Abstract
Fructose, which is produced by the isomerization of glucose isomerase, is a crucial precursor for the biosynthesis of rare sugars. In this study, thermophilic glucose isomerases (GI) from Caldicellulosiruptor acetigenus (CAGI), Thermoanaerobacter thermocopriae (TTGI), and Thermotoga petrophila (TPGI) were screened from GenBank database by a virtual probe and were successfully expressed in Escherichia coli BL21(DE3). The results of characterization demonstrated that the optimal pH for CAGI and TTGI were 8.0 and were maintained at 80% in a slightly acidic environment. The relative residual activities of CAGI and TTGI were found to be 40.6% and 52.6%, respectively, following an incubation period of 24 h at 90 ℃. Furthermore, CAGI and TTGI exhibited superior catalytic performance that their reaction equilibrium both reached only after an hour at 85 ℃ with 200 g/L glucose, and the highest conversion rates were 54.2% and 54.1%, respectively. This study identifies competitive enzyme candidates for fructose production in the industry with appreciable cost reduction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
D-Fructose is an isomer of D-glucose that is abundant in fruit juice and honey [1] and widely utilized in the food [2], pharmaceutical [3], and other fields [4]. D-Fructose has the advantages of easy glycogen formation, rapid metabolism, and energy conversion in the human body [5], without increasing the glycemic index [6]. Notably, D-fructose is an essential raw material for the biosynthesis of functional sugar [7, 8]. One of the current research hotspots is how to efficiently and economically produce fructose. In the industrial production of fructose, glucose isomerase (GI, EC 5.3.1.5), a class of homotetramer proteins containing metal ions[9], is utilized. It is generally believed that GI falls into two categories, with Class II inserting 20 to 35 amino acids at the N-terminus [10]. It is generally believed that GI falls into two categories, with Class II inserting 20 to 35 amino acids at the N-terminus [11]. The GI monomer is primarily composed of two structural regions. The N-terminal enzyme-catalyzed pocket and two metal-binding sites are folded into a (α/β)8 bucket, while the C-terminal domain is folded into a ring [12, 13]. The overlap of the two subunits forms a tightly bound dimer [14, 15]. At 60–65 ℃, D-glucose is isomerized by GI into D-fructose during industrial production [16]. It has been reported that increasing the temperature of industrial fructose production improves both efficiency and cost [17]. Anoxybacillus geoensis [18], Caldanaerobacter subterraneus [19], Caldicoprobacter algeriensis [20], Caldicellulosiruptor bescii [21], and Streptomyces sp. SK [22] produced thermophilic GIs. Due to poor thermostability, however, these wild-type enzymes cannot meet industrial demand. Consequently, it is essential to discover new GI with increased catalytic efficiency and enhanced thermostability [23].
The mining and screening of enzymes from organisms that inhabit extreme environments have become one of the solutions to the challenges encountered in the practical applications of enzymes [24]. By far, more than 67,000 GI genes have been added to the GenBank database. Whereas the majority of GI genes originated from Streptococcus pneumoniae, Escherichia coli, and Staphylococcus aureus, extremophiles contributed a small amount. In this study, GIs derived from Caldicellulosiruptor acetigenus (CAGI), Thermoanaerobacter thermocopriae (TTGI), and Thermotoga petrophila (TPGI) were successfully expressed using virtual probes. Their biological activities including thermal stability and catalytic efficiency were measured and compared for the evaluation of their ability in the industrial production of fructose.
Materials and Methods
Culture Medium, Chemicals, Strains, and Plasmids
All of the reagents used in this study were of analytical grade and available on the market. Escherichia coli BL21 (DE3) and pET-28b(+) (Novagen, Darmstadt, Germany) were used to express genes synthesized by Tsingke Biotechnology Ltd. (Hangzhou, China). Recombinant E. coli was grown in Luria–Bertani (LB) medium with 50 µg/mL kanamycin (kan) at 37 ℃.
Database Mining of GI Gene
FoldX and Rosseta software were used to scan the whole protein energy of the reported Class II GI [25,26,27,28]. A virtual probe was designed according to conserved sequences and predicted sites, and Basic Local Alignment Search Tool (BLASTp) on the Genbank database (https://www.ncbi.nlm.nih.gov/protein) was used for thermophilic GI genes mining. Secondary structures and hydrogen bond numbers were calculated by VADAR [29]. SWISS-MODEL was used to simulate the three-dimensional (3D) structure models (https://swissmodel.expasy.org) [30], and the models were evaluated using PROCHECK (https://saves.mbi.ucla.edu/) [31]. AutoDock 4.2 [32] was used for molecular docking and calculation of binding affinity. Caldicellulosiruptor bescii GI (accession no. ACM59729), Caldicoprobacter algeriensis GI (accession no. MH172473), Thermoanaerobacter siderophilus GI (accession no. WP_006569355), Thermotoga neapolitana GI (accession no. L38994), Thermus Caldophilus GI (PDB: 1BXC), and Thermus oshimai (accession no. WP_016329521) were used as templates.
Heterologous Expression and Purification of GIs
The GIs genes after codons optimization were synthesized by Tsingke Biotechnology Co., Ltd. (Beijing, China) and subcloned into plasmid pET-28b. After being transfected into E. coli BL21 (DE3), the recombinant plasmid was grown to an OD600 nm of 0.6–0.8 in LB medium with 50 µg/mL kan. A final concentration of 0.1 mmol/L of isopropyl-D-thiogalacto-pyranoside was added, and the expression was then induced at 28 ℃ for 12 h. Recombinant cells expressing the protein were harvested and resuspended in phosphate buffer saline (PBS) buffer, then lysed by sonication (200W, SCIENTZ, China). The crude enzyme was collected by cryogenic centrifuge at 8500 × g for 10 min. Ni2+ affinity chromatography was used to purify protein, as described previously [33]. Bio-Gel P-6 Desalting Cartridges (Bio-Rad, California, USA) was used to desalt. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE, GenScript, Nanjing, China) was used to detect the target protein, and the total protein concentration was determined by the BCA method [34]. The molecular weight of the purified enzyme was determined by SEC-HPLC. The chromatographic column was AdvanceBio SEC 300 Å (7.8 × 300 mm, 2.7 µm particle size), the mobile phase was PBS buffer, the flow rate was 1 mL/min, and the determined wavelength is 205 nm. The peak time of AdvanceBio SEC 300 Å protein standard was taken as the horizontal coordinate, corresponding to the logarithm of protein molecular weight was the ordinate, and the standard curve was drawn to calculate the molecular weight of the purified enzyme.
Biochemical Characterization and Cellular Catalysis of GIs
Determination of Activity Enzyme Activities of GIs
The standard reaction system in a total volume of 500 µL was constituted of 50 µL diluted GI, 50 mmol/L NaH2PO4-Na2HPO4 (pH 7.0), 5 g/L glucose as substrate, 1 mmol/L Co2+, and 10 mmol/L Mg2+. The mixture was incubated at 80 ℃ for 10 min and terminated by adding 500 µL of 50 mmol/L H2SO4. Hyper XP Carbohydrate H+ (300 mm × 7.7 mm; Thermo Fisher Scientific, Waltham, USA) was used to analyze the concentration of glucose and fructose using HPLC (Waters 1515, Milford, USA) equipped with a refractive index detector (Waters 2414, Milford, USA) with a flow rate of 0.6 mL/min at 60 ℃ and 5 mmol/L H2SO4 as the mobile phase. One unit (U) of isomerase activity was defined as the enzyme amount needed to produce 1 µmol of D-fructose per minute under the assay conditions.
Effect of pH on GI Enzyme Activity
The optimum pH was measured at 80 ℃ in 50 mmol/L Mcilvaine buffer (pH 5.0–8.0), Tris–HCl (pH 8.0–9.0), and Glycine–NaOH (pH 9.0–10.0) with a span of 0.5. The highest enzyme activity was defined as 100%. The pH stability of the three enzymes was calculated by measuring their residual enzyme activity after they were placed in a wider pH range (3.0–12.0) in the buffer for 1 h at 25 ℃, with the highest activity defined as 100%.
Effect of Temperature on GI Enzyme Activity
The thermal activity of GI was determined by measuring viability at temperatures of 50–95 ℃ (water bath) and 95–120 ℃ (oil bath). The highest activity of GI was taken as 100% at different temperatures. The thermal stability of the GI was determined in two ways: (1) the purified enzyme was incubated for 1 h at 70, 80, and 90 ℃; (2) the purified enzyme was incubated for 24 h with metal ions added to the standard reaction. The activity of the non-heated enzyme was set as 100%. GI of 0.1 mg/mL was determined from 25 to 95 ℃ with a step of 1 mm on ChirascanTm Circular Dichroism Spectrometer (Applied Photophysics Ltd., United Kingdom), and Tm values were obtained by fitting the data with Global 3.
Effect of Metal Ions and Chemical Reagents on GI Activity and Kinetic Parameters Determination
In order to investigate the effects of metal ions and chemical agents on the activity, the reaction was tested in a buffer solution of NaH2PO4-Na2HPO4 buffer (pH 7.0) with 10 mmol/L (final concentration) EDTA, SDS, β-mercaptoethanol (β-ME), CoCl2, BaCl2, FeSO4, CaCl2, MgCl2, ZnCl2, MnCl2, CuCl2, or NiCl2 at 80 ℃. In addition, 1 mmol/L Co2+ was employed to investigate the synergistic effect of metal ions and chemical reagents. The activity of adding CoCl2 is defined as 100%. Kinetic parameters were determined in the range of 10 − 60 mmol/L for GI using glucose as substrate. The obtained data were analyzed by the Lineweaver − Burk plot method [35].
Cellular Catalysis of GI
The bioconversion of recombinant cells was tested at 85 ℃ (oil bath), and cooling devices were added to prevent evaporation. A total volume of 20 mL reaction system includes 50 g/L wet cells, 200 g/L D-glucose, 1 mmol/L Co2+, 10 mmol/L Mg2+, 50 mmol/L NaH2PO4-Na2HPO4 buffer (pH 7.0) for 4 h. Samples were withdrawn at different time intervals, and the conversion rate was measured. The ratio of the generated D-fructose to the leftover D-glucose was used to compute the conversion.
Results and Discussion
Mining and Screening of Thermostable GIs
To improve the efficacy of industrial fructose production, it is necessary to extract a GI gene with a high level of activity and thermal stability. Due to the low cost, high throughput, and high precision of new DNA sequencing technologies, a large number of organisms have been sequenced [36]. As more extreme microorganisms are sequenced, genome mining is becoming a powerful tool for mining industrial enzymes applied to extreme conditions. The mining of genes from gene libraries not only reduces the workload of conventional screening, but also facilitates the targeting of enzymes with particular properties. In the present study, thermophilic glucose isomerases were extracted via database mining. According to previous reports, the free N-terminal and C-terminal of proteins play a crucial role in protein stability. Consequently, the energy results obtained from FoldX and Rosetta calculations (Fig S1) were utilized to design GeneBank-mining virtual probes (Fig S2) at the N-terminal and C-terminal ends. As a result, nine potential thermophilic glucose isomerase genes were mined from the database (Fig S3). The amount of remaining glucose was detected by a glucose oxidase kit for preliminary screening. Three GIs including CAGI, TTGI, and TPGI were ultimately selected for subsequent analysis.
Gene Cloning, Expression, and Protein Purification
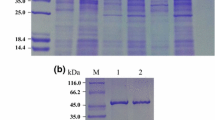
To determine the expression level of GI genes, the recombinant plasmids pET-28b carrying various GIs were transformed into E. coli BL21 (DE3). The purified and desalted protein was verified by SDS-PAGE, and the theoretical molecular weights of CAGI, TTGI, and TPGI were 51.04 kDa, 50.96 kDa, and 51.79 kDa, respectively (Fig. 1). The molecular weights of three GIs under physiological conditions was of 130 kDa as determined by SEC-HPLC (Fig S5). Combined with the theoretical molecular weight of GI, GIs exists primarily in the form of homologous dimer (theoretical molecular weight of 102 kDa) under physiological conditions. In addition, some proteins with small molecular weights might be the results of improper folding during the expression process. The failure to assemble correctly could theoretically impact the enzyme activity. The concentrations of purified enzymes determined by BCA Kit were 4.94 mg/mL, 7.95 mg/mL, and 1.42 mg/mL for CAGI, TTGI, and TPGI, respectively.
Biochemical Characterization of CAGI, TTGI, and TPGI
Effect of pH on GI Activity
Different pH affects the conformational change of the enzyme, resulting in varying activities [37]. The enzyme activities of all three enzymes CAGI, TTGI, and TPGI reached the highest at pH 8.0 (Fig. 2a). At pH 6.5–7.0, CAGI can retain greater than 88% of its relative activity, and TTGI can also retain approximately 80% of its activity. The activity of TPGI decreased significantly in an acidic environment and was lost at pH 6.0. The pH stability of GI was subsequently determined. The three enzymes were more stable in an acidic environment, as demonstrated by the results (Fig. 2b). Nonetheless, TPGI maintained 88% relative activity at pH 3.0 while CAGI and TTGI exhibited no detectable activity at the same condition. Following a 1 h incubation at pH 4.0–6.0 and 25 ℃, the residual enzyme activities of all three enzymes were higher than 97%. After 1 h of incubation at an alkaline pH (8.0–11.0), the residual enzyme activity of the TTGI and TPGI was still above 63%. These results indicate that GIs can be stored over a broad pH range without significant enzyme activity loss. All three enzymes are highly active in an environment with a slightly acidic pH. A slightly acidic environment minimizes the interaction between glucose and the ε-amino group of lysine residues, thereby preventing GI inactivation and lowering production costs. However, the optimal pH range for commercial GI is typically 7.5–9.0, and they show low activity in acidic environments [21]. This suggests that the mined enzyme in this study is a potential GI candidate for commercialization.
Enzymatic properties of purified GIs. a Effects of pH on the GIs determined at 80 ℃ from pH 5.0 to 10.0. b pH stability assay. After pre-incubation of the GIs at pH 3.0–12.0 and 25 ℃ for 2 h, the residual activities of GIs were determined at optimal condition. c and d Thermoactivities of the GIs at 50–95 ℃ (water bath) and 95–120 ℃ (oil bath). e and f Thermostabilities assays of GIs and GIs with metal ions. The GIs were pre-incubated at 70, 80, or 90 ℃, and aliquots were removed at specific. Error bars represent the means ± SD (n = 3)
Effect of Temperature on GI Activity
The thermal activity of GI was measured at 50–120 ℃. The optimal temperature for TPGI was 90 ℃, and CAGI and TTGI still maintained high activities in the temperature range of 90–120 ℃ (Fig. 2c and d). After 1-h incubation at 90 ℃, the residual enzyme activities of TTGI and CAGI were 6.9% and 3.4%, while TPGI was completely inactivated (Fig. 2e). The thermal stability of GI would be significantly enhanced by the presence of metal ions according to the previous studies [38, 39]. Our results confirmed that the auxiliary effect of metal ions on CAGI and TTGI activities as they preserved around 90% of their initial activities after 12-h incubation at 90 ℃, and TPGI retained 52.8% (Fig. 2f). Extending the incubation time to 24 h, the residual enzyme activity of TTGI was 52.6%, and CAGI was 40.1%. CAGI and TTGI demonstrated superior thermal stability. The Tm values of CAGI, TTGI, and TPGI were 67.9 ± 0.4 ℃, 64.9 ± 0.3 ℃, and 62.0 ± 0.4 ℃, respectively (Fig. 3). The higher Tm value of CAGI was consistent with the highest thermal stability within 12 h. The thermal stability of the selected enzyme was superior to what had been previously reported. The optimal temperatures for reported GIs from Geobacillus caldoxylosilyticu [40], Caldicellulosiruptor bescii [21], Anoxybacillus gonensisG2T [18], and Caldicoprobacter algeriensis [20] were at 80, 80, 50, and 90 ℃, respectively. The residual enzyme activity of Caldicoprobacter algeriensis GI [20] was approximately 28% of the initial activity after 90 ℃ incubation for 2 h. A GI from Bacillus sp [41] retained around 20% of its initial activity at 80 ℃ after 30 min of incubation. A GI of Caldicellulosiruptor bescii [21] lost about 20% of its initial activity at 90 ℃ after 2 h of incubation.
Molecular dynamics (MD) simulation has been widely used to analyze the thermal stability of enzymes recently. The thermal stability mechanisms of fructosidases and amylases have been analyzed by MD simulation. MD can provide contribution of region to thermal stability via the root mean square fluctuation (RMSF) and guide subsequent experiments [42]. The conformational changes of CAGI and TTGI were simulated at 90 ℃ for 50 ns for investigation of thermal stability mechanisms. It was found that TTGI exhibits greater flexibility at loop-helix (aa 66–85) (Fig S7). Previous studies have demonstrated that the conformation of proteins with greater flexibility has a higher entropy, resulting in a greater free energy required for denaturation and thereby preserving their structural stability. The thermal stability of TTGI after 12 h is superior to that of CAGI, confirming the above analysis. Therefore, we hypothesize that the flexible terminal of GI with a certain degree of flexibility typically contributes more to the thermal stability.
The difference in thermal stability is not due to a single factor, but rather to the cumulative effect of numerous weak interactions. In general, β-folds are generally considered to be more stable than α-helices. It was discovered that the intrinsic dipole of the α-helix is stabilized by a positively charged residue at the C-terminal and a negatively charged residue at the N-terminal, which results in an antiparallel arrangement of adjacent helices and provides the protein with stabilizing energy of 5000–7000 kcal/mol [43]. TTGI contained 17% β-fold and demonstrated greater thermal stability than CAGI and TPGI, both of which contained 16%. The aromatic interactions including π-π and cation-π interaction were also one of the factors that influence the structural stability of the protein. A pair of aromatic contacts on the surface of the protein or in close proximity to the active site can increase protein stability by 0.6–1.3 kcal/mol [42]. It was determined that there were 235 aromatic H-bonds in CAGI and 232 in TTGI and none in TPGI (Table 1). Similar results were found for π-π and cation-π, CAGI was 36 and 16, and TTGI was 44 and 8, respectively. TPGI did not produce any aromatic interaction. Consequently, we speculate that the aromatic interactions might be one of the factors influencing the thermal stability of GI.
Effect of Metal Ions and Chemical Reagents on Activity and Kinetic Parameters
There are two metal binding sites in the active center of GIs named M1 and M2, known as structural and catalytic metal sites, respectively [44]. GIs showed no activity when no metal ions were present (Fig. 4). All three enzymes were activated in the presence of Co2+, while Mg2+ had a slight activating effect on CAGI and TTGI. In the presence of Co2+, the addition of Mg2+ increased TTGI activity by up to 115.9%. The addition of additional Mg2+ or Co2+ had no effect on CAGI or TPGI. Meal ion Fe2+ has a slight activating effect on TTGI in the presence of Co2+, and the relative vitality was 107.8%, which differs from previous studies [21, 35]. Comparatively, other metal ions and chemical reagents inhibited the activities of GIs, which was consistent with previous reports [11, 20, 21, 39]. It has been demonstrated that metal ions with a radius greater than 0.8 Å have softer electrostatic interactions with GI, which inhibits protein activity [45]. This was also supported by the fact that the radii of Ba2+ (1.35 Å) and Ca2+ (0.99 Å) were greater than 0.8 Å, inhibiting the GI activity to varying degrees.
The effects of ions or reagents on purified GIs. CK is to add 1 mmol/L Co2+. Hollow columns indicate the addition of 10 mmol/L metal ions or chemical reagents. Solid columns indicate the addition of 10 mmol/L metal ions or chemical reagents. Solid columns indicate the addition of 1 mmol/L Co.2+ and 10 mmol/L metal ions or chemical reagents. Error bars represent the means ± SD (n = 3)
Kinetic parameters of the GI were determined toward D-glucose at 80 ℃. The Km and Vmax of CAGI were estimated to be 64.0 ± 2.3 µmol/L and 59.2 ± 2.3 U/mg, respectively, while those of TTGI were 103.4 ± 14.3 µmol/L and 72.8 ± 7.0 U/mg (Table 2). The Km of TPGI was 85.7 ± 9.0 µmol/L, and Vmax was 131.9 ± 9.3 U/mg. The maximum velocity of the mined GI was significantly greater than what was reported (Table 2). The significance of comparing Km value alone is obscure. The turnover number (kcat/Km) of CAGI, TTGI, and TPGI were 46.2 L/min mmol, 35.3 L/min mmol, and 77.0 L/min mmol, respectively. This indicated that the catalytic efficiency of these three enzymes were high, while those of GIs from Thermoanaerobacter saccharolyticum [17] and Bacillus stearothermophilus [46] were only 2.6 L/min mmol and 1.5 L/min mmol, respectively. These indicated that the three GIs discovered in this study have a higher catalytic efficiency.
Cellular Catalysis
Biocatalysis has entered a golden age with the advent of bioinformatics, high-throughput screening, and enzyme-directed evolution. Currently, immobilized GIs are used in the industrial production of fructose. Typically, the cell membrane prevents substrate and product from penetrating, resulting in a weak cellular catalytic effect. However, the extraction of enzymes and their greater fragility compared to intact cells would increase the production cost. Here, recombinant cells of CAGI and TTGI with excellent thermal stability and high catalytic performance were used as catalysts to reduce the cost of fructose production. Within 30 min, the yield of D-fructose by CAGI and TTGI reached 47.2% and 48.5%, respectively. The reaction equilibrium was achieved after 1 h, and a bioconversion rate of 54.0% was reached until the 4-h reaction (Fig. 5). Neifar et al. [20] used a purified enzyme solution to reach 54.7% after 120 min at 85 ℃ with the substrate concentration of 0.8 mol/L. The conversion by purified GI from Thermotoga naphthophila RKU-10 reached 52.2% at 95 ℃ [23]. Deng et al. [50] reported that an equilibrium was reached after 4 h at 70 ℃ with a maximum conversion of 53% by a GI of Thermobifida Fusca WSH03-11. Jia et al. [39] achieved 52% conversion after 5 h using recombinant GI cells. In comparison, CAGI and TTGI screened in this study exhibited higher catalytic efficiency and shorter times to reach reaction equilibrium (Table S2). These advantages indicated that the GIs mined in this study are ideal candidates for low-cost industrial fructose production.
Conclusion
In conclusion, bioinformatics software-designed virtual probes were used to successfully mine thermophilic GIs, which were then heterologously expressed in E. coli, and the enzyme properties of three GIs were measured. The TTGI still retained 52.6% of the enzyme activity after 24 h of incubation at 90 ℃, much higher than previously reported GIs, indicating that TTGI is superior for industrial production of D-fructose with excellent catalytic efficiency at high temperature. After half an hour of catalysis by TTGI, a maximum conversion of 54.1% was achieved. This research provides not only an attractive candidate GI for low-cost fructose production, but also a theoretical foundation for computer simulation studies of GIs.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Riby, J. E., Fujisawa, T., & Kretchmer, N. (1993). Fructose absorption. American Journal of Clinical Nutrition, 58, 748–753.
Jang, C., Hui, S., Lu, W., Cowan, A. J., Morscher, R. J., Lee, G., Liu, W., Tesz, G. J., Birnbaum, M. J., & Rabinowitz, J. D. (2018). The small intestine converts dietary fructose into glucose and organic acids. Cell metabolism, 27, 351–361.
Shinn, A. K., & Greenfield, S. F. (2010). Topiramate in the treatment of substance-related disorders: A critical review of the literature. The Journal of Clinical Psychiatry, 71, 634–648.
Li, H., Yang, S., Saravanamurugan, S., & Riisager, A. (2017). Glucose isomerization by enzymes and chemo-catalysts: Status and current advances. Acs Catalysis, 7, 3010–3029.
Campos, V. C., & Tappy, L. (2016). Physiological handling of dietary fructose-containing sugars: Implications for health. International Journal of Obesity, 40, S6–S11.
Moulin, S., Seematter, G., & Seyssel, K. (2017). Fructose use in clinical nutrition: Metabolic effects and potential consequences. Current Opinion in Clinical Nutrition and Metabolic Care, 20, 272–278.
Araya, E., Urrutia, P., Romero, O., Illanes, A., & Wilson, L. (2019). Design of combined crosslinked enzyme aggregates (combi-CLEAs) of beta-galactosidase and glucose isomerase for the one-pot production of fructose syrup from lactose. Food Chemistry, 288, 102–107.
Wu, R. C. Y., Botts, S. R., Johnson-Henry, K. C., Landberg, E., Abrahamsson, T. R., & Sherman, P. M. (2022). Variations in the composition of human milk oligosaccharides correlates with effects on both the intestinal epithelial barrier and host inflammation: A pilot study. Nutrients, 14, 1014.
Munshi, P., Snell, E. H., van der Woerd, M. J., Judge, R. A., Myles, D. A. A., Ren, Z., & Meilleur, F. (2014). Neutron structure of the cyclic glucose-bound xylose isomerase E186Q mutant. Acta Crystallographica Section D: Structural Biology, 70, 414–420.
Bhosale, S. H., Rao, M. B., & Deshpande, V. V. (1996). Molecular and industrial aspects of glucose isomerase. Microbiological Reviews, 60, 280–300.
Liu, Z. Q., Zheng, W., Huang, J. F., Jin, L. Q., Jia, D. X., Zhou, H. Y., Xu, J. M., Liao, C. J., Cheng, X. P., Mao, B. X., & Zheng, Y. G. (2015). Improvement and characterization of a hyperthermophilic glucose isomerase from Thermoanaerobacter ethanolicus and its application in production of high fructose corn syrup. Journal of Industrial Microbiology & Biotechnology, 42, 1091–1103.
Xu, H., Shen, D., Wu, X. Q., Liu, Z. W., & Yang, Q. H. (2014). Characterization of a mutant glucose isomerase from Thermoanaerobacterium saccharolyticum. Journal of Industrial Microbiology & Biotechnology, 41, 1581–1589.
Cho, J. W., Han, B. G., Park, S. Y., Kim, S. J., Kim, M. D., & Lee, B. I. (2013). Overexpression, crystallization and preliminary X-ray crystallographic analysis of a putative xylose isomerase from Bacteroides thetaiotaomicron. Acta Crystallographica, Section F: Structural Biology and Crystallization Communications, 69, 1127–1130.
Ben Hlima, H., Bejar, S., Riguet, J., Haser, R., & Aghajari, N. (2013). Identification of critical residues for the activity and thermostability of Streptomyces sp SK glucose isomerase. Applied Microbiology and Biotechnology, 97, 9715–9726.
Jenkins, J., Janin, J., Rey, F., Chiadmi, M., Vantilbeurgh, H., Lasters, I., Demaeyer, M., Vanbelle, D., Wodak, S. J., Lauwereys, M., Stanssens, P., Mrabet, N. T., Snauwaert, J., Matthyssens, G., & Lambeir, A. M. (1992). Protein engineering of xylose (glucose) isomerase from Actinoplanes-missouriensis.1. crystallography and site-directed mutagenesis of metal-binding sites. Biochemistry, 31, 5449–5458.
Vuilleumier, S. (1993). Worldwide production of high-fructose syrup and crystalline fructose. American Journal of Clinical Nutrition, 58, 733–736.
Lee, C., & Zeikus, J. G. (1991). Purification and characterization of thermostable glucose-isomerase from Clostridium-thermosulfurogenes and Thermoanaerobacter strain b6a. Biochemical Journal, 273, 565–571.
Karaoglu, H., Yanmis, D., Sal, F. A., Celik, A., Canakci, S., & Belduz, A. O. (2013). Biochemical characterization of a novel glucose isomerase from Anoxybacillus gonensis G2(T) that displays a high level of activity and thermal stability. Journal of Molecular Catalysis b-enzymatic, 97, 215–224.
Kim, B. C., Yu, S. N., Kim, K. Y., Lee, J. S., Pyun, Y. R., & Ahn, S. C. (2010). Cloning, expression and characterization of xylose isomerase, XylA, from Caldanaerobacter subterraneus subsp yonseiensis. Biotechnology Letters, 32, 929–933.
Neifar, S., Ben Hlima, H., Mhiri, S., Mezghani, M., Bouacem, K., Ibrahim, A. H., Jaouadi, B., Bouanane-Darenfed, A., & Bejar, S. (2019). A novel thermostable and efficient Class II glucose isomerase from the thermophilic Caldicoprobacter algeriensis: Biochemical characterization, molecular investigation, and application in high fructose syrup production. International Journal of Biological Macromolecules, 129, 31–40.
Dai, C. X., Miao, T. T., Hai, J. P., Xiao, Y. Y., Li, Y., Zhao, J. R., Qiu, H. L., & Xu, B. (2020). A novel glucose isomerase from Caldicellulosiruptor bescii with great potentials in the production of high-fructose corn syrup. BioMed Research International, 2020(7), 1871934.
Srih-Belghith, K., & Bejar, S. (1998). A thermostable glucose isomerase having a relatively low optimum pH: Study of activity and molecular cloning of the corresponding gene. Biotechnology Letters, 20, 553–556.
Fatima, B., Aftab, M. N., & Ikram-ul, H. (2016). Cloning, purification, and characterization of xylose isomerase from Thermotoga naphthophila RKU-10. Journal of Basic Microbiology, 56, 949–962.
Rigoldi, F., Donini, S., Redaelli, A., Parisini, E., & Gautieri, A. (2018). Review: Engineering of thermostable enzymes for industrial applications. APL Bioengineering, 011501, 2–17.
Beerens, K., Mazurenko, S., Kunka, A., Marques, S. M., Hansen, N., Musil, M., Chaloupkova, R., Waterman, J., Brezovsky, J., Bednar, D., Prokop, Z., & Damborsky, J. (2018). Evolutionary analysis as a powerful complement to energy calculations for protein stabilization. ACS Catalysis, 8, 9420–9428.
Cheng, Z., Lan, Y., Guo, J., Ma, D., Jiang, S., Lai, Q., Zhou, Z., & Peplowski, L. (2020). Computational design of nitrile hydratase from Pseudonocardia thermophila JCM3095 for improved thermostability. Molecules, 25, 4806.
Dvorak, P., Bednar, D., Vanacek, P., Balek, L., Eiselleova, L., Stepankova, V., Sebestova, E., Bosakova, M. K., Konecna, Z., Mazurenko, S., Kunka, A., Vanova, T., Zoufalova, K., Chaloupkova, R., Brezovsky, J., Krejci, P., Prokop, Z., Dvorak, P., & Damborsky, J. (2018). Computer-assisted engineering of hyperstable fibroblast growth factor 2. Biotechnology and Bioengineering, 115, 850–862.
Musil, M., Stourac, J., Bendl, J., Brezovsky, J., Prokop, Z., Zendulka, J., Martinek, T., Bednar, D., & Damborsky, J. (2017). FireProt: Web server for automated design of thermostable proteins. Nucleic Acids Research, 45, W393–W399.
Willard, L., Ranjan, A., Zhang, H. Y., Monzavi, H., Boyko, R. F., Sykes, B. D., & Wishart, D. S. (2003). VADAR: A web server for quantitative evaluation of protein structure quality. Nucleic Acids Research, 31, 3316–3319.
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., Heer, F. T., de Beer, T. A. P., Rempfer, C., Bordoli, L., Lepore, R., & Schwede, T. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Research, 46, W296–W303.
Colovos, C., & Yeates, T. O. (1993). Verification of protein structures - Patterns of nonbonded atomic interactions. Protein Science, 2, 1511–1519.
Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30, 2785–2791.
Shen, J. D., Cai, X., Ni, Y. W., Jin, L. Q., Liu, Z. Q., & Zheng, Y. G. (2021). Structural insights into the thermostability mechanism of a nitrile hydratase from Caldalkalibacillus thermarum by comparative molecular dynamics simulation. Proteins-Structure Function and Bioinformatics, 89, 978–987.
Khramtsov, P., Kalashnikova, T., Bochkova, M., Kropaneva, M., Timganova, V., Zamorina, S., & Rayev, M. (2021). Measuring the concentration of protein nanoparticles synthesized by desolvation method: Comparison of Bradford assay, BCA assay, hydrolysis/UV spectroscopy and gravimetric analysis. International Journal of Pharmaceutics, 599, 18.
Lineweaver, H., & Burk, D. (1934). The determination of enzyme dissociation constants. Journal of the American Chemical Society, 56, 658–666.
Chu, L., Huang, J., Muhammad, M., Deng, Z., & Gao, J. (2020). Genome mining as a biotechnological tool for the discovery of novel marine natural products. Critical reviews in biotechnology, 40, 571–589.
Liu, Z. Q., Dong, S. C., Yin, H. H., Xue, Y. P., Tang, X. L., Zhang, X. J., He, J. Y., & Zheng, Y. G. (2017). Enzymatic synthesis of an ezetimibe intermediate using carbonyl reductase coupled with glucose dehydrogenase in an aqueous-organic solvent system. Bioresource Technology, 229, 26–32.
Bandlish, R. K., Hess, J. M., Epting, K. L., Vieille, C., & Kelly, R. M. (2002). Glucose-to-fructose conversion at high temperatures with xylose (glucose) isomerases from Streptomyces murinus and two hyperthermophilic Thermotoga species. Biotechnology and Bioengineering, 80, 185–194.
Jia, D. X., Zhou, L., & Zheng, Y. G. (2017). Properties of a novel thermostable glucose isomerase mined from Thermus oshimai and its application to preparation of high fructose corn syrup. Enzyme and Microbial Technology, 99, 1–8.
Kim, B. C., Yu, S. N., Kim, K. Y., Lee, J. S., Pyun, Y. R., & Ahn, S. C. (2010). Cloning, expression and characterization of xylose isomerase, xylA, from Caldanaerobacter subterraneus subsp yonseiensis. Biotechnol Letters, 32, 929–933.
Chauthaiwale, J., & Rao, M. (1994). Production and purification of extracellular D-xylose isomerase from an alkaliphilic, Thermophilic bacillus sp. Applied and Environmental Microbiology, 60, 4495–4499.
Chen, Q. M., Zhang, W. L., & Mu, W. M. (2021). Molecular Dynamics simulation for food enzyme engineering: Why this technique should be encouraged to learn. Journal of Agricultural and Food Chemistry, 69, 4–6.
Renugopalakrishnan, V., Garduno-Juarez, R., Narasimhan, G., Verma, C. S., Wei, X., & Li, P. Z. (2005). Rational design of thermally stable proteins: Relevance to bionanotechnology. Journal of Nanoscience and Nanotechnology, 5, 1759–1767.
Whitlow, M., Howard, A. J., Finzel, B. C., Poulos, T. L., Winborne, E., & Gilliland, G. L. (1991). A metal-mediated hydride shift mechanism for xylose isomerase based on the 1.6 Å Streptomyces-rubiginosus structures with xylitol and D-xylose. Proteins-Structure Function and Genetics, 9, 153–173.
Kovalevsky, A. Y., Hanson, L., Fisher, S. Z., Mustyakimov, M., Mason, S. A., Forsyth, V. T., Blakeley, M. P., Keen, D. A., Wagner, T., Carrell, H. L., Katz, A. K., Glusker, J. P., & Langan, P. (2010). Metal ion roles and the movement of hydrogen during reaction catalyzed by D-xylose isomerase: A joint X-ray and neutron diffraction study. Structure, 18, 688–699.
Kaneko, T., Saito, K., Kawamura, Y., & Takahashi, S. (2001). Molecular cloning of acid-stable glucose isomerase gene from Streptomyces olivaceoviridis E-86 by a simple two-step PCR method, and its expression in Escherichia coli. Bioscience Biotechnology and Biochemistry, 65, 1054–1062.
Sriprapundh, D., Vieille, C., & Zeikus, J. G. (2000). Molecular determinants of xylose isomerase thermal stability and activity: Analysis of thermozymes by site-directed mutagenesis. Protein Engineering, 13, 259–265.
Vieille, C., Hess, J. M., Kelly, R. M., & Zeikus, J. G. (1995). Xyla cloning and sequencing and biochemical-characterization of xylose isomerase from Thermotoga-neapolitana. Applied and environmental microbiology, 61, 1867–1875.
Brown, S. H., Sjøholm, C., & Kelly, R. M. (1993). Purification and characterization of a highly thermostable glucose isomerase produced by the extremely thermophilic eubacterium, Thermotoga maritima. Biotechnology and Bioengineering, 41, 878–886.
Deng, H., Chen, S., Wu, D., Chen, J., & Wu, J. (2014). Heterologous expression and biochemical characterization of glucose isomerase from Thermobifida fusca. Bioprocess and Biosystems Engineering, 37, 1211–1219.
Funding
This work was supported by the Innovation Demonstration Special Project of Henan Province in China (grant number 201111110100) and Major Science and Technology Projects for Public Welfare of Henan Province in China (grant number 201300110300).
Author information
Authors and Affiliations
Contributions
Yu-Qi Dong: methodology, software, validation, investigation, and writing—original draft. Ji-Dong Shen: methodology, software, validation, investigation, and writing—original draft. Long Pan: validation and investigation. Ji-Hong Huang: visualization and supervision. Zhi-Qiang Liu: visualization and supervision. Yu-Guo Zheng: supervision.
Corresponding authors
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Yes. All authors agreed to participate in this research.
Consent for Publication
Yes. All authors have approved the last version of the manuscript for its submission.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dong, YQ., Shen, JD., Pan, L. et al. Mining and Characterization of Thermophilic Glucose Isomerase Based on Virtual Probe Technology. Appl Biochem Biotechnol 195, 4399–4413 (2023). https://doi.org/10.1007/s12010-023-04349-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-023-04349-5