase observed for kM after immobilization indicates a decrease in the affinity of the enzyme for the substrate, which may stem from conformational changes, reduced enzyme flexibility, and diffusion constraints of substrate and products [4]. In the immobilization of HRP on Ca alginate-starch beads, the enzyme is surrounded by the support, which partially blocks the active site, also reducing access to the substrate [83]. Additionally, lower kM values for guaiacol indicate a higher enzyme affinity for guaiacol compared to hydrogen peroxide. The decrease in other parameters after immobilization happens in conjunction with the increase in kM. Vmax indicates the rate at which the enzyme becomes saturated by substrate, and its decrease implies a lower substrate concentration required for all catalytic sites to be filled at a given time. As with kcat, changes in Vmax are commonly observed in enzyme immobilizations, and are linked to conformational changes in the enzyme, electrostatic interactions between the enzyme and the medium, the nature of the support, and the microenvironment [87].
kcat corresponds to the number of substrate molecules that an enzyme can convert into product per unit time [88]. A reduction in its value, as observed after the immobilization of HRP, results in a lower rate of product formation per unit time. In addition, kcat/kM is related to the catalytic efficiency of the enzyme [89], and a reduction in its value suggests that HRP immobilization has a lower overall catalytic potential than free HRP.
Thermal Inactivation and Thermodynamic Parameters of Free and Immobilized HRP
Thermal stability plays a significant role in many industrial applications, where high temperatures are required to catalyze the reaction rate [90]. In this regard, we evaluated the thermal stability of both free and immobilized HRP by thermal inactivation, and the results obtained are shown in Table 3 (for more details, see Fig. S2 and Fig. S3).
The kd values increased for the same enzymatic form (free or immobilized) as the temperature increased, indicating lower thermal stability at higher temperatures. However, after immobilization, lower kd values were obtained at the same working temperature, demonstrating that immobilization contributed to an increase in thermal stability. In addition, immobilized HRP presented higher values of t1/2 and D-value. The t1/2 corresponds to the time required to reduce the enzyme activity to 50% of the initial activity [57], while the D-value is the exposure time of the enzyme at a given temperature capable of preserving 10% of the initial enzyme activity [55]. An increase in the values of these parameters after immobilization indicates that there was an improvement in thermal stability, as the immobilized enzyme requires a longer exposure time to achieve the same percentage of inactivation as free HRP at the evaluated temperatures. The higher stability conferred to immobilized HRP may be related to a decrease in protein–protein interactions and a reduction in the intensity of macromolecular vibrations when subjected to high temperatures. This leads to an increase in enzyme rigidity caused by proper attachment to the support and protection of the enzymes by the alginate matrix [14, 58, 65, 89].
Finally, the SF values indicate how many times the t1/2 of immobilized HRP was higher than that of free HRP. Because immobilized HRP was more thermally stable than free HRP at all working temperatures, SF values > 1 were observed, which was more pronounced at lower temperatures. A reduction in t1/2 values and D-value was observed as the temperature increased for both free and immobilized HRP, indicating that the higher the exposure temperature, the less time the enzyme could withstand without loss of catalytic activity. These results are consistent with those observed in other works involving HRP immobilization, where a higher thermal stability after immobilization was also observed [83, 86, 90].
Thermodynamic parameters were also evaluated, and the results obtained are displayed in Table 4. Upon immobilization of HRP, an increase in all evaluated parameters was observed at the same temperature. Different thermodynamic parameters, such as Gibbs free energy (ΔG), enthalpy (ΔH), and entropy (ΔS), provide a comparative measure of enzyme functionality and stability in different operating environments [91]. In the present study, all ΔG values were positive, revealing that the processes were endergonic and not spontaneous [92]. In this regard, for all temperatures evaluated, there was an increase in the ΔG, ΔH, and ΔS values of HRP after immobilization. An increase in ΔG values after immobilization indicates that there is less enzyme susceptibility to denaturation, providing insight into the additional amount of energy required for HRP denaturation [91]. When comparing the ΔG values at different temperatures, there was a brief increase for free HRP, while immobilized HRP showed a decrease, indicating that immobilization provided lower resistance to protein unfolding caused by heat, possibly due to the decrease in active site availability resulting from protein denaturation and conformational changes [58, 93].
The observed increase in ΔH of HRP after immobilization indicates high thermal stability. The ΔH value is related to the degree of breaking of the transition state stabilization bonds (active stable state to a denatured inactive state), and higher ΔH values indicate a higher energy demand for the thermal denaturation of HRP [90, 94]. This increase can be attributed to the formation of covalent bonds between the enzyme and the support, resulting in decreased conformational flexibility and preventing thermal unfolding and denaturation. Additionally, the positive value of ΔH indicates that the catalytic reaction is endothermic [92]. Comparing the ΔH values with increasing temperature, there was a decrease in the values for both free and immobilized HRP, indicating a lower amount of energy required for enzyme denaturation at elevated temperatures.
ΔS is related to the thermal unfolding of the enzyme, and thermal deactivation causes the unfolding of the enzyme structure, leading to enzyme disorder or deactivation entropy [92]. The higher values observed for immobilized HRP indicate greater disorder in the system, with no enzyme aggregation, whereas the negative values observed for ΔS of free HRP are related to a decrease in disorder or randomness of the enzyme system, as well as aggregation. Furthermore, an increase in temperature caused a decrease in the ΔS values for both free and immobilized HRP because of greater enzyme denaturation at higher temperatures and enzyme aggregation. The enzyme inactivation energy (Ed) is the lowest energy required to initiate enzyme denaturation [91]. The present study showed that the enzyme Ed increased after the immobilization process, indicating a higher thermal stability for the immobilized HRP; therefore, additional energy is required to break down the immobilized form of HRP compared to its native free form. These results suggest obtaining a stable immobilized biocatalyst that is less sensitive to temperature changes than free enzymes [95], due to the protection conferred by the support and the bonds formed towards the enzyme.
Storage Time
Immobilization is one of the most promising approaches to improve enzyme stability, operability, and reusability. Aiming at large-scale applications, obtaining reusable and stable biocatalysts is of great interest because it directly affects operational costs [83]. In this sense, the stability of free and immobilized HRP was evaluated for a total period of 120 days at two different temperatures (4 °C and 25 °C), and the observed results are shown in Fig. 3.
Enzyme activity during storage time (120 days, H2O2 concentration 0.3 mM in solution, guaiacol concentration 3.3 mM in solution, pH 6.0): (black triangle (▲)) free horseradish peroxidase (4 °C); (white square (□)) immobilized horseradish peroxidase (4 °C); (black circle (●)) free horseradish peroxidase (25 °C) and (white triangle (Δ)) immobilized horseradish peroxidase (25 °C). Each point corresponds to the mean ± standard deviation of three independent experiments performed in triplicate. Note: free horseradish peroxidase (4 °C), 100% relative activity at day 0 corresponds to 1.47 U/mL; immobilized horseradish peroxidase (4 °C), 100% relative activity at day 0 corresponds to 0.52 U/mL; free horseradish peroxidase (25 °C), 100% relative activity at day 0 corresponds to 1.71 U/mL; immobilized horseradish peroxidase (25 °C), 100% relative activity at day 0 corresponds to 0.50 U/mL
The immobilized HRP stored at 4 °C demonstrated greater storage stability until day 45, retaining 93.71% of its initial activity, compared to free HRP (4 °C), which retained 89.63%. However, after 45 days, there was a significant decrease in the residual enzymatic activity of immobilized HRP (4 °C), and this phenomenon was not observed in such intensity for the free HRP stored at the same temperature. The loss of catalytic activity can be attributed to conformational changes that may have occurred during the storage period [65]. The decrease in peroxidase activity could be related to enzyme unfolding and subsequent denaturation [96]. In the comparison between free and immobilized HRP stored at 25 °C, it was observed that the free enzyme had higher stability, retaining 66.61% of the initial catalytic activity, whereas the immobilized HRP retained only 23.56% activity at the end of 120 days. It also became evident that storage at 4 °C allows superior maintenance of enzymatic activity for up to 45 days as compared to storage at 25 °C. After this period, the enzymatic activity of free HRP approaches the activity observed for the same enzymatic form at 25 °C, while for the immobilized HRP, the catalytic activity decreases to values lower than the immobilized HRP at 25 °C. The loss of catalytic activity over time can be attributed to possible conformational changes in the enzyme structure, preventing the formation of a catalytically active complex between substrates and active sites [46, 65].
Morphology and Structure of Immobilized HRP
The surface morphologies of the alginate, Ca alginate-starch support, and immobilized HRP beads were evaluated by SEM, and the images obtained are shown in Fig. 4. The formation of alginate beads without the presence of starch in their composition (Fig. 4A and B) resulted in a smoother surface when compared to the surface of the beads containing starch (Fig. 4C and D). Thus, the addition of starch results in an increase in the surface area of the beads, which is still present on the surface of the beads containing immobilized HRP (Fig. 4E and F). However, the surface of the beads containing the immobilized HRP underwent smoothing compared to the surface of the beads without enzyme. Morphological changes on the surface of alginate beads after enzyme immobilization have been reported in previous studies, suggesting that the interaction between the enzyme and support can cause surface structural modifications [97,98,99,100].
To study the surface chemistry and the functional groups involved in alginate, starch, free extracted HRP, hybrid support, and immobilized HRP, the samples were subjected to FT-IR-ATR spectroscopy. The obtained spectra are shown in Fig. 5. For alginate, a characteristic broad peak was found to be present at 3234 cm−1, which was caused by the stretching vibrations of the −OH groups [101]. This peak was also observed in the spectra of starch (3294 cm−1), free extracted HRP (3286 cm−1), hybrid support (3236 cm−1), and immobilized HRP (3256 cm−1). In the alginate spectrum, there was also a peak at 1597 cm−1, which can be attributed to the carbonyl (C = O) of the COO− group (carboxylate anion). The peaks at 1406 and 1314 cm−1 are attributed to the symmetric stretching of COO−, the peak at 1084 cm−1 can be attributed to C–O–C stretching [102], and the peak at 1025 cm−1 is attributed to the OH group of guluronate [103].
In the case of starch, two additional peaks to the one at 3294 cm−1 were observed: one at 2934 cm−1 and another at 2886 cm−1, resulting from C-H stretching vibrations of CH2 or CH3 [104]. Peaks between 1500 and 1300 cm−1 are commonly assigned to multiple vibration modes containing -CH (-CH and -CH2 groups) and OH deformation. A peak attributed to the stretching vibrations of carbonyl groups (C = O) was observed around 1643 cm−1 [24], at 1078 cm−1 to C–C and C-O stretching, and at 996 cm−1 to C-O–H deformation vibration [104, 105]. The peak at 3286 cm−1 in the free extracted HRP spectra can be assigned to the OH stretching of hydroxyl groups (−OH) or NH stretching vibration [21]. The peaks at 2920 and 2852 cm−1 were assigned to alkyl chains (-CH2), and those at 1635 and 1517 cm−1 were assigned to amide I (α-helix structure) and amide II, respectively [106]. The peak at 1397 cm−1 was attributed to various NH modes (amide III) and that at 1075 cm−1 to the C-N stretching vibration of aliphatic amines [107].
The spectrum of the hybrid support showed similarities to that of alginate, which is the main component of the support. As a result, the starch peaks overlap with those of alginate. However, a small peak shift was observed, which may be due to the formation of intermolecular hydrogen bonds between starch and alginate [24]. Comparing the spectrum of the immobilized HRP with that of the hybrid support, no significant differences were observed, as the ratio of enzyme to the amount of alginate was small, and the alginate bands were predominant. Nevertheless, a slight increase in peak intensity was observed, possibly due to interactions between the enzyme and the support, as well as interactions between the amino acid constituents of the enzyme. Furthermore, an increase in the absorption bands observed at 1416 and 1010 cm−1 can be attributed to the C-N and C-O vibrational stretching of the amino acids of HRP [21].
Biodegradation of Phenol Red Dye by Free and Immobilized HRP
Horseradish peroxidase is capable of degrading phenols and substituted phenols through a free-radical polymerization mechanism [35]. The biodegradation of phenol red dye using free and immobilized HRP was investigated in a batch system for a total time of 90 min. Aliquots were collected throughout the degradation experiment, and the phenolic content was quantified using the previously obtained calibration curve (y = 0.07092x + 0.01458, R2 = 0.9966, Fig. S4). The results of the biodegradation experiment, conducted with an initial dye concentration of 100 mg/L and employing 0.0015 U/mL of either free or immobilized enzyme, are presented in Fig. 6. The UV–Vis spectra of the phenol red dye after 90 min of treatment under different conditions are shown in Fig. S5.
Biodegradation of phenol red dye (dye concentration 100 mg/L, pH 6.0, temperature 22 ± 3 °C, H2O2 concentration 0.1 M, free or immobilized enzyme amount 0.0015 U/mL) over time by free (black circle (●)) and immobilized (white square (□)) horseradish peroxidase. Each point corresponds to the mean ± standard deviation of three independent experiments performed in triplicate
We found that the free HRP showed a lower biodegradation potential than the immobilized HRP, degrading only 6.62% of the dye in 90 min, while the immobilized HRP was able to degrade 63.57% in the same period. Thus, immobilized HRP was able to degrade a concentration 9.6 times higher than the free HRP in 90 min. The higher biodegradation capacity of immobilized HRP can be attributed to multipoint interactions between the enzyme and the support, which provide intramolecular forces that prevent changes in the active conformational structure of the enzymes [46, 54]. Therefore, enzyme immobilization on the Ca alginate-starch hybrid support can restrict the denaturation or unfolding of the 3D structure of HRP. Several other studies have also reported increased biodegradability of other molecules and substances by HRP after immobilization due to the protection afforded to the 3D structure of the enzyme [32, 54, 86, 89]. Furthermore, alginate beads can partially adsorb phenol red dye. As shown in Fig. S6, the alginate beads had adsorbed approximately 4.68% of the initial dye concentration after 90 min. Consequently, it can be inferred that of the measured 63.57% biodegradation of phenol red dye by immobilized HRP, approximately 4.7% can be attributed to dye adsorption, whereas the predominant factor of 58.9% corresponds to enzymatic biodegradation. These results imply that the biodegradation of phenol red dye by immobilized HRP occurs via two mechanisms: adsorption and biocatalysis. Notably, biocatalysis exhibited a significantly more prominent role, consistent with the findings reported by Urrea et al. [31]. However, it is important to note that despite achieving 63.57% removal of phenol red dye, further optimization of parameters such as pH, temperature, H2O2 concentration, and catalyst concentration (whether free or immobilized enzyme) can effectively contribute to a higher percentage of degradation.
Reusability of Immobilized HRP
The reusability of immobilized enzymes is a very desirable characteristic for their consideration in industrial applications [3]. Thus, the recyclability of immobilized HRP on a Ca alginate-starch hybrid support in the biodegradation of phenol red dye was investigated. Immobilized HRP beads were subjected to 15 sequential reuse cycles (Fig. 7), with biodegradation in the first cycle considered 100%, and the others calculated as relative percentages.
Reuse of immobilized horseradish peroxidase for the biodegradation of phenol red dye (dye concentration 100 mg/L, pH 6.0, temperature 22 ± 3 °C, H2O2 concentration 0.1 M, amount of free or immobilized enzyme 0.0015 U/mL). Each value corresponds to the mean ± standard deviation of three independent experiments performed in triplicate. Note: a phenol red relative biodegradation of 100% corresponds to 63.3% removal of the dye, and a residual enzymatic activity of 100% corresponds to 0.00145 U/mL
After six cycles of reuse, immobilized HRP retained 62.04% of its initial biodegradation capacity, and after ten cycles, 43.06%, with a residual enzymatic activity of 42.36% ± 0.98. However, after the tenth cycle, a significant reduction in biodegradation activity was observed, with only 8.34% of the initial biodegradation and 8.80% ± 0.66 of the initial enzymatic activity remaining after 15 cycles. The loss of catalytic activity over several reuse cycles may occur due to leakage of unfixed enzyme fractions or entrapment of the enzyme catalytic site due to the accumulation of highly active free radicals [3, 86].
Morphological Characterization of Immobilized HRP After Phenol Red Dye Biodegradation
To verify possible morphological changes in the immobilized HRP beads after use in the biodegradation process, a portion of the beads submitted to the reuse test was collected after 15 cycles of phenol red dye biodegradation and subjected to morphological analysis by SEM (Fig. 8) and elemental analysis by EDS (Table 5).
Morphological analysis showed a reduction in surface prominence after the beads were used for the biodegradation of the phenol red dye. Comparing Fig. 8C and E (before use) with Fig. 8D and F (after use), it can be observed that there were fewer surface protuberances. This change in morphology contributes to lower catalytic activity and may be linked to the leakage of unbound enzyme fractions or structural damage due to the accumulation of biodegradation by-products. In addition, there was a change in the elemental composition of the immobilized HRP when comparing the beads before and after immobilization. As shown in Table 5, there was a decrease in C content, an increase in O and Ca contents, the disappearance of Cl, and the appearance of small levels of K. These results are possibly due to changes in composition caused by the use of the beads in the biodegradation process.
Conclusion
HRP has shown promising characteristics for industrial applications, such as in the biodegradation of effluents and dyes. In this study, two purification steps (precipitation with ammonium sulfate and dialysis) were successfully used to purify HRP extracted from fresh horseradish roots, resulting in a 3.74-fold purification level. The immobilization of HRP (50 mg protein per g support) resulted in a yield of 88.33% ± 0.98, an efficiency of 56.89% ± 2.20, and an activity recovery of 50.26% ± 2.25. Immobilized HRP showed higher catalytic activity than free HRP at pH values of 4.0 to 6.0 and 8.5 to 9.0, as well as at different temperatures, preserving 90.15% of its catalytic activity at 85 °C, demonstrating a better thermal stability profile. The immobilization of HRP resulted in modifications in the kinetic and thermodynamic parameters, with considerable improvements in t1/2 and Ed compared to free HRP, owing to the greater thermal stability conferred. The immobilized HRP maintained 93.71% of its initial catalytic activity after 45 days of storage at 4 °C. Regarding the biodegradation of phenol red, immobilized HRP exhibited 63.57% biodegradation of the dye after 90 min. Moreover, immobilized HRP preserved 43.06% of the initial biodegradation activity after 10 cycles of reuse. After 15 cycles of reuse, there was a reduction in the surface protuberances of the alginate granules and a modification in their elemental composition. The results obtained in this study suggest that the immobilization of HRP on a Ca alginate-starch hybrid support may be a promising alternative for increasing enzymatic thermal stability and the possibility of reuse. Thus, this work highlights the potential of immobilized HRP for applications in biotechnological treatments for pollutant degradation.
Data Availability
All data generated or analyzed during this study are included in this article (and its supplementary information files).
References
Li, Z. L., Cheng, L., Zhang, L. W., Liu, W., Ma, W. Q., & Liu, L. (2017). Preparation of a novel multi-walled-carbon-nanotube/cordierite composite support and its immobilization effect on horseradish peroxidase. Process Safety and Environment Protection, 107, 463–467. https://doi.org/10.1016/j.psep.2017.02.021
Bilal, M., Zhao, Y., Rasheed, T., Iqbal, H. M. N., & Cui, J. (2018). “Smart” chemistry and its application in peroxidase immobilization using different support materials. International Journal of Biological Macromolecules, 119, 278–290. https://doi.org/10.1016/j.ijbiomac.2018.07.134
Bilal, M., Rasheed, T., Zhao, Y., & Iqbal, H. M. N. (2019). Agarose-chitosan hydrogel-immobilized horseradish peroxidase with sustainable bio-catalytic and dye degradation properties. International Journal of Biological Macromolecules, 124, 742–749. https://doi.org/10.1016/j.ijbiomac.2018.11.220
Zahirinejad, S., Hemmati, R., Homaei, A., Dinari, A., Hosseinkhani, S., Mohammadi, S., & Vianello, F. (2021). Nano-organic supports for enzyme immobilization: Scopes and perspectives. Colloids and Surfaces. B, Biointerfaces, 204, 111774. https://doi.org/10.1016/j.colsurfb.2021.111774
Basso, A., & Serban, S. (2019). Industrial applications of immobilized enzymes – A review. Molecular Catalysis, 479, 110607. https://doi.org/10.1016/j.mcat.2019.110607
Fopase, R., Paramasivam, S., Kale, P., & Paramasivam, B. (2020). Strategies, challenges and opportunities of enzyme immobilization on porous silicon for biosensing applications. Journal of Environmental Chemical Engineering, 8(5), 104266. https://doi.org/10.1016/j.jece.2020.104266
Rodríguez-Restrepo, Y. A., & Orrego, C. E. (2020). Immobilization of enzymes and cells on lignocellulosic materials. Environmental Chemistry Letters, 18(3), 787–806. https://doi.org/10.1007/s10311-020-00988-w
Wang, Z., Liu, Y., Li, J., Meng, G., Zhu, D., Cui, J., & Jia, S. (2022). Efficient immobilization of enzymes on amino funtionalized MIL-125-NH2 metal organic framework. Biotechnology and Bioprocess Engineering, 27, 135–144. https://doi.org/10.1007/s12257-020-0393-y
Cui, J., Zhao, Y., Liu, R., Zhong, C., & Jia, S. (2016). Surfactant-activated lipase hybrid nanoflowers with enhanced enzymatic performance. Science and Reports, 6, 27928. https://doi.org/10.1038/srep27928
Cui, J., Cui, L., Jia, S., Su, Z., & Zhang, S. (2016). Hybrid cross-linked lipase aggregates with magnetic nanoparticles: A robust and recyclable biocatalysis for the epoxidation of oleic acid. Journal of Agriculture and Food Chemistry, 68, 7179–7187. https://doi.org/10.1021/acs.jafc.6b01939
Cui, J., Ren, S., Lin, T., Feng, Y., & Jia, S. (2018). Shielding effects of Fe3+-tannic acid nanocoatings for immobilized enzyme on magnetic Fe3O4@silica core shell nanosphere. Chemical Engineering Journal, 343, 629–637. https://doi.org/10.1016/j.cej.2018.03.002
Cui, J., Feng, Y., & Jia, S. (2018). Silica encapsulated catalase@ metal-organic framework composite: A highly stable and recyclable biocatalyst. Chemical Engineering Journal, 351(506–514), 2018. https://doi.org/10.1016/j.cej.2018.06.121
Feng, Y., Du, Y., Kuang, G., Zhong, L., Hu, H., Jia, S., & Cui, J. (2022). Hierarchical micro- and mesoporous ZIF-8 with core-shell superstructures using colloidal metal sulfates as soft templates for enzyme immobilization. Journal of Colloid and Interface Science, 610, 709–718. https://doi.org/10.1016/j.jcis.2021.11.123
Zhang, S., Bilal, M., Zdarta, J., Cui, J., Kumar, A., Franco, M., Ferreira, L. F. R., & Iqbal, H. M. N. (2021). Biopolymers and nanostructured materials to develop pectinases-based immobilized nanobiocatalytic systems for biotechnological applications. Food Research International, 140, 109979. https://doi.org/10.1016/j.foodres.2020.109979
Gao, Y., Shah, K., Kwok, I., Wang, M., Rome, L. H., & Mahendra, S. (2022). Immobilized fungal enzymes: Innovations and potential applications in biodegradation and biosynthesis. Biotechnology Advances, 57, 107936. https://doi.org/10.1016/j.biotechadv.2022.107936
Liu, S., Bilal, M., Rizwan, K., Gul, I., Rasheed, T., & Iqbal, H. M. N. (2021). Smart chemistry of enzyme immobilization using various support matrices – A review. International Journal of Biological Macromolecules, 190, 396–408. https://doi.org/10.1016/j.ijbiomac.2021.09.006
Wen, H.., Zhang, L., Du, Y., Wang, Z., Jiang, Y., Bian, H., Cui, J. & Jia, S. (2020). Bimetal based inorganic-carbonic anhydrase hybrid hydrogel membrane for CO2 capture. Journal of CO2 Utilization, 39, 101171. https://doi.org/10.1016/j.jcou.2020.101171
Ren, S., Li, C., Tan, Z., Hou, Y., Jia, S., & Cui, J. (2019). Carbonic anhydrase@ZIF-8 hydrogel composite membrane with improved recycling and stability for efficient CO2 capture. Journal of Agriculture and Food Chemistry, 67, 3372–3379. https://doi.org/10.1021/acs.jafc.8b06182
Bian, H., Cao, M., Wen, H., Tan, Z., Jia, S., & Cui, J. (2019). Biodegradation of polyvinyl alcohol using cross-linked enzyme aggregates of degrading enzymes from Bacillus niacini. International Journal of Biological Macromolecules, 124, 10–16. https://doi.org/10.1016/j.ijbiomac.2018.11.204
Zdarta, J., Meyer, A. S., Jesionowski, T., & Pinelo, M. (2018). A general overview of support materials for enzyme immobilization: Characteristics, properties, practical utility. Catalysts, 8(92), 1–27. https://doi.org/10.3390/catal8020092
Bilal, M., & Iqbal, H. M. N. (2019). Lignin peroxidase immobilization on Ca-alginate beads and its dye degradation performance in a packed bed reactor system. Biocatalysis and Agricultural Biotechnology, 20, 101205. https://doi.org/10.1016/j.bcab.2019.101205
Matto, M., & Husain, Q. (2009). Calcium alginate-starch hybrid support for both surface immobilization and entrapment of bitter gourd (Momordica charantia) peroxidase. Journal of Molecular Catalysis. B, Enzymatic, 57(1–4), 164–170. https://doi.org/10.1016/j.molcatb.2008.08.011
Prokopijevic, M. (2021). Natural polymers: Suitable carriers for enzyme immobilization. Biologia Serbica, 43(1), 43–49. https://doi.org/10.5281/zenodo.5512407
Rohman, S., Kaewtatip, K., Kantachote, D., & Tantirungkij, M. (2020). Encapsulation of Rhodopseudomonas palustris KTSSR54 using beads from alginate/starch blends. Journal of Applied Polymer Science, 138(12), 1–9. https://doi.org/10.1002/app.50084
Schoebitz, M., Simonin, H., & Poncelet, D. (2012). Starch filler and osmoprotectants improve the survival of rhizobacteria in dried alginate beads. Journal of Microencapsulation, 26(6), 532–538. https://doi.org/10.3109/02652048.2012.665090
Kailasapathy, K., Perera, C., & Phillips, M. (2006). Evaluation of alginate-starch polymers for preparation of enzyme microcapsules. International Journal of Food Engineering, 2(2), 1–13. https://doi.org/10.2202/1556-3758.1109
Andrades, D., Graebin, N. G., Kadowaki, M. K., Ayub, M. A. Z., Fernandez-Lafuente, R., & Rodrigues, R. C. (2019). Immobilization and stabilization of different β-glucosidases using the glutaraldehyde chemistry: Optimal protocol depends on the enzyme. International Journal of Biological Macromolecules, 129, 672–678. https://doi.org/10.1016/j.ijbiomac.2019.02.057
Magro, L. D., Kornecki, J. F., Klein, M. P., Rodrigues, R. C., & Fernandez-Lafuente, R. (2020). Pectin lyase immobilization using the glutaraldehyde chemistry increases the enzyme operation range. Enyzme and Microbial Technology, 132, 109397. https://doi.org/10.1016/j.enzmictec.2019.109397
Chun, H. J., Park, C. H., Kwon, I. K. & Khang, G. (2018). Cutting-edge enabling technologies for regenerative medicine. Singapore: Springer Singapore, 1078, (Advances in Experimental Medicine and Biology).
Sharma, A., Thatai, K. S., Kuthiala, T., Singh, G., & Arya, S. K. (2021). Employment of polysaccharides in enzyme immobilization. Reactive & Functional Polymers, 167, 105005. https://doi.org/10.1016/j.reactfunctpolym.2021.105005
Urrea, D. A. M., Gimenez, A. V. F., Rodriguez, Y. E., & Contreras, E. M. (2021). Immobilization of horseradish peroxidase in Ca-alginate beads: Evaluation of the enzyme leakage on the overall removal of an azo-dye and mathematical modeling. Process Safety and Environment Protection, 156, 134–143. https://doi.org/10.1016/j.psep.2021.10.006
Keshta, B. E., Gemeay, A. H., & Khamis, A. A. (2022). Impacts of horseradish peroxidase immobilization onto functionalized superparamagnetic iron oxide nanoparticles as a biocatalyst for dye degradation. Environmental Science and Pollution Research, 29, 6633–6645. https://doi.org/10.1007/s11356-021-16119-z
El-Naggar, M. E., Abdel-Aty, A. M., Wassel, A. R., Elaraby, N. M., & Mohamed, S. A. (2021). Immobilization of horseradish peroxidase on cationic microporous starch: Physico-bio-chemical characterization and removal of phenolic compounds. International Journal of Biological Macromolecules, 181, 734–742. https://doi.org/10.1016/j.ijbiomac.2021.03.171
Petronijevic, M., Panic, S., Savic, S., Agbaba, J., Jazic, J. M., Milanovic, M., & Durisic-Mladenovic, N. (2021). Characterization and application of biochar-immobilized crude horseradish peroxidase for removal of phenol from water. Colloids and Surfaces. B, Biointerfaces, 208, 112038. https://doi.org/10.1016/j.colsurfb.2021.112038
Bayramoglu, G., Altintas, B., & Arica, M. Y. (2012). Cross-linking of horseradish peroxidase adsorbed on polycationic films: Utilization for direct dye degradation. Bioprocess and Biosystems Engineering, 35, 1355–1365. https://doi.org/10.1007/s00449-012-0724-2
Samsami, S., Mohamadizaniani, M., Sarrafzadeh, M.-H., Rene, E. R., & Firoozbahr, M. (2020). Recent advances in the treatment of dye-containing wastewater from textile industries: Overview and perspectives. Process Safety and Environment Protection, 143, 138–163. https://doi.org/10.1016/j.psep.2020.05.034
Sun, H., Qin, J., Yi, L., Ruan, Y., Wang, J., & Fang, D. (2022). A new process for degradation of Auramine O dye and heat generation based on orifice plate hydrodynamic cavitation (HC): Parameter optimization and performance analyses. Process Safety and Environment Protection, 161, 669–683. https://doi.org/10.1016/j.psep.2022.03.058
Shah, P., Unnarkat, A., Patel, F., Shah, M., & Shah, P. (2022). A comprehensive review on spinel based novel catalysts for visible light assisted dye degradation. Process Safety and Environment Protection, 161, 703–722. https://doi.org/10.1016/j.psep.2022.03.030
Gautam, A., Rawat, S., Verma, L., Singh, J., Sikarwar, S., Yadav, B. C., & Kalamdhad, A. S. (2018). Green synthesis of iron nanoparticle from extract of waste tea: An application for phenol red removal from aqueous solution. Environmental Nanotechnology, Monitoring & Management, 10, 377–387. https://doi.org/10.1016/j.enmm.2018.08.003
Kumar, G., & Dutta, R. K. (2022). Sunlight mediated photo-Fenton degradation of tetracycline antibiotic and methylene blue dye in aqueous medium using FeWO4/Bi2MoO6 nanocomposite. Process Safety and Environment Protection, 159, 862–873. https://doi.org/10.1016/j.psep.2022.01.063
Cuprys, A., Thomson, P., Ouarda, Y., Suresh, G., Rouissi, T., Brar, S. K., Drogui, P., & Surampalli, R. Y. (2020). Ciprofloxacin removal via sequential electro-oxidation and enzymatic oxidation. Journal of Hazardous Materials, 389(121890), 1–9. https://doi.org/10.1016/j.jhazmat.2019.121890
Zhang, C., You, S., Zhang, J., Qi, W., Su, R., & He, Z. (2020). An effective in-situ method for laccase immobilization: Excellent activity, effective antibiotic removal rate and low potential ecological risk for degradation products. Bioresource Technology, 308, 1–10. https://doi.org/10.1016/j.biortech.2020.123271
Bilal, M., Iqbal, H. M. N., Shah, S. Z. H., Hu, H., Wang, W., & Zhang, X. (2016). Horseradish peroxidase-assisted approach to decolorize and detoxify dye pollutants in a packed bed bioreactor. Journal of Environmental Management, 183(3), 836–842. https://doi.org/10.1016/j.jenvman.2016.09.040
Ali, M., & Husain, Q. (2018). Guar gum blended alginate/agarose hydrogel as a promising support for the entrapment of peroxidase: Stability and reusability studies for the treatment of textile effluents. International Journal of Biological Macromolecules, 116, 463–471. https://doi.org/10.1016/j.ijbiomac.2018.05.037
Alatawi, F. S., Elsayed, N. H., & Monier, M. (2020). Immobilization of horseradish peroxidase on modified nylon-6 fibers. ChemistrySelect, 5(23), 6841–6850. https://doi.org/10.1002/slct.202000818
Bayramoglu, G., Akbulut, A., & Arica, M. Y. (2021). Utilization of immobilized horseradish peroxidase for facilitated detoxification of a benzidine based azo dye. Chemical Engineering Research and Design, 165, 435–444. https://doi.org/10.1016/j.cherd.2020.11.017
Muenchen, D. K., Martinazzo, J., Brezolin, A. N., De Cezaro, A. M., Rigo, A. A., Mezarroba, M. N., Manzoli, A., Leite, F. L., Steffens, J., & Steffens, C. (2018). Cantilever functionalization using peroxidase extract of low cost for glyphosate detection. Applied Biochemistry and Biotechnology, 186(4), 1061–1073. https://doi.org/10.1007/s12010-018-2799-y
Fernandes, M., Souza, D. H., Henriques, R. O., Alves, M. V., Skoronski, E., & Junior, A. F. (2020). Obtaining soybean peroxidase from soybean hulls and its application for detoxification of 2,4-dichlorophenol contaminated water. Journal of Environmental Chemical Engineering, 8(3), 1–7. https://doi.org/10.1016/j.jece.2020.103786
Zeraik, A. E., Souza, F. S., Fatibello-Filho, O., & Leite, O. D. (2008). Desenvolvimento de um spot test para o monitoramento da atividade da peroxidase em um procedimento de purificação. Quimica Nova, 31, 731–734. https://doi.org/10.1590/S0100-40422008000400003
Köktepe, T., Altin, S., Tohma, H., Gülçin, İ. & Köksal, E. (2017). Purification, characterization and selected inhibition properties of peroxidase from haricot bean (Phaseolus vulgaris L.). International Journal of Food Properties, 20(S2), 1944–1953. https://doi.org/10.1080/10942912.2017.1360903
Queiroz, M. L. B., Conceição, K. C., Melo, M. N., Sánchez, O. C., Alvarez, H. M., Soares, C. M. F. & Fricks, A. T. (2018). Imobilização de peroxidase de raiz forte em bagaço de cana-de-açúcar. Química Nova, 41(9), 1019–1024. https://doi.org/10.21577/0100-4042.20170279
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72(1–2), 248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Sheldon, R. A., & van Pelt, S. (2013). Enzyme immobilisation in biocatalysis: Why, what and how. Chemical Society Reviews, 42(15), 6223–6235. https://doi.org/10.1039/c3cs60075k
Bilal, M., Iqbal, H. M. N., Hu, H., Wang, W., & Zhang, X. (2017). Enhanced bio-catalytic performance and dye degradation potential of chitosan-encapsulated horseradish peroxidase in a packed bed reactor system. Science of the Total Environment, 575, 1352–1360. https://doi.org/10.1016/j.scitotenv.2016.09.215
Zeyadi, M., & Almulaiky, Y. Q. (2020). A novel peroxidase from Ziziphus jujuba fruit: Purification, thermodynamics and biochemical characterization properties. Science and Reports, 10(1), 1–11. https://doi.org/10.1038/s41598-020-64599-9
Rigo, D., Santos, P. N. A., Fischer, B., Vendruscolo, M. D., Fernandes, I. A., Fricks, A. T., Dallago, R. M. & Zeni, J. (2021). Covalent immobilization of lipase in residual yerba mate stick (Ilex paraguariensis A. St.-Hil.). Biointerface Research in Applied Chemistry, 11(6), 14564–14579. https://doi.org/10.33263/BRIAC116.1456414579
Yandri, Y., Tiarsa, E. R., Suhartati, T., Satria, H., Irawan, B., & Hadi, S. (2022). The stability improvement of α-amylase enzyme from Aspergillus fumigatus by immobilization on a bentonite matrix. Biochemistry Research International, 2022, 1–7. https://doi.org/10.1155/2022/3797629
Gennari, A., Mobayed, F. H., Rafael, R. S., Rodrigues, R. C., Sperotto, R. A., Volpato, G., & Souza, C. F. V. (2018). Modification of Immobead 150 support for protein immobilization: Effects on the properties of immobilized Aspergillus oryzae β-galactosidase. Biotechnology Progress, 34(4), 934–943. https://doi.org/10.1002/btpr.2652
Cunha, B. S., Bataglioli, R. A., Taketa, T. B., Lopes, L. M., & Beppu, M. M. (2019). Ionic liquid functionalization of chitosan beads for improving thermal stability and copper ions uptake from aqueous solution. Journal of Environmental Chemical Engineering, 7(13), 10381. https://doi.org/10.1016/j.jece.2019.103181
Taketa, T. B., Mahl, C. R. A., Calais, G. B., & Beppu, M. M. (2021). Amino acid-functionalized chitosan beads for in vitro copper ions uptake in the presence of histidine. International Journal of Biological Macromolecules, 188, 421–431. https://doi.org/10.1016/j.ijbiomac.2021.08.017
Alver, E., & Metin, A. Ü. (2017). Chitosan based metal-chelated copolymer nanoparticles: Laccase immobilization and phenol degradation studies. International Biodeterioration and Biodegradation, 125, 235–242. https://doi.org/10.1016/j.ibiod.2017.07.012
Souza, P. D. B. & Corbellini, V. A. (2019). Síntese e avaliação de azoderivado de 2-fenilbenzoxazol como substrato fluorogênico na diferenciação de Candida spp. Revista Jovens Pesquisadores, 9(1), 28–39. https://doi.org/10.17058/rjp.v9i1.13384
Nazari, G., Abolghasemi, H., Esmaieli, M., & Pouya, E. S. (2016). Aqueous phase adsorption of cephalexin by walnut shell-based activated carbon: A fixed-bed column study. Applied Surface Science, 375, 144–153. https://doi.org/10.1016/j.apsusc.2016.03.096
Zhang, C., & Cai, X. (2018). Immobilization of Horseradish peroxidase on Fe3O4/nanotubes composites for biocatalysis-degradation of phenol. Composite Interfaces, 26(5), 379–396. https://doi.org/10.1080/09276440.2018.1504265
Vineh, M. B., Saboury, A. A., Pootschi, A. A., & Ghasemi, A. (2020). Biodegradation of phenol and dyes with horseradish peroxidase covalently immobilized on functionalized RGO-SiO2 nanocomposite. International Journal of Biological Macromolecules, 164, 4403–4414. https://doi.org/10.1016/j.ijbiomac.2020.09.045
Yaseen, D. A., & Scholz, M. (2019). Textile dye wastewater characteristics and constituents of synthetic effluents: A critical review. International Journal of Environmental Science and Technology, 16, 1193–1226. https://doi.org/10.1007/s13762-018-2130-z
Guo, J., Liu, X., Zhang, X., Wu, J., Chai, C., Ma, D., Chen, Q., Xiang, D., & Ge, W. (2019). Immobilized lignin peroxidase on Fe3O4@SiO2@polydopamine nanoparticles for degradation of organic pollutants. International Journal of Biological Macromolecules, 138, 433–440. https://doi.org/10.1016/j.ijbiomac.2019.07.105
Joel, E. B., Mafulul, S. G., Adamu, H. E., Goje, L. J., Tijjani, H., Igunnu, A. & Malomo, S. O. (2020). Peroxidase from waste cabbage (Brassica oleracea capitata L.) exhibits the potential to biodegrade phenol and synthetic dyes from wastewater. Scientific African, 10, e00608. https://doi.org/10.1016/j.sciaf.2020.e00608
Zhou, J., Chen, J., Zhuang, N., Zhang, A., Chen, K., Xu, N., Xin, F., Zhang, W., Dong, W. & Jiang, M. (2020). Immobilization and purification of enzymes with the novel affinity tag ChBD-AB from Chitinolyticbacter meiyuanensis SYBC-H1. Frontiers in Bioengineering and Biotechnology, 8, 579. https://doi.org/10.3389/fbioe.2020.00579
Al-Bagmi, M. S., Khan, M. S., Ismael, M. A., Al-Senaidy, A. M., Bacha, A. B., Husain, F. M., & Alamery, S. F. (2019). An efficient methodology for the purification of date palm peroxidase: Stability comparison with horseradish peroxidase (HRP). Saudi Journal of Biological Sciences, 26(2), 301–307. https://doi.org/10.1016/j.sjbs.2018.04.002
Li, X., & Kushad, M. M. (2005). Purification and characterization of myrosinase from horseradish (Armoracia rusticana) roots. Plant Physiology and Biochemistry, 43(6), 503–511. https://doi.org/10.1016/j.plaphy.2005.03.015
Bertóti, R., Böszörményi, A., Alberti, A., Béni, S., M-Hamvas, M., Szöke, E., Vasas, G. & Gonda, S. (2019). Variability of bioactive glucosinolates, isothiocyanates and enzyme patterns in horseradish hairy root cultures initiated from different organs. Molecules, 24(15), 2828.https://doi.org/10.3390/molecules24152828
Bilal, M., & Asgher, M. (2015). Dye decolorization and detoxification potential of Ca-alginate beads immobilized manganese peroxidase. BMC Biotechnology, 15(111), 1–14. https://doi.org/10.1186/s12896-015-0227-8
Melo, M. N., Pereira, F. M., Rocha, M. A., Ribeiro, J. G., Diz, F. M., Monteiro, W. F., Ligabue, R. A., Severino, P., & Fricks, A. T. (2020). Immobilization and characterization of horseradish peroxidase into chitosan and chitosan/PEG nanoparticles: A comparative study. Process Biochemistry, 98, 160–171. https://doi.org/10.1016/j.procbio.2020.08.007
Shen, C., Zhao, Y., Liu, R., Mao, Y., & Morgan, D. (2018). Adsorption of phosphorus with calcium alginate beads containing drinking water treatment residual. Water Science and Technology, 78(9), 1980–1989. https://doi.org/10.2166/wst.2018.473
Zafeiri, I., Beri, A., Linter, B., & Norton, I. (2021). Mechanical properties of starch-filled alginate gel particles. Carbohydrate Polymers, 255, 117373. https://doi.org/10.1016/j.carbpol.2020.117373
Bilal, M., Iqbal, M., Hu, H., & Zhang, Z. (2016). Mutagenicity and cytotoxicity assessment of biodegraded textile effluent by Ca-alginate encapsulated manganese peroxidase. Biochemical Engineering Journal, 109, 153–161. https://doi.org/10.1016/j.bej.2016.01.020
Ahenkorah, I., Rahman, M., Karim, R., Beecham, S., & Saint, C. (2021). A review of enzyme induced carbonate precipitation (EICP): The role of enzyme kinetics. Sustainable Chemistry, 2(1), 92–114. https://doi.org/10.3390/suschem2010007
Guzik, U., Hupert-Kocurek, K. & Wojcieszynska, D. (2014). Immobilization as a strategy for improving enzyme properties – application to oxidoreductases. Molecules, 19(7), 8995–9018. https://doi.org/10.3390/molecules19078995
Khan, M. R. (2021). Immobilized enzymes: A comprehensive review. Bulletin of the National Research Centre, 45(207), 1–13. https://doi.org/10.1186/s42269-021-00649-0
Bayramoglu, G., & Arica, M. Y. (2019). Biodegradation of methylene blue and carbaryl by Trametes versicolor laccase preparations in the presence of a mediator compound. Journal of Macromolecular Science, Part A, 56(3), 277–285. https://doi.org/10.1080/10601325.2019.1565549
Mohamed, S. A., Elaraby, N. M., Abdel-Aty, A. M., Shaban, E., Abu-Saied, M. A., Kenawy, E.-R., & El-Naggar, M. E. (2021). Improvement of enzymatic properties and decolorization of azo dye: Immobilization of horseradish peroxidase on cationic maize starch. Biocatalysis and Agricultural Biotechnology, 38, 102208. https://doi.org/10.1016/j.bcab.2021.102208
Zdarta, J., Degórska, O., Jankowska, K., Rybarczyk, A., Piasecki, A., Ciesielczyk, F., & Jesionowski, T. (2022). Removal of persistent sulfamethoxazole and carbamazepine from water by horseradish peroxidase encapsulated into poly(vinyl chloride) electrospun fibers. International Journal of Molecular Sciences, 23(1), 272. https://doi.org/10.3390/ijms23010272
Akharume, F. U., Aluko, R. E., & Adedeji, A. A. (2021). Modification of plant proteins for improved functionality: A review. Comprehensive Reviews in Food Science and Food Safety, 20(1), 198–224. https://doi.org/10.1111/1541-4337.12688
Qamar, S. A., Asgher, M., & Bilal, M. (2020). Immobilization of alkaline protease from Bacillus brevis using Ca-alginate entrapment strategy for improved catalytic stability, silver recovery, and dehairing potentialities. Catalysis Letters, 150, 3572–3583. https://doi.org/10.1007/s10562-020-03268-y
Kalsoom, U., Khalid, N., Ibrahim, A., Ashraf, S. S., Bhatti, H. N., Ahsan, Z., Zdarta, J., & Bilal, M. (2023). Biocatalytic degradation of reactive blue 221 and direct blue 297 dyes by horseradish peroxidase immobilized on iron oxide nanoparticles with improved kinetic and thermodynamic characteristics. Chemosphere, 312, 137095. https://doi.org/10.1016/j.chemosphere.2022.137095
Park, C. (2022). Visual interpretation of the meaning of kcat/kM in enzyme kinetics. Journal of Chemical Education, 99(7), 2556–2562. https://doi.org/10.1021/acs.jchemed.1c01268
Ademakinwa, A. N. (2021). A heat-resistant intracellular laccase immobilized via cross-linked enzyme aggregate preparation: Characterization, application in bisphenol A removal and phytotoxicity evaluation. Journal of Hazardous Materials, 419, 126480. https://doi.org/10.1016/j.jhazmat.2021.126480
Aldhahri, M., Almulaiky, Y. Q., El-Shishtawy, R. M., Al-Shawafi, W. M., Salah, N., Alshahrie, A., & Alzahrani, H. A. H. (2021). Ultra-Thin 2D CuO nanosheet for HRP immobilization supported by encapsulation in a Polymer matrix: Characterization and dye degradation. Catalysis Letters, 151, 232–246. https://doi.org/10.1007/s10562-020-03289-7
Wu, J., Ma, X., Li, C., Zhou, X., Han, J., Wang, L., Dong, H., & Wang, Y. (2022). A novel photon-enzyme cascade catalysis system based on hybrid HRP-CN/Cu3(PO4)2 nanoflowers for degradation of BPA in water. Chemical Engineering Journal, 427, 131808. https://doi.org/10.1016/j.cej.2021.131808
Ladole, M. R., Pokale, P. B., Patil, S. S., Belokar, P. G., & Pandit, A. B. (2020). Laccase immobilized peroxidase mimicking magnetic metal organic frameworks for industrial dye degradation. Bioresource Technology, 317, 124035. https://doi.org/10.1016/j.biortech.2020.124035
Bindu, V. U., & Mohanan, P. V. (2020). Thermal deactivation of α-amylase immobilized magnetic chitosan and its modified forms: A kinetic and thermodynamic study. Carbohydrate Research, 498, 108185. https://doi.org/10.1016/j.carres.2020.108185
Campello, G. S., Trindade, R. A., Rêgo, T. V., Burkert, J. F. M., & Burkert, C. A. V. (2012). Immobilization of β-galactosidase from Kluyveromyces lactis on Eupergit C and properties of the biocatalyst. International Journal of Food Engineering, 8, 3. https://doi.org/10.1515/1556-3758.2760
Muley, A. B., Thorat, A. S., Singhal, R. S., & Babu, K. H. (2018). A tri-enzyme co-immobilized magnetic complex: Process details, kinetics, thermodynamics and applications. International Journal of Biological Macromolecules, 118, 1781–1795. https://doi.org/10.1016/j.ijbiomac.2018.07.022
Giannakopoulou, A., Patila, M., Spyrou, K., Chalmpes, N., Zarafeta, D., Skretas, G., Gournis, D., & Stamatis, H. (2019). Development of a four-enzyme magnetic nanobiocatalyst for multi-step cascade reactions. Catalysts, 9(12), 995. https://doi.org/10.3390/catal9120995
Yang, Q., Yan, Y., Yang, X., Liao, G., Wang, D., & Xia, H. (2019). Enzyme immobilization in cage-like 3D-network PVA-H and GO modified PVA-H(GO@PVA-H) with stable conformation and high activity. Chemical Engineering Journal, 372, 946–955. https://doi.org/10.1016/j.cej.2019.04.216
Guo, R., Zheng, X., Wang, Y., Yang, Y., Ma, Y., Zou, D., & Liu, Y. (2021). Optimization of cellulase immobilization with sodium alginate-polyethylene for enhancement of enzymatic hydrolysis of microcrystalline cellulose using response surface methodology. Applied Biochemistry and Biotechnology, 193, 2043–2060. https://doi.org/10.1007/s12010-021-03517-9
Rai, S. K., Kaur, H., Singh, A., Kamboj, M., Jain, G., & Yadav, S. K. (2021). Production of D-tagatose in packed bed reactor containing and immobilized L-arabinose isomerase on alginate support. Biocatalysis and Agricultural Biotechnology, 38, 102227. https://doi.org/10.1016/j.bcab.2021.102227
Almulaiky, Y. Q., & Al-Harbi, S. A. (2022). Preparation of a calcium alginate-coated polypyrrole/silver nanocomposite for site-specific immobilization of polygalacturonase with high reusability and enhanced stability. Catalysis Letters, 152, 28–42. https://doi.org/10.1007/s10562-021-03631-7
Almulaiky, Y. Q., & Almaghrabi, O. (2022). Polyphenol oxidase from Coleus forskohlii: Purification, characterization, and immobilization onto alginate/ZnO nanocomposite materials. Catalysis Letters, 152, 3089–3099. https://doi.org/10.1007/s10562-022-03916-5
Djelad, A., Mokhtar, A., Khelifa, A., Bengueddach, A., & Sassi, M. (2019). Alginate-whey an effective and green adsorbent for crystal violet removal: Kinetic, thermodynamic and mechanism studies. International Journal of Biological Macromolecules, 139, 944–954. https://doi.org/10.1016/j.ijbiomac.2019.08.068
Yang, N., Wang, R., Rao, P., Yan, L., Zhang, W., Wang, J., & Chai, F. (2019). The fabrication of calcium alginate beads as a green sorbent for selective recovery of Cu(II) from metal mixtures. Crystals, 9(5), 255. https://doi.org/10.3390/cryst9050255
Salachna, P., Grzeszczuk, M., Meller, E., & Soból, M. (2018). Oligo-alginate with low molecular mass improves growth and physiological activity of Eucomis autumnalis under salinity stress. Molecules, 23(4), 812. https://doi.org/10.3390/molecules23040812
Zhang, K., Zhang, Z., Zhao, M., Milosavljevic, V., Cullen, P. J., Scally, L., Sun, D.-W., & Tiwari, B. K. (2022). Low-pressure plasma modification of the rheological properties of tapioca starch. Food Hydrocolloids, 125, 107380. https://doi.org/10.1016/j.foodhyd.2021.107380
Lomelí-Ramírez, M. G., Barrios-Guzmán, A. J., García-Enriquez, S., Rivera-Prado, J. J. & Manríquez-González, R. (2014). Chemical and mechanical evaluation of bio-composites based on thermoplastic starch and wood particles prepared by thermal compression. Bioresources, 9(2), 2960–2974. https://doi.org/10.15376/biores.9.2.2960-2974
Ivanov, Y. D., Pleshakova, T. O., Shumov, I. D., Kozlov, A. F., Ivanova, I. A., Valueva, A. A., Ershova, M. O., Tatur, V. Y., Stepanov, I. N., Repnikov, V. V., & Ziborov, V. S. (2021). AFM study of changes in properties of horseradish peroxidase after incubation of its solution near a pyramidal structure. Science and Reports, 11, 9907. https://doi.org/10.1038/s41598-021-89377-z
Tavares, T. S., da Rocha, E. P., Nogueira, F. G. E., Torres, J. A., Silva, M. C., Kuca, K., & Ramalho, T. N. (2020). Δ-FeOOH as support for immobilization peroxidase: Optimization via a chemometric approach. Molecules, 25(2), 259. https://doi.org/10.3390/molecules25020259
Funding
This study was partially funded by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—CAPES – Finance code 001, Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Universidade do Vale do Taquari—Univates.
Author information
Authors and Affiliations
Contributions
Conceptualization: A.C.W., E.M.E., and L.H.; investigation: A.C.W., B.E.S., S.G.C., and V.A.C.; methodology: A.C.W., S.G.C., G.S.H., and V.A.C.; writing—original draft: A.C.W., G.S.H., and J.S.H.S.; formal analysis: B.E.S., B.C., and J.S.H.S.; resources: V.A.C., E.M.E., and L.H.; supervision: L.H.; project administration: L.H.
Corresponding author
Ethics declarations
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
All the authors approved the version to be published.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Weber, A.C., da Silva, B.E., Cordeiro, S.G. et al. Immobilization of Horseradish Peroxidase on Ca Alginate-Starch Hybrid Support: Biocatalytic Properties and Application in Biodegradation of Phenol Red Dye. Appl Biochem Biotechnol 196, 4759–4792 (2024). https://doi.org/10.1007/s12010-023-04772-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-023-04772-8