Abstract
Various detection methods have been developed to date for identification of animal species. New techniques based on PCR approach have raised the hope of developing better identification methods, which can overcome the limitations of the existing methods. PCR-based methods used the mitochondrial DNA (mtDNA) as well as nuclear DNA sequences. In this study, by targeting nuclear DNA, multiplex PCR and real-time PCR methods were developed to assist with qualitative and quantitative analysis. The multiplex PCR was found to simultaneously and effectively distinguish four species (fox, dog, mink, and rabbit) ingredients by the different sizes of electrophoretic bands: 480, 317, 220, and 209 bp. Real-time fluorescent PCR’s amplification profiles and standard curves showed good quantitative measurement responses and linearity, as indicated by good repeatability and coefficient of determination R 2 > 0.99. The quantitative results of quaternary DNA mixtures including mink, fox, dog, and rabbit DNA are in line with our expectations: R.D. (relative deviation) varied between 1.98 and 12.23% and R.S.D. (relative standard deviation) varied between 3.06 and 11.51%, both of which are well within the acceptance criterion of ≤ 25%. Combining the two methods is suitable for the rapid identification and accurate quantification of fox-, dog-, mink-, and rabbit-derived ingredients in the animal products.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wal-Mart Stores Inc., the world’s largest retailer, has recalled donkey meat in its stores in eastern China after it declared that tests identified fox DNA in the product in 2014. The carcasses of fur animals (such as mink and fox), as byproducts of the fur industry, are generally inedible because of their particularly gamey flavor. However, slaughterhouses must dispose of the byproducts of the many fur animals killed every year, and they have found a maximally profitable solution by selling them to food and feedstuff manufacturers at a low price, such that these carcasses circulate in the animal products market as unclearly labeled ingredients [1]. Meanwhile, prohibited food items, such as fox, mink, and even euthanized dog meat, in various dishes have put Muslim consumers on red alert to determine the halal status of marketed foods [2]. In addition, highly valued mink furs and fox furs are often dyed, renovated, and replaced by cheaper rabbit fur. Many unscrupulous traders profiteer from refurbishing second-hand fur or adulterating fur using advanced processing technology, because of the high price of fur products [3]. Consumers should be protected from these malicious practices by the quick, precise, and specific identification of animal species.
Several analytical methods have been employed to identify animal species, based on anatomical, histological, microscopic, organoleptic, chemical, electrophoretic, chromatographic, or immunological principles [4]. However, because of the inherent limitations of these methods, DNA-based methods have been considered as more powerful tools [5]. DNA has a higher thermal stability, is present in the majority of cells, and its sequence data can be analyzed at different levels of specificity, from groups or species to even individuals, regardless of the tissue origin [6, 7]. During the last decade, several polymerase chain reaction (PCR) techniques using species-specific primers have been developed and successfully applied to identify and authenticate animal species [8]. Most reported PCR applications for animal species identification have focused on domestic animals, such as pig, cattle, sheep, goat, and chicken [9,10,11,12]. PCR-based methods have used mitochondrial DNA (mtDNA) and nuclear DNA sequences as molecular markers for species identification [13, 14]. In general, PCR amplification of mtDNA is used extensively in species determination, and its strength is demonstrated by the abundance of the mitochondrial genome in cells [15]. However, the copy number of mitochondrial DNA varies by species, and in different tissue types within the same species. Therefore, mitochondrial DNA is not suitable for quantification; however, nuclear DNA has a great potential [6, 15].
In contrast to traditional PCR technology, multiplex PCR achieves the simultaneous amplification of multiple DNA targets in a single reaction vessel, offering the distinct advantage of increased speed, and lower costs and reagent use [16,17,18,19]. Multiplex PCR uses a mixture of primer pairs to amplify multiple oligonucleotide targets in a single reaction tube, and its products are identified on agarose gels based on size differences. Multiplex PCR assays have been developed by several researchers to identify ruminant, poultry, fish, and pork ingredients using multiple primer pairs that targeted nuclear and mitochondrial DNA to generate species-specific PCR fragments [20,21,22]. Over time, multiplex PCR has been recognized as a robust, cost-effective, sensitive, and reliable method to discriminate banned species from permitted products [23].
In contrast to conventional PCR techniques, real-time PCR approaches allow discrimination and measurement of even minute traces of DNA in products containing animal ingredients. The simplest, least expensive, and most direct fluorescent system adapted to real-time PCR involves the incorporation of SYBR Green I dye, which binds to the minor groove of double-stranded DNA. The intensity of the fluorescence is proportional to the amount of generated double-stranded DNA, permitting the quantification of DNA. In addition, the specificity of the amplified products can be verified, and false positive signals, caused by non-specific amplification or primer-dimers, can be identified by analyzing real-time PCR melting curves [1]. Additionally, the use of fluorescent DNA-intercalating dyes is more flexible, because it eliminates the need for individual probe design [24]. The use of probes results in greater sensitivity and specificity, but at a higher cost. Considering its advantages, real-time PCR has been employed to conduct quantitative research by many experts. In particular, since the horsemeat scandal of 2013, real-time PCR has been widely used to detect animal DNA in food products. These studies have mostly concentrated on identifying or measuring pork, beef, rabbit meat, and horsemeat [15, 25,26,27], whereas studies on fur animal species are rare. Therefore, in the present study, we used real-time fluorescent PCR to accurately evaluate the presence and quantity of animal DNA.
In this study, the multiplex PCR and real-time fluorescent PCR were employed to achieve rapid, accurate, and cost-effective species identification for fox, dog, mink, and rabbit based on species-specific nuclear DNA sequences. A multiplex PCR was developed for the qualitative identification, and real-time PCR was performed for quantitative determination. This approach enabled us to identify mink, fox, rabbit, and dog ingredients and detect the proportion of the four adulterant species in meat products and feeds. Furthermore, this method could be used to distinguish mink fur and fox fur from adulterants and other unknown animal-labeled products.
Materials and Methods
Preparation of Samples
Skin samples of fox and mink were collected from local municipal slaughterhouses, and blood samples from dogs and rabbits were obtained from the Veterinary Hospital of Huazhong Agricultural University. To protect the DNA from enzymatic degradation, all fresh samples were stored frozen at − 20 °C.
DNA Extraction
DNA was extracted from samples of the four species (fox, dog, mink, and rabbit) using the phenol-chloroform method [28]. The DNA concentration was estimated using UV absorption spectrophotometry at 260 nm with a NanoDrop2000 spectrophotometer. The isolated DNA stock solution was stored frozen at − 20 °C.
The Selection of Species-Specific Sequences
DNA sequences of fox, mink, dog, and rabbit were aligned with those of other species (mouse, rat, hamster, guinea pig, cattle, buffalo, sheep, goat, horse, pig, chicken, duck, goose, fox, dog, donkey, mink, rabbit, and raccoon dog) available in GenBank, using basic local alignment search tool (http://www.ncbi.nlm.nih.gov/BLAST). The published DNA sequences of the different species were downloaded from the National Center for Biotechnology Information (NCBI) GenBank. After alignment, unique sequences were selected.
Primer Design
The principle used to screen the primers was that each pair of primers had intraspecies universality and interspecies specificity. The fox, dog, mink, and rabbit species-specific primer pairs were designed using Primer Premier 5.0 software (PREMIER Biosoft, Palo Alto, CA, USA), avoiding self-dimers, cross-dimers, and hairpin structures. The specificity of the species-specific primers was confirmed using the Primer-BLAST program (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). The primer pairs were synthesized by Tsingke Biological Technology, Co., Ltd. (Beijing, China). The four sets of primers used in our investigation are listed in Table 1. The four species-specific primer pairs have already been patented in China (CN105296477A, CN105296648A, CN105349653A, under substantive examination).
Verification of Species Specificity
The species specificity of the fox, dog, mink, and rabbit primers was then assessed using DNA from a wide variety of species, including mouse, rat, hamster, guinea pig, cattle, buffalo, sheep, goat, horse, pig, chicken, duck, goose, fox, dog, donkey, mink, rabbit, and raccoon dog, which were provided by Liu Bang’s Group (Key Laboratory of Agricultural Animal Genetics, Breeding and Reproduction of Ministry of Education & Key Laboratory of Pig Genetics and Breeding of Ministry of Agriculture, Huazhong Agricultural University, Wuhan, China). Conventional PCR and real-time PCR assays using the fox, dog, mink, and rabbit species-specific primers were processed among the above 19 species to confirm primer specificity.
Conventional PCR
All the PCRs were conducted in a total volume of 20 μL, containing 10 μL of 2× Taq PCR MasterMix (Aidlab, Beijing) for PCR; 0.2, 0.15, 0.3, and 0.15 μΜ of four primer pairs (fox, dog, mink, and rabbit, respectively); a DNA pool of 50 ng DNA from each of the four species; and sterile water to make up the volume. PCR amplifications were run using a MyCycler™ Thermal cycler under the following cycling conditions: denaturation at 94 °C for 5 min, 30 cycles of 94 °C for 30 s, 64 °C for 30 s, and 72 °C for 30 s. The PCR products were stored at 4 °C for further analysis. The predicted species-specific product sizes from fox, dog, mink, and rabbit were 480, 317, 220, and 209 bp, respectively. Each targeted species-specific primer was validated by a PCR assay, with DNA from 18 species (excluding the target species) and water as negative control. The specificity of the conventional PCR products was analyzed by 2% agarose gel electrophoresis containing gel-red in 1× Tris-acetate-EDTA (TAE) buffer for 30 min at 100 V. The gels were visualized and photographed in a Bio-Rad GelDoc 1000 gel documentation system (Bio-Rad, USA). The PCR products were sequenced (Tsingke, China) to verify the specificity of the primers.
Qualitative Real-Time PCR
Real-time PCRs were performed in a volume of 20 μL, containing 10 μL of SYBR® Premix Ex Taq II (Takara, Japan), 0.3, 0.2, 0.3, and 0.2 μΜ of the four primer pairs (fox, dog, mink, and rabbit, respectively), and 2 μL of the DNA templates. The amplifications were conducted using the Bio-Rad CFX96TM Optics Module, and the operational protocol was as follows: 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s, 64 °C for 15 s, and 72 °C for 30 s. A melting curve analysis was then programmed in such a way that its slope was formed from 65 to 95 °C by increasing each step by 0.5 °C. The specificity of the real-time PCR products was identified by analyzing melting curves using the software Bio-Rad CFX Manger 3.1.
Multiplex PCR Assay of Fox, Dog, Mink, and Rabbit
In practice, cheaper materials tend to be adulterated into expensive products. Therefore, we designed 11 PCR reaction tubes with different DNA templates. The four pairs of primers (fox, dog, mink, and rabbit) were mixed at a certain ratio of 4:3:6:3 to a total of 0.8 μΜ. PCR products were analyzed using 3% agarose gel electrophoresis containing gel-red in 0.5× Tris-borate-EDTA (TBE) buffer for 3 h at 100 V.
Quantitative Analysis of Fox, Dog, Mink, and Rabbit Using Real-Time PCR
Construction of Standard Curves
To construct standard curves, the concentration of the pure genomic DNA from the four species (fox, dog, mink, and rabbit) was measured by Nanodrop2000, and the DNA was diluted to 62.5, 12.5, 2.5, 0.5, and 0.1 ng/μL as templates of real-time PCR. Four separate standard curves involving the relationship of the cycle threshold (Ct) value and the common logarithm of DNA template concentration were generated for fox, rabbit, dog, and mink respectively, according to the above description and were used for the quantification detection of the DNA from the four species.
Quantification of Quaternary DNA Mixtures
To achieve quantification of fox, dog, mink, and rabbit DNA, the four species’ DNA was diluted to 30 ng/μL, and three quaternary mixtures in varying proportions were produced (Table 2) and used as templates of quantitative real-time PCR. Combining the formulas from the four standard curves, the relative amount of the target DNA could be calculated by analyzing the real-time PCR data. The formula for the standard curves is y = a x + b, and the process to calculate the DNA content is:
In the formula, N% represents the mink, fox, rabbit, or dog DNA content, Ct value is taken from the real-time PCR for a particular sample, a and b are from the formula of the corresponding standard curve.
Results and Discussion
Verification of Species Specificity
The designed primers were checked for their specificity using the following species: mouse, rat, hamster, guinea pig, cattle, buffalo, sheep, goat, horse, pig, chicken, duck, goose, fox, dog, donkey, mink, rabbit, and raccoon dog. Using the specific primers for fox, dog, mink, and rabbit, no non-specific PCR products were generated in any of the abovementioned target species (Fig. 1). No cases of cross-contamination were observed. The sizes of the specific PCR products of fox, mink, dog, and rabbit were in accordance with the expected sizes: 480, 317, 220, and 209 bp. The melting curves of the amplified products from real-time PCR of mink, fox, rabbit, and dog showed a single melting peak, and there was no melting peak in the melting curves of the blank and negative controls, which revealed that the targeted product of each reaction was a single ingredient, and there was no non-specific amplification or primer-dimers (Fig. 2). Each test was repeated four times, giving reproducible results. The sequenced results of all PCR products were consistent with the targeted sequences. Therefore, the four pairs of primers were considered species-specific and reliable for further study.
Verification of species specificity by conventional PCR. a, b, c, d The amplification by conventional PCR of fox, dog, mink, and rabbit DNA, respectively, to confirm the species specificity. Lane M: BM2000 marker; the DNA templates of PCR products are used in Lane 1: empty; Lane 2: fox; Lane 3: dog; Lane 4: mink; Lane 5: rabbit; Lane 6: mouse; Lane 7: rat; Lane 8: hamster; Lane 9: cavy; Lane 10: cow; Lane 11: buffalo; Lane 12: sheep; Lane 13: goat; Lane 14: horse; Lane 15: pig; Lane 16: chicken; Lane 17: duck; Lane 18: donkey; Lane 19: goose; Lane 20: raccoon dog
Verification of species specificity by real-time fluorescent PCR. a, b, c, d The melting curves of real-time PCR for fox (dark green), dog (orange), mink (red), and rabbit (blue) DNA to confirm the species specificity. The light green curves and pink curves at the bottom are for the blank control and negative control. The DNA templates for the negative control were DNA mixtures of mouse, rat, hamster, cavy, cow, buffalo, sheep, goat, horse, pig, chicken, duck, donkey, goose, raccoon dog, and the studied animals (fox, dog, mink, rabbit), excluding the targeted animal itself in each case. Deionized water instead of DNA template was used in the blank control
Multiplex PCR Assay of Fox, Dog, Mink, and Rabbit
A multiplex PCR approach, which is more rapid and economical in terms of time and labor, can provide simultaneous detection to overcome the challenge observed when using only single gene target PCR formats. By optimizing the PCR conditions, the direct multiplex assay for the simultaneous detection of fox, dog, mink, and rabbit DNA was successfully developed. The number and size of the electrophoretic bands were in accordance with the number and species of the added DNA mixture (Fig. 3). For single species DNA templates (lanes 2–5), the electrophoresis results showed single bands; for DNA templates from two species (lanes 6–9), the results showed two bands; using DNA from three species generated three electrophoretic bands (lanes 10 and 11); and a mixture of all four DNA templates of fox, dog, mink, and rabbit produced four bands on the agarose gel (lane 12). Clear and sharp bands of the expected sizes of 480, 317, 220, and 209 bp for fox, dog, mink, and rabbit, respectively, were observed on an agarose gel electrophoresis of the PCR products. Repeated tests showed that the results were reproducible. The multiplex PCR method we employed is an efficient method to simultaneously identify these four species from DNA mixtures. This method saves experimental time and cost and is likely to show enhanced efficiency to detect animal-derived ingredients in food and feedstuff compared with traditional simplex PCR. The development of the multiplex PCR method will strengthen the labeling management of food and feed, guarantee the benefits to consumers, and promote import and export trade. Moreover, the method could also be applied to distinguish fox, mink, rabbit, and dog fur from any adulterants. Using this method, it would be possible to verify whether the meat, feedstuff or fur product contain one or more of the four animal ingredients, according to the number and size of bands on agarose gel electrophoresis of the multiplex PCR products. At the same time, the presence of any of these species-specific bands in unlabeled ingredients would suggest adulteration.
Multiplex PCR amplification of fox, dog, mink, and rabbit DNA. Lane M: BM2000 marker; Lane 1: negative control; the DNA templates used in Lane 2–5 were fox, dog, mink, and rabbit DNA, respectively; Lanes 6–9: fox/dog, fox/rabbit, mink/dog, mink/rabbit DNA mixtures; Lanes 10–11: fox/dog/rabbit and mink/dog/rabbit DNA mixtures; Lane 12: fox/dog/mink/rabbit DNA mixture
Quantitative Analysis of Fox, Dog, Mink, and Rabbit by Real-Time PCR
Standard Curves of the Four Animal Species
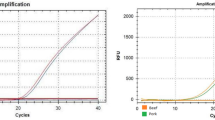
Each standard curve was based on five plots (62.5, 12.5, 2.5, 0.5, 0.1 ng/μL of pure genomic DNA), performed with three technical replicates. Figure 4 shows typical amplification profiles and standard curves associated with the methodology, and highlights good quantitative measurement responses and linearity. The quantitative method here performed presented good repeatability and coefficient of determination (R 2 > 0.99; Table 3). In the early experiments to construct the standard curves, the results indicated that a high DNA concentration (≥ 100 ng/μL) inhibited the amplification of the target fragment. Therefore, choosing a reasonable DNA concentration range is important to construct standard curves, and samples to be detected should be diluted to an appropriate concentration.
Real-time PCR amplification curves. Typical amplification curves (left) and the corresponding standard curves (right) for fox, dog, mink, and rabbit (a, b, c, d). For each standard curve, the five plots’ DNA template concentrations were 62.5, 12.5, 2.5, 0.5, and 0.1 ng/μL, respectively, performed with three technical replicates
Quantification Results of the Quaternary DNA Mixtures
The real-time quantitative PCRs were based on the four species’ serially diluted DNA (62.5 to 0.1 ng/μL), the four pairs of species-specific primers, and three independently prepared quaternary DNA mixtures as templates, with a PCR technical level of six replications across three replicate PCR plates; therefore, each sample was represented by 18 PCR replicates. Combining the formulas from the four corresponding standard curves, the relative amount of the four species’ genomic DNA was calculated by analyzing the real-time PCR data. Table 4 shows the four species’ detected mean DNA content value and closely associated accuracy and precision levels. The relative deviation (R.D.) varied between 1.98 and 12.23%, and the relative standard deviation (R.S.D.) varied between 3.06 and 11.51%; the maximum bias was related to the 5% rabbit DNA level. All R.D. and R.S.D. values in this study lay well within the acceptance criterion of ≤ 25% [29], which showed good comparability between the estimated and expected DNA percentages. These results demonstrated that the quantitative method based on real-time fluorescent PCR could be used to accurately determine the relative content of fox, rabbit, dog, and mink DNA in unknown samples. If the DNA of fox, dog, mink, or rabbit is identified in food, feedstuff, or fur products by qualitative detection, we could obtain the percentage of the DNA of the identified species in the sample by inputting the Ct value gained from the amplification curve into related species’ formula. The amplification curve was generated using amplification of real-time fluorescent PCR with the species-specific primer of identified species. The reliability of the real-time fluorescent quantitative PCR used for species identification relied on the selection of DNA markers is supported by many previous studies [6, 30, 31]. Compared with nuclear DNA, mtDNA is generally regarded as not suitable for quantitative purposes because of the variability in the number of mitochondrial copies among species and between tissue types within the same species [15, 30]. Real-time PCR has been reported to detect meat species in food products frequently in recent years. Some studies simply applied the technique for the qualitative detection of particular species, without achieving quantification [25, 26], whereas other studies combined the technique with specific probes to strengthen the specificity and successfully quantified the target species DNA, but at a high cost [27, 32]. Thus, the real-time quantitative PCR method used in this study will provide a low-cost, highly accurate, precise, and specific way to quantitate DNA from fox-, dog-, mink-, and rabbit-derived products.
Conclusion
In conclusion, we established conventional PCR and real-time qualitative PCR methods to specifically identify fox, dog, mink, and rabbit DNA. The multiplex PCR system represents a relatively inexpensive, simultaneous, and flexible method to identify the origin of fox-, dog-, mink-, and rabbit-derived adulteration, whereas the quantitative PCR was suitable to accurately analyze the ingredients in detail. Our approach, combining multiplex PCR with real-time PCR, uses the same set of primers for species identification among fox, mink, dog, and rabbit. This study presents a useful tool for assessment programs to enforce labeling regulations of animal products.
References
Fajardo, V., Gonzalez, I., Rojas, M., Garcia, T., & Martin, R. (2010). A review of current PCR-based methodologies for the authentication of meats from game animal species. Trends in Food Science & Technology, 21, 408–421.
Mohamad, N. A., Sheikha, A. F. E., Mustafa, S., & Mokhtar, N. F. K. (2013). Comparison of gene nature used in real-time PCR for porcine identification and quantification: a review. Food Research International, 50(1), 330–338.
Subramanian, S., Karthik, T., & Vijayaraaghavan, N. N. (2005). Single nucleotide polymorphism for animal fibre identification. Journal of Biotechnology, 116(2), 153–158.
Kumar, A., Kumar, R. R., Sharma, B. D., Gokulakrishnan, P., Mendiratta, S. K., & Sharma, D. (2015). Identification of species origin of meat and meat products on the DNA basis: a review. Critical Reviews in Food Science and Nutrition, 55(10), 1340–1351.
Santos, C. G., Melo, V. S., Amaral, J. S., Estevinho, L., Oliveira, M. B. P. P., & Mafra, I. (2012). Identification of hare meat by a species-specific marker of mitochondrial origin. Meat Science, 90(3), 836–841.
Ballin, N. Z., Vogensen, F. K., & Karlsson, A. H. (2009). Species determination—can we detect and quantify meat adulteration? Meat Science, 83(2), 165–174.
Fajardo, V., González, I., Martín, I., Rojas, M., Hernández, P. E., García, T., & Martín, R. (2008). Real-time PCR for detection and quantification of red deer (Cervus elaphus), fallow deer (Dama dama), and roe deer (Capreolus capreolus) in meat mixtures. Meat Science, 79(2), 289–298.
Bhat, M. M., Salahuddin, M., Mantoo, I. A., Adil, S., Jalal, H., & Pal, M. A. (2016). Species-specific identification of adulteration in cooked mutton Rista (a Kashmiri Wazwan cuisine product) with beef and buffalo meat through multiplex polymerase chain reaction. Veterinary world, 9(3), 226.
Calvo, J. H., Zaragoza, P., & Osta, R. (2001). Technical note: a quick and more sensitive method to identify pork in processed and unprocessed food by PCR amplification of a new specific DNA fragment. Journal of Animal Science, 79(8), 2108.
Pascoal, A., Prado, M., Calo, P., Cepeda, A., & Barros-Velázquez, J. (2005). Detection of bovine DNA in raw and heat-processed foodstuffs, commercial foods and specific risk materials by a novel specific polymerase chain reaction method. European Food Research and Technology, 220(3–4), 444–450.
Martín, I., García, T., Fajardo, V., Lópezcalleja, I., Rojas, M., Pavón, M. A., et al. (2007). Technical note: detection of chicken, turkey, duck, and goose tissues in feedstuffs using species-specific polymerase chain reaction. Journal of Animal Science, 85(2), 452–458.
Mane, B. G., Mendiratta, S. K., & Tiwari, A. K. (2009). Polymerase chain reaction assay for identification of chicken in meat and meat products. Food Chemistry, 116(3), 806–810.
Robin, E. D., & Wong, R. (1988). Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. Journal of Cellular Physiology, 136(3), 507–513.
Dai, Z., Qiao, J., Yang, S., Hu, S., Zuo, J., Zhu, W., et al. (2015). Species authentication of common meat based on pcr analysis of the mitochondrial coi gene. Applied Biochemistry and Biotechnology, 176(6), 1770–1780.
Nixon, G. J., Wilkes, T. M., & Burns, M. J. (2015). Development of a real-time PCR approach for the relative quantitation of horse DNA. Analytical Methods, 7(20), 8590–8596.
Henegariu, O., Heerema, N. A., Dlouhy, S. R., Vance, G. H., & Vogt, P. H. (1997). Multiplex PCR: critical parameters and step-by-step protocol. BioTechniques, 23(3), 504–511.
Safdar, M., Junejo, Y., Arman, K., & Abasıyanık, M. F. (2014). Rapid bovine and caprine species identification in ruminant feeds by duplex real-time PCR melting curve analysis using EvaGreen fluorescence dye. Molecular Biotechnology, 56(8), 770–776.
Bottero, M. T., & Dalmasso, A. (2011). Animal species identification in food products: evolution of biomolecular methods. The Veterinary Journal, 190(1), 34–38.
Safdar, M., & Abasıyanık, M. F. (2013). Simultaneous identification of pork and poultry origins in pet foods by a quick multiplex real-time pcr assay using evagreen florescence dye. Applied Biochemistry and Biotechnology, 171(7), 1855–1864.
Dalmasso, A., Fontanella, E., Piatti, P., Civera, T., Rosati, S., & Bottero, M. T. (2004). A multiplex PCR assay for the identification of animal species in feedstuffs. Molecular and Cellular Probes, 18(2), 81–87.
Ghovvati, S., Nassiri, M. R., Mirhoseini, S. Z., Moussavi, A. H., & Javadmanesh, A. (2009). Fraud identification in industrial meat products by multiplex PCR assay. Food Control, 20(8), 696–699.
Safdar, M., & Junejo, Y. (2015). A multiplex-conventional PCR assay for bovine, ovine, caprine and fish species identification in feedstuffs: Highly sensitive and specific. Food Control, 50, 190–194.
Ali, M. E., Razzak, M. A., & Hamid, S. B. A. (2014). Multiplex PCR in species authentication: probability and prospects—a review. Food Analytical Methods, 7(10), 1933–1949.
Farrokhi, R., & Jafari Joozani, R. (2011). Identification of pork genome in commercial meat extracts for Halal authentication by SYBR green I real-time PCR. International Journal of Food Science & Technology, 46(5), 951–955.
Ali, M. E., Hashim, U., Mustafa, S., Man, Y. C., Dhahi, T. S., Kashif, M., et al. (2012). Analysis of pork adulteration in commercial meatballs targeting porcine-specific mitochondrial cytochrome b gene by TaqMan probe real-time polymerase chain reaction. Meat Science, 91(4), 454–459.
Cammà, C., Di Domenico, M., & Monaco, F. (2012). Development and validation of fast Real-Time PCR assays for species identification in raw and cooked meat mixtures. Food Control, 23(2), 400–404.
Iwobi, A., Sebah, D., Kraemer, I., Losher, C., Fischer, G., Busch, U., & Huber, I. (2015). A multiplex real-time PCR method for the quantification of beef and pork fractions in minced meat. Food Chemistry, 169, 305–313.
Sambrook J, Fritsch EF, & Maniatis T. (1989). Molecular cloning: a laboratory manual, 2nd ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
European Network of Genetically Modified Organism Laboratories (ENGL) Document. (2008). Definition of minimum performance requirements for analytical methods of GMO testing. http://gmo-crl.jrc.ec.europa.eu/doc/Min_Perf_Requirements_Analytical_methods.pdf.
Floren, C., Wiedemann, I., Brenig, B., Schütz, E., & Beck, J. (2015). Species identification and quantification in meat and meat products using droplet digital PCR (ddPCR). Food Chemistry, 173, 1054–1058.
Soares, S., Amaral, J. S., Oliveira, M. B. P., & Mafra, I. (2013). A SYBR Green real-time PCR assay to detect and quantify pork meat in processed poultry meat products. Meat Science, 94(1), 115–120.
Huang, Q., Xu, T., Wang, G. Y., Huang, J. F., Xia, H., Yin, R., et al. (2012). Species-specific identification of ruminant components contaminating industrial crude porcine heparin using real-time fluorescent qualitative and quantitative PCR. Analytical and Bioanalytical Chemistry, 402(4), 1625–1634.
Acknowledgements
This study was supported by Major International Cooperation NSFC (31210103917).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Rights and permissions
About this article
Cite this article
Wu, Q., Xiang, S., Wang, W. et al. Species Identification of Fox-, Mink-, Dog-, and Rabbit-Derived Ingredients by Multiplex PCR and Real-Time PCR Assay. Appl Biochem Biotechnol 185, 1–12 (2018). https://doi.org/10.1007/s12010-017-2621-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-017-2621-2