Abstract
Cold-active enzymes are extremozymes produced by psychrophiles, and have attracted much attention as biocatalysts due to their capacity to resist extreme reaction conditions in industrial processes. This study aimed to isolate bacterial strains from Arctic fjord sediments and screen them for lipase enzyme production. Out of 73 isolates, 8 were identified as good lipase producers. Bacillus cereus I13 exhibited the highest activity of 11.42 U/mL on the 4th day of incubation at 20 °C, along with the large zone of clearance of 27 mm. Optimum conditions for lipase production were identified as pH 7.0, a temperature of 20 °C, and 96 h of incubation, with glucose as a carbon source, yeast as a nitrogen source, and olive oil as a substrate. Bacillus cereus I13 has the potential to be used for the industrial production of cold-active lipase, and this study provides evidence that potent isolates can effectively increase lipase production.
Graphical Abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The microorganisms that inhabit polar regions possess diverse and unique physiological and biochemical properties, making them important. Psychrophilic microorganisms are categorized into psychrophiles and psychrotrophs based on their growth temperature. Psychrophiles can grow at temperatures ranging from -20 °C to 10 °C, but not above 10 °C. Psychrotrophs, on the other hand, can thrive below 0 °C, with an optimum growth temperature between 10 °C and 25 °C (Mukhtar et al., 2022). The cold-adapted enzymes and biomolecules produced by polar microorganisms offer numerous opportunities in biotechnology, including enzyme technology, antifreeze proteins, bioremediation, pharmaceuticals, and other health-related applications (Rampelotto 2014).
Cold-active enzymes are valuable biocatalysts that exhibit high catalytic efficiency at low and moderate temperatures, unlike mesophilic enzymes. Research on cold-adapted enzymes has revealed several unique characteristics, including Increased surface hydrophobicity, fewer electrostatic interactions, fewer disulfide bridges, reduced core hydrophobicity, higher glycine residue, increased formation of loop structures, fewer proline residues (Siddiqui 2015). Enzymes such as amylase, protease, xylanase, β-galactosidase, lipase, pectinase, alkaline phosphatase, DNA ligase, nuclease, and laccase have been reported from cold regions (Verma et al., 2021). However, their short half-life and low thermal stability limit their industrial use.To enhance the thermal and solvent stability of cold-active enzymes, different immobilization methods have been suggested by researchers (Pulicherla et al., 2011). In addition to immobilization, various molecular techniques such as protein engineering, recombinant DNA technology, and metagenomic approaches could also contribute to the development of unique and commercially-viable cold-active enzymes. Therefore, novel approaches could play a significant role in the field of biotechnology, rather than relying solely on traditional methods of cold-active enzyme production (Kuddus 2018).
Lipase (EC 3.1.1.3) is a hydrolase enzyme that catalyzes the hydrolysis of long-chain triglycerides. It is considered the most versatile enzyme used in biotechnological applications (Patel et al., 2021). Microorganisms are potential producers of extracellular lipases, which can be easily cultivated at an industrial scale for the mass production of more stable lipases (Adlercreutz 2017). These lipases can be genetically manipulated and are capable of large-scale cultivation. They have been used in the degradation of lipid waste and polyurethane, bioremediation, and bioaugmentation (Lee et al., 2015). Microbial lipases are widely used in the processing of fats and oils, detergents, food processing, synthesis of fine chemicals, pharmaceuticals, cosmetics, and the paper industry, among others (Melani et al., 2020).
Cold-active lipases are enzymes that can efficiently catalyze the hydrolysis of lipids at low temperatures. These enzymes are produced by bacteria that inhabit cold regions, where temperatures remain consistently cold at around 0 ± 2° C. Various bacteria that produce cold-active lipases have been identified, including: Moraxella sp. TA144 (Feller et al., 1991), Psychrobacter immobilis B10 (Arpigny et al., 1997), Psychrobacter sp. Ant300 (Kulakovaa et al., 2004), Psychrobacter sp. 7195 (Zhang et al., 2007), Pseudoalteromonas haloplanktis TAC125 (De Pascale et al., 2008) and Halomonas sp. BRI 8 (Jadhav et al., 2013). Besides the polar regions, cold-active lipase-producing microorganisms have also been identified in other cold regions, such as glaciers and high mountain tops. These include Acinetobacter sp.6 from Siberian tundra soil (Novototskaya-Vlasova et al., 2013), Pseudomonas sp. B11-1 from Alaskan soil (Choo et al., 1998), Pseudomonas sp. from Signy islands of Antarctica (Salwoom et al., 2019), Bacillus pumilus from Antarctica (Arifin et al., 2013), Halomonas sp. from Antarctic seawater (Jadhav et al., 2013) Stenotrophomonas maltophilia from Arctic sediment (Neethu et al., 2015), Pseudomonas mandelii from Batura Glaciers of Pakistan (Shaheen et al., 2020) and Actinobacteria and Proteobacteria from West Spitsbergen (Rasol et al., 2014). The ability of these microorganisms to produce cold-active lipases is thought to be an adaptation to the cold temperatures of their environment. Cold-active lipases have potential applications in a variety of industries, including the food, detergent, and pharmaceutical industries.
Bacillus species are known for producing lipolytic enzymes with significant biotechnological value. Some of the commonly known bacterial lipase producers include B. alcalophilus, B. licheniformis, B. pumilus, and B. subtilis (Lindsay et al., 2000; Awad et al., 2015). The catalytic properties and potential applications of Bacillus lipases have been extensively reviewed by Guncheva and Zhiryakova (2011). Besides, cold-active enzymes such as lipase, L-asparaginase, β-galactosidase, protease, amylase, xylanase, and chitinase have been reported from Arctic sediment previously (Saleena et al., 2022). This study aims to isolate, screen, and identify the best lipase-producing bacteria from Arctic sediment. Additionally, the nutritional and physicochemical factors influencing lipase production by the identified strain were also investigated.
Materials and methods
Primary screening of bacterial isolates for lipase production
Sediment samples were collected from Kongsfjorden and Krossfjorden, located on the west side of Spitsbergen in the Svalbard archipelago (79°N 12 E) in the Arctic Ocean during an Arctic expedition in October 2016 on board the research vessel RV Teisten. Sixteen locations of Arctic fjords were sampled using a Van Veen grab. Sediment samples were collected from the surface (0–2 cm) and subsurface (3–9 cm) stations using a sterile glass corer with a one-inch diameter. The samples were collected aseptically and transported as a cold shipment to the microbiology laboratory at Cochin University of Science and Technology in India for further studies.220 bacterial strains were isolated from surface and subsurface sediment samples from Kongsfjorden and Krossfjorden in the Arctic. The isolates were enriched in Zobell Marine Broth at 20 °C and screened for their ability to produce lipase using various media including Tributyrin agar plates, Rhodamine B olive oil agar plates, Phenol red olive oil agar plates, and Tween 80 agar plates. All of the media were obtained from HiMedia.
The initial screening to identify cold-active lipase producers among the isolates was performed using tributyrin agar plates containing 1% tributyrin (Sigma, USA). The tributyrin agar medium was prepared by adding peptone (5 g/L), yeast extract (3 g/L), NaCl (20 g/L), agar (15 g/L), and tributyrin (10 mL/L) to distilled water with a pH of 7.5. After sterilization, the autoclaved medium was poured onto a sterile Petriplate and allowed to solidify. A loopful of each pure culture was streaked onto the tributyrin agar plates and incubated at 20 °C for 48 h. The bacterial strains showing the largest zones of hydrolysis were selected for further study (Katiyar et al., 2017).
The Rhodamine B olive oil agar medium (pH 7.0) was prepared by adding Peptone (8 g/L), Yeast extract (4 g/L), NaCl (3 g/L) and Agar (20 g/L) to distilled water. After sterilization, filter-sterilized olive oil (31.25 mL/L) and Rhodamine B dye solution (10 mL/L) with a concentration of 1.0 mg/mL were added with vigorous shaking. The mixture was then poured into sterile Petri dishes and allowed to solidify. The bacterial culture was streaked onto the ROA plates and incubated at 20 °C for 48 h. The hydrolysis of the substrate was indicated by the formation of orange fluorescent halos around bacterial colonies visible upon UV irradiation (Bharathi et al., 2019).
The Phenol red olive oil agar medium (pH 7.4) was prepared using peptone (5 g/L), yeast extract (3 g/L), CaCl2(1 g/L) and agar (20 g/L). After sterilization, filter-sterilized phenol red dye (1 mg/mL) and olive oil substrate (10 mL/L) were added. The bacterial culture was streaked onto POA plates and incubated at 20 °C for 48 h. A change in color from orange to pink indicated the release of fatty acids due to lipolysis (Lee et al., 2015).
The Tween 80 agar medium (pH 7.0) was prepared by dissolving Peptone (10 g/L), NaCl (5 g/L), CaCl2.2H2O (0.1 g/L) and Agar (20 g/L) in distilled water. After sterilization, Tween 80 (10 mL/L) was sterilized separately and added to the medium. The mixture was then poured into sterile Petri dishes and allowed to solidify. The bacterial culture was streaked onto Tween 80 agar plates and incubated at 20 °C for 48 h. The presence of lipase production was indicated by the formation of a white precipitate around the colonies (Lee et al., 2015).
Secondary screening of lipase producing bacteria
The secondary screening was conducted using a liquid broth medium (pH 7.0) containing Peptone (5 g/L), Yeast extract (5 g/L), NaCl (0.5%) and Olive oil (10 mL/L). After sterilization, a 1.0 mL overnight culture of the lipase-producing bacterial strains was inoculated into 100 mL of the production medium and incubated at 20 °C with shaking at 150 rpm for 5 days. After incubation samples were taken every 24 h and the culture was centrifuged at 10,000 rpm for 10 min at 4 °C. The supernatant containing crude lipase was used for quantitative analysis.
Titrimetric assay of lipids
Lipase activity was determined through titration using olive oil as the substrate, and the amount of free fatty acids released during the catalytic activity of lipases on triacylglycerols was measured (Stoytcheva et al., 2012). A buffer solution was prepared with 50 mM Na2HPO4/NaH2PO4 at pH 7.0. The reaction cocktail was prepared by adding 5% olive oil emulsified in 5% gum acacia in 100 mM sodium phosphate buffer at pH 7.0. Next, 100 µl of the crude supernatant from each bacterial isolate and control was added to the reaction cocktail (10 mL) and incubated for 15 min at 20 °C at 100 rpm in a shaking incubator. The reaction was terminated, and fatty acids were extracted by adding 1 mL of acetone: ethanol solution (1:1) and mixing thoroughly. Then, 2–3 drops of phenolphthalein indicator were added to each of the reaction mixtures. The contents of each reaction mixture were titrated against 0.05 M NaOH solution to an endpoint of pink color at pH 10.
The equation to calculate lipase activity is:
Where,
- V1:
-
volume of NaOH solution used in blank titration
- V2:
-
volume of NaOH solution used in the titration of a sample
- N:
-
normality of NaOH solution
- F:
-
factor (equivalent weight of fatty acid/2)
- Vt:
-
volume of enzyme used
- t:
-
incubation time in minutes
The result is expressed as micromoles of free fatty acids liberated per mL of crude enzyme per minute (µmol/mL/min).
Agar well diffusion assay of lipase crude enzyme
Tributyrin agar plates were prepared by adding 1% (v/v) tributyrin to the medium. The tributyrin agar medium (pH 7.5) was prepared using Peptone (5 g/L), Yeast extract (3 g/L), NaCl (20 g/L) and Agar (15 g/L) in distilled water. Tributyrin was emulsified using a homogenizer. Sterile agar plates were aseptically punched with a sterile cork borer to obtain a 4 mm diameter well in 2 halves of the plate. The wells were loaded with 50 µl of the supernatant containing crude lipase enzymes separately. Plates were then incubated at 20 °C for 48 h. A clear zone of hydrolysis developed due to hydrolysis and the zone was measured. All experiments were done in triplicates. The collected data were used for further study (Bhavani et al., 2012).
Spectrophotometric assay of lipase
A spectrophotometric lipase assay was performed using p-NPP (para-nitrophenyl palmitate) as a substrate. The p-NPP was freshly prepared in isopropanol at a concentration of 0.3% (w/v) (solution A) and emulsified for 3 min using a continuous pulse. Solution B was prepared using 0.1% (w/v) gum acacia and 0.4% (v/v) Triton X 100 in distilled water. The substrate stock was prepared by combining 0.5 mL of solution A with 9.5 mL of solution B. For the assay, a reaction mixture (1.0 mL) was prepared by adding 900 µl of substrate emulsion, 50 µl of Tris buffer (pH 7.0), and 50 µl of crude lipase enzyme. The reaction mixture was incubated at 40 °C for 15 min, and the reaction was stopped by adding 2.0 mL of Na2CO3. The liberated p-nitrophenol was quantified spectrophotometrically at 410 nm. Lipase activity was expressed in International units (IU), defined as the amount of enzyme required to liberate 1 µmol of p-nitrophenol per minute under standard conditions (Willerding et al., 2011).
Identification of the potent isolate
Isolation of genomic DNA, 16S rRNA amplification and phylogenetic analysis
The bacterial strains exhibiting potential lipase activity were identified using molecular techniques. DNA was extracted using the Origin DNA extraction kit, and PCR amplification (Applied Biosystems) was conducted using 16S rRNA primers: 8F (5’- AGAGTTTGATCCTGGCTCAG -3’) and 1510R (5’-GGTTACCTTGTTACGACTT-3’). The PCR conditions consisted of an initial denaturation at 95 °C for 2 min, followed by 30 cycles of denaturation at 95 °C for 2 min, annealing at 58 °C for 1 min, and extension at 72 °C for 2 min. A final extension was performed at 72 °C for 10 min. The PCR products were sequenced at Agrigenome (Kochi, India) and the sequences were submitted to BLAST (http://www.ncbi.nim.noh.gov/BLAST) to check their homology with already available 16S rRNA gene sequences in the GenBank. Multiple alignments of the sequences were also carried out using ClustalW software, and the phylogenetic tree was constructed by neighbor-joining method using Mega 6 software.
Optimization of media parameters for cold-active lipase activity
The effect of various physicochemical and nutritional factors on extracellular lipase production was studied. Parameters including pH, temperature, incubation period, agitation, carbon source, nitrogen source, and different substrates were considered for this study. The bacterial inoculum was obtained from a 48-h culture with an OD value of OD600 = 1.0 and incubated in an orbital shaker. The effect of each parameter was analyzed. All experiments were done in triplicates.
Effect of different pH on lipase activity
To determine the optimum pH for enzyme production, the pH of the tributyrin broth was varied from 4 to 10 while keeping other parameters constant. The culture broth was harvested at 24 h intervals by centrifugation at 10,000 g for 30 min at 4 °C, and the resulting supernatant was used as a crude enzyme solution for the assay of lipase activity. Spectrophotometric measurements were taken at 410 nm to determine the lipase activity.
Effect of different temperatures on lipase activity
For the selection of optimum temperature for the production of lipases, the temperature varying from 5 °C to 35 °C was selected by keeping the remaining parameters the same. The culture broth was harvested at 24 h intervals by centrifugation at 10,000 g for 30 min at 4 °C. The supernatant collected was used as a crude enzyme solution and was assayed for enzyme activity. Lipase activity was measured spectrophotometrically at 410 nm.
Effect of different agitation speeds on lipase activity
To determine the optimal agitation speed for peak enzyme activity, Isolate I13 was cultured in an orbital shaking incubator at 20 °C with varying agitation speeds ranging from static conditions to 220 rpm. The culture was grown in a broth containing yeast extract, NaCl, peptone, sucrose, and 1% (w/v) olive oil, with an agitation speed of 120 rpm at 20 °C in an orbital shaker. The culture broth was harvested at 24 h intervals by centrifugation at 10,000 g for 30 min at 4 °C, and the supernatant was collected as a crude enzyme solution and assayed for enzyme activity. Lipase activity was measured spectrophotometrically at 410 nm.
Effect of different Incubation periods on lipase activity
In this study, the incubation period for lipase production was optimized by conducting fermentation for different time intervals, namely 0 h, 24 h, 48 h, 72 h, 96 h, and 120 h. Isolate I13 was cultured in a broth containing yeast extract, NaCl, peptone, sucrose, and 1% (w/v) olive oil at 20 °C in an orbital shaker at an agitation speed of 120 rpm. The culture broth was harvested at 24 h intervals by centrifugation at 10,000 g for 30 min at 4 °C. The collected supernatant was used as a crude enzyme solution and was assayed for enzyme activity. Lipase activity was measured spectrophotometrically at 410 nm.
Effect of different carbon sources on lipase activity
The effect of various carbon sources on lipase production was studied by replacing sucrose in the growth medium with lactose, glucose, maltose, or fructose at a final concentration of 1% (w/v) while keeping the other parameters constant. The culture broth was harvested by centrifugation at 10,000 g for 30 min at 4 °C, and the resulting supernatant was used as a crude enzyme solution for the measurement of enzyme activity. Lipase activity was determined spectrophotometrically at 410 nm.
Effect of different nitrogen sources on lipase activity
To investigate the impact of nitrogen sources on lipase production, the effects of different nitrogen sources including beef extract, tryptone, peptone, yeast extract, sodium nitrate, and potassium nitrate were examined. These nitrogen sources were added to the broth at a final concentration of 1% (w/v), and individually tested by replacing the peptone present in the growth medium. The culture broth was harvested by centrifugation at 10,000 g for 30 min at 4 °C. The supernatant was collected and used as a crude enzyme solution for enzyme activity assays, with lipase activity measured spectrophotometrically at 410 nm.
Effect of different oils as a Substrate for lipase production
Different oils such as gingelly oil, sunflower oil, coconut oil, waste cooking oil, and diesel were used to replace olive oil in the growth media at a final concentration of 1% (w/v) to investigate their effect on lipase production. All other parameters were kept constant. The culture broth was harvested by centrifugation at 10,000 g for 30 min at 4 °C, and the supernatant was collected as a crude enzyme solution and assayed for enzyme activity. Lipase activity was measured spectrophotometrically at 410 nm.
Results
Isolation and screening of lipase-producing bacterial strain
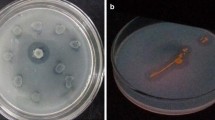
Bacterial isolates from Arctic fjord sediments were chosen for the study. 220 isolates in total were tested for lipase synthesis, and 151 (69%) of them demonstrated a distinct hydrolysis zone in tributyrin, with 73 (33%) demonstrating very strong competence (Fig. 1a). On the basis of their diameter in the hydrolysis zone, these isolates were further selected. Lipolytic activity was observed on bacterial isolates streaked on Rhodamine B olive oil agar plates (ROA), Phenol red olive oil agar plates, and Tween 80-based agar plates. The formation of yellow color in phenol red olive oil agar plates confirmed the production of lipase, due to the liberation of fatty acid and subsequent pH change (Fig. 1b). The formation of orange fluorescent halos was observed on ROA plates under UV light, resulting from complex formation between cationic rhodamine-B and uranyl fatty acid ion (Fig. 2) (Lanka and Latha 2015). The Rhodamine B test was found to be more convenient for distinguishing pure lipase producers (Alhamdani and Alkabbi 2016).
Following qualitative screenings, 24 isolates were identified as pure lipase producers. The culture supernatants of these isolates were prepared, and their extracellular lipase activities were measured using the p-NPP assay method. Eight bacteria with lipase activity above 1.0 U/mL were selected for further quantitative screening, which involved the titrimetric assay of lipase and agar well diffusion method. Among the selected isolates, I13 exhibited the maximum activity of 11.42 U/mL after 96 h of incubation (Table 1) and also showed the highest zone of hydrolysis of 27 mm in the agar well diffusion assay (Table 2). Lipase activity increased with time, reaching maximum activity after 96 h of incubation before decreasing. The potent isolate was molecularly characterized through 16S rRNA phylogenetic analysis. Phylogenetic analysis of the bacterial isolate I13 sequences against the GenBank database at NCBI revealed that their closest relative is Bacillus cereus, with 100% sequence similarity. The Bacillus cereus I13 sequences have been assigned the accession number OR544946 (Fig. 3). This marks the first report of Bacillus cereus strains producing lipases isolated from the sediments of Arctic fjord.
Determination of optimum fermentation parameters for the potent isolate
Effect of pH
Based on the studies, it is suggested that various biotic and abiotic factors can significantly impact enzyme hydrolysis. Lipases, in particular, have been found to function optimally across a wide range of pH and temperature conditions, with bacterial lipases exhibiting the highest effectiveness under alkaline conditions. In the present study, maximum lipase production was observed at pH 7.0 (Fig. 4), which can be attributed to the cold-adapted characteristics of isolate I13.
Effect of temperature
Temperature is a critical parameter that can affect enzyme production, and its optimal range varies across organisms. The physical properties of the cell membrane can be altered by temperature, which in turn influences the secretion of extracellular enzymes. To determine the optimal temperature for lipase production by the potent isolate I13, it was incubated at various temperatures ranging from 5 °C to 40 °C. The highest lipase activity was observed at 20 °C (Fig. 5).
Effect of the incubation period
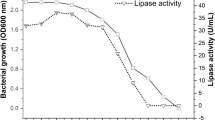
Determining the optimum incubation period for maximum enzyme activity is a crucial parameter, as different organisms exhibit significant variation in enzyme activity at different incubation periods. In this study, the optimum incubation period for the highest lipase activity was observed after 96 h of incubation. However, the activity of the lipase enzyme decreased with further incubation (Fig. 6). This decline in lipase activity is attributed to the denaturation or decomposition of the lipase enzyme resulting from its interactions with other compounds in the fermented medium.
Effect of agitation speed
Lipase production is higher under shaking conditions compared to static conditions in all bacterial strains. Medium agitation enhances bacterial growth and lipase production by improving the rate of oxygen transfer into the production medium and facilitating the dispersal of oil micelles into the microbial cell. This study's results showed a similar trend to previous studies, where the highest lipase activity was observed under agitation at 150 rpm for 96 h at 20 0C (Fig. 7).
Effect of carbon source
The addition of different carbon sources to the production media led to an increase in lipase production. Various carbon sources, such as lactose, sucrose, glucose, maltose, and fructose, were added to study their effect on lipase production. Among these, glucose was found to be the best carbon source for achieving the highest lipase activity, followed by fructose (Fig. 8).
Effect of nitrogen source
The effect of nitrogen sources was studied by adding various nitrogen sources, such as beef extract, tryptone, peptone, yeast extract, sodium nitrate, and potassium nitrate, to the production medium. The maximum lipase production was noted in the medium with yeast extract (Fig. 9).
Effect of substrate
The type and concentration of lipid substrates can influence the synthesis of lipase enzymes. To study the effect of various substrates on the lipase production medium, different oil substrates such as gingelly oil (1), olive oil (2), sunflower oil (3), coconut oil (4), waste cooking oil(5), and diesel (6) were added to the medium. The results showed that olive oil and waste cooking oil were the best substrates for lipase production, while sunflower oil caused a significant decline in the production of lipase enzymes. Therefore, olive oil is considered the best and most cost-effective substrate for lipase production (Fig. 10).
Discussion
Cold-adapted lipases have gained significant interest due to their unique properties such as higher flexibility, which enables them to interact with substrates more efficiently, and lower activation energy compared to meso/thermophilic microorganisms (Al-Ghanayem and Joseph, 2020). Enzymes from organisms living in extreme conditions have also attracted attention in recent years (Fact et al., 2020). Lipases, which are mostly of microbial origin, are widely used for various industrial and clinical research applications and are currently dominating the market. The global market for lipase enzymes is expected to reach $590.5 Million by 2023, with a compound annual growth rate (CAGR) of 6.5% ( Remonatto et al., 2022).
In addition to biodiesel and detergent production, microbial lipases find use in various other industries such as the food industry for the production of cheese, butter, and other dairy products, in the textile industry for the degreasing of fabrics, and the pharmaceutical industry for the synthesis of drugs and drug intermediates (Javed et al., 2018). The ability of microbial lipases to function under a wide range of temperature and pH conditions makes them suitable for various industrial applications. Furthermore, the increasing demand for sustainable and eco-friendly products has led to a rise in the use of microbial lipases in green chemistry processes such as bioremediation, biocatalysis, and organic synthesis. Overall, microbial lipases are versatile enzymes with a wide range of applications in different industries and have huge potential for future research and development. Studies have reported that lipases from Thermophilic Anoxybacillus sp., Acinetobacter venetianus, and Burkholderia ubonensis can be used in the production of biodiesel, and the lipase transesterification process reduces downstream processing costs (Zhu et al., 2020). In detergent production, lipases from Pseudomonas ADT3, Bacillus sonorensis, Bacillus licheniformis, Bacillus flexus, and Bacillus pumilus have been used in numerous studies. More recently, cold-adapted lipases have been discovered in Arctic and Antarctic microorganisms. The psychrotrophic Arctic bacterium Arthrobacter gangotriensis, obtained from soil samples, exhibits lipolytic activity with a wide pH range and good stability (Ramle and Rahim, 2016). Previous studies on lipases have also reported lipolytic activity in the bacterial strain KS 46 Stenotrophomonas maltophilia, isolated from Arctic sediments (Neethu et al., 2015). The Bacillus pumilus ArcL 5, with lipolytic activity, was isolated from the Chukchi Sea in the Arctic Ocean (Wi et al., 2014), and Bacillus pumilus producing lipases were reported from the Antarctic region (Arifin et al., 2013).
Lipases from Bacillus sp. are considered to be lidless and small (actually the smallest lipases known). Numerous studies have reported lipases from Bacillus species, and Bacillus cereus is considered to be a potential one. Al-Zazee et al. (2022) reported extracellular lipase production by Bacillus cereus MS6 from the effluent of a local Sewerage Yemen Company of ghee & soap industry City of Taiz, Yemen. Demirkan et al. (2021) isolated many Bacillus sp. strains from agricultural soils of Turkey, and the potent extracellular lipase producer was Bacillus cereus ATA179. The potential of the enzyme in the detergent industry was also studied. Akhter et al. (2022) isolated an alkaliphilic lipase from a novel Bacillus cereus NC7401 strain isolated from diesel fuel polluted soil adhered to the roots of Tagetes minuta grown in Muzaffarabad city of Azad Kashmir, Pakistan. Hassan et al. (2018) conducted a study among marine bacterial isolates obtained from the Mediterranean Sea and screened them for their lipase production. The most promising lipase producer that exhibited the highest lipolytic hydrolysis was identified as Bacillus cereus. In this study, they conducted economic treatment of oily wastewater. Wang et al. (2019) reported that Bacillus cereus XN12 displayed higher dietary oil degradation efficiency than Bacillus subtilis and Enterobacter cloacae. Ghaima et al. (2014) also reported lipase from Bacillus cereus of diesel fuel polluted soil samples of Iraq. These studies demonstrate the potential of Bacillus cereus for lipase production. The enzyme can be used in a variety of applications, including the detergent industry, the treatment of oily wastewater, and the degradation of dietary oil waste. Lipases from Bacillus cereus are considered to be versatile enzymes for industry, especially in the field of biotechnology. The scientific and industrial communities have been working hard over the past few decades to find innovative cold-active enzymes with potential use in a variety of biotechnological processes. The thermostability and activity of cold-active enzymes have proven significant for the implementation in protein engineering technologies. In our study, we found that lipase from Bacillus cereus is a cold-active enzyme that is more stable than other meso- or thermophilic counterparts. Therefore, it is considered to be a potential enzyme in the relevant field.
To optimize lipase production, organisms with the highest lipase activity were identified and isolated, and different growth parameters were varied using a one-variable-at-a-time approach. Starting with the pH of the production medium, the highest lipase activity was recorded at pH 7.0, after which a noticeable decline occurred. The cold-adapted characteristics of isolate I13 explain its maximum lipase production at pH 7. Abbas et al., 2017 also reported maximum lipase production at pH 7.0 by Bacillus subtilis. Many cold-adapted lipase-producing bacterial strains, such as Pseudomonas antarctica sp., P. meridiana sp., Acinetobacter sp., Aeromonas sp., etc., show lipase activity in neutral to alkaline pH (Salwoom et al., 2019). The isolates' lipase activity was drastically reduced at acidic pH and was stable at alkaline pH. In contrast to our study, some Bacillus sp. shows maximum lipase production in alkaline pH. Ghori et al., 2011 reported lipase activity of Bacillus sp. at pH 9.0, and Saraswat et al., 2018 reported lipase-producing Bacillus subtilis at pH 8.0, which strongly indicates the alkaline nature of the enzyme.
The lipase producer I13 was tested for its ability to produce lipase at different temperatures ranging from 5 °C to 40 °C to determine the optimum temperature for lipase production. The highest lipase activity was observed at 20 °C, indicating that the organism is psychrotrophic. This finding is consistent with previous reports of cold-active lipase production at 10 °C by Bacillus cereus isolated from sediments of the Mediterranean Sea (Hassan et al., 2018) and by Bacillus sp. isolated from the sewage of olive oil at 20 °C (Yasemin et al., 2017). Joseph et al., 2011 also reported cold-active lipase production at 10 °C by the bacterial species Micrococcus roseus, which was isolated from the Gangotri glaciers of the Himalayas. While some lipases from Bacillus sp. have been reported to show maximum production between 35 °C to 55 °C (Balaji et al., 2020), our findings indicate that the lipase from I13 is a cold-active enzyme.
In summary, lipases from different microorganisms have been used for various applications such as biodiesel and detergent production. Cold-adapted lipases have been found in Arctic and Antarctic microorganisms, which exhibit lipolytic activity at low temperatures. Optimization of lipase production parameters such as pH, temperature, and incubation time has been extensively studied, with varying results depending on the microorganism used. Bacillus sp. is a common lipase producer, and maximum lipase production is usually observed between 24–72 h of incubation. In our studies, the highest lipase activity was observed after 96 h of incubation, and activity of the lipase enzyme got decreased on further incubation. Nadaf and Thimmappa, 2020 reported lipase production by Bacillus subtillis at 48 h of incubation, whereas Prasad and Sethi, 2013 reported lipase production by Bacillus sp. at 60 h of incubation. In the study of Mazhar et al., 2017, the best lipase production by Bacillus subtillis was observed at 72 h of incubation. But in the report of Bendary et al., 2015, maximum lipase production by Bacillus sp. was observed on the 5th day of incubation and further incubation shows a reduction in enzyme activity.
Agitation is indeed an important factor for proper aeration and nutrient availability during bacterial culture, and it can have a significant effect on lipase production. The optimal agitation rate may vary depending on the bacterial strain and the culture conditions. In the case of strain I13, it was found that too high an agitation rate led to a sharp decline in bacterial growth and lipase production. This is consistent with the findings of other studies, such as Pham et al., 2021, which reported that maximum lipase production by Bacillus sp. was observed at 150 rpm, and Sirisha et al., 2010, which found that Bacillus pumilus produced the highest lipase activity at 160 rpm. However, there are also reports of bacterial strains that show maximum lipase production at lower agitation rates, such as Bacillus cereus, which was found to produce the best lipase activity at 150 rpm in the study mentioned.
Overall, carbon and nitrogen sources are important factors for the growth and production of lipase enzymes. Glucose and fructose have been reported as the best carbon sources for lipase production from Bacillus sp. Organic nitrogen sources such as yeast extract are considered better for lipase production than inorganic nitrogen sources. Olive oil is a commonly used inducer for lipase enzyme production, and sunflower oil has also been reported to enhance lipase production by Bacillus sp. It is important to note that the choice of carbon and nitrogen sources may vary depending on the microorganism used for lipase production. Das et al., 2014 and Bharathi et al., 2019 also suggest that yeast is the best nitrogen source for the production of lipase from Bacillus sp. In contrast to our findings, tryptone act as the best nitrogen source for maximum lipase production by Bacillus vallismortis (Mondal et al., 2019). As lipase is an inducible enzyme, the most important nutrient for lipase activity is its substrates, such as oils or other inducers. The use of olive oil in a medium enhances lipase enzyme production compared to other lipid substrates. Many studies also suggest olive oil as the best substrate for lipase enzyme production (Ilesanmi et al., 2020). In the report of Isiaka Adetunji and Olufolahan Olaniran 2018, sunflower oil enhances lipase production compared to other inducer oils by Bacillus aryabhattai.
Bacillus cereus is a Gram-positive, rod-shaped bacterium that is known to produce a variety of extracellular enzymes, including lipases. Lipase is an enzyme that catalyzes the hydrolysis of lipids, and it has a wide range of potential applications, including in the food, cosmetics, and pharmaceutical industries. Lipases from Bacillus sp. are considered to be lidless and small (actually the smallest lipases known). Numerous studies have reported lipases from Bacillus species, and Bacillus cereus is considered to be a potential one. Al-Zazee et al. (2022) reported extracellular lipase production by Bacillus cereus MS6 from the effluent of a local Sewerage Yemen Company of ghee & soap industry City of Taiz, Yemen. Demirkan et al. (2021) isolated many Bacillus sp. strains from agricultural soils of Turkey, and the potent extracellular lipase producer was Bacillus cereus ATA179. The potential of the enzyme in the detergent industry was also studied. Akhter et al. (2022) isolated an alkaliphilic lipase from a novel Bacillus cereus NC7401 strain isolated from diesel fuel polluted soil adhered to the roots of Tagetes minuta grown in Muzaffarabad city of Azad Kashmir, Pakistan. Hassan et al. (2018) conducted a study among marine bacterial isolates obtained from the Mediterranean Sea and screened them for their lipase production. The most promising lipase producer that exhibited the highest lipolytic hydrolysis was identified as Bacillus cereus. In this study, they conducted economic treatment of oily wastewater. Wang et al. (2019) reported that Bacillus cereus XN12 displayed higher dietary oil degradation efficiency than Bacillus subtilis and Enterobacter cloacae. Ghaima et al. (2014) also reported lipase from Bacillus cereus of diesel fuel polluted soil samples of Iraq. These studies demonstrate the potential of Bacillus cereus for lipase production.The scientific and industrial communities have been actively searching for innovative cold-active enzymes with potential use in a variety of biotechnological processes for decades. Lipases from Bacillus cereus are considered to be versatile enzymes for industry, especially in the field of biotechnology. The thermostability and activity of cold-active enzymes have proven to be significant for the implementation of protein engineering technologies. In this study, we isolated a new strain of Bacillus cereus that produces a cold-active lipase with high activity and specificity. We also optimized the growth conditions for lipase production, and we characterized the physical and biochemical properties of the lipase. Our study provides a new platform for the production of lipases with industrial applications. The lipase produced by our new strain of Bacillus cereus has the potential to be used in a variety of industries. For example, it could be used to produce food products with a longer shelf life, to develop new cosmetics, or to create new pharmaceutical products. Our study also provides insights into the genetic regulation of cold active lipase production in Bacillus cereus, which could be used to further improve the production of lipases in this bacterium. We found that lipase from Bacillus cereus is a cold-active enzyme that is more stable than other meso- or thermophilic counterparts. Therefore, it is considered to be a promising enzyme in the relevant field.
Conclusion
Overall, the study highlights the potential of Bacillus cereus I13 as a cold-active lipase producer, isolated from bacterial strains of the Arctic fjord. The study also emphasizes the importance of optimizing growth and physiological parameters to achieve high yields of the enzyme, which could have significant industrial and economic benefits. Further research in this area could focus on scaling up production and exploring different applications for this extracellular lipase. The main limitation of this study is that it only tested a few bacterial strains from the Arctic fjord. It is possible that other strains of bacteria may produce even higher yields of lipase. The study of culturable bacteria is even more limited than that of non-culturable bacteria from the Arctic fjord. However, among the culturable bacteria, the number of positive isolates for lipase enzyme is high. Therefore, this study cannot fully explain the lipase-producing capability of the Arctic microbial community.
The high yield of lipase obtained from the optimization of growth and physiological parameters of Bacillus cereus I13 indicate its potential for large-scale production. Further studies can be conducted to optimize the production process and purify the enzyme for industrial applications. The lipase produced by Bacillus cereus I13 have a number of potential applications, such as food processing, detergent manufacturing, bioremediation. It is important to determine how stable the lipase produced by Bacillus cereus I13 is under different conditions. This information will be important for determining the best way to store and transport the enzyme, as well as the range of industrial applications where it can be used. The molecular structure of the lipase produced by Bacillus cereus I13 can be characterized using a variety of techniques, such as X-ray crystallography and nuclear magnetic resonance spectroscopy. This information can be used to design new enzymes with improved properties, such as higher activity or stability.
Data Availability
The data supporting the findings of this study are available from the corresponding authors upon reasonable request.
Abbreviations
- BLAST:
-
Basic Local Alignment Search Tool
- CAGR:
-
Compound Annual Growth Rate
- DNA:
-
Deoxyribonucleic acid
- IU:
-
International Unit
- MTCC:
-
Microbial Type Culture Collection
- mM:
-
Millimolar
- PCR:
-
Polymerase Chain Reaction
- ROA:
-
Rhodamine Olive oil Agar
- POA:
-
Phenol red Olive oil Agar
- p-NPP:
-
Para-nitro phenyl palmitate
- rRNA:
-
Ribosomal Ribonucleic acid
- UV:
-
Ultra violet
References
Abbas N, Javed J, Abbas Z, Choudry S, Ali S (2017) Lipase Production from Bacillus subtilis using various Agricultural waste. Int J Adv Eng Manag Sci 3(5):239830. https://doi.org/10.24001/ijaems.3.5.1
Adlercreutz P (2017) Comparison of lipases and glycoside hydrolases as catalysts in synthesis reactions. Appl Microbiol Biotechnol 101(2):513–519. https://doi.org/10.1007/s00253-016-8055-x
Akhter K, Karim I, Aziz B, Bibi A, Khan J, Akhtar T (2022) Optimization and characterization of alkaliphilic lipase from a novel Bacillus cereus NC7401 strain isolated from diesel fuel polluted soil. PLoS ONE 17(8):e0273368. https://doi.org/10.1371/journal.pone.0273368
Al-Ghanayem AA, Joseph B (2020) Current prospective in using cold-active enzymes as eco-friendly detergent additive. Appl Microbiol Biotechnol 104(7):2871–2882. https://doi.org/10.1007/s00253-020-10429-x
Alhamdani MA, Alkabbi HJJ (2016) Isolation and identification of lipase producing bacteria from oil-contaminant soil. J Biol, Agric Healthcare 6(20):1–7
Al-Zazaee MMA, Abdu MM, Mahmoud DA, Abdu AAAM (2022) Extraction and Characterization of Lipase Enzymes from Bacillus cereus (MS6) and their Medical & Industrial Applications. Int J Innov Sci Res Technol Special Issue-(2nd ICTSA–2022)
Arifin AR, Kim SJ, Yim JH, Suwanto A, Kim HK (2013) Isolation and biochemical characterization of Bacillus pumilus lipases from the Antarctic. J Microbiol Biotechnol 23(5):661–667. https://doi.org/10.4014/jmb.1212.12040
Arpigny JL, Lamotte J, Gerday C (1997) Molecular adaptation to cold of an Antarctic bacterial lipase. J Mol Catal B Enzym 3(1–4):29–35. https://doi.org/10.1016/S1381-1177(96)00041-0
Awad GE, Mostafa H, Danial EN, Abdelwahed NA, Awad HM (2015) Enhanced production of thermostable lipase from Bacillus cereus ASSCRC-P1 in waste frying oil based medium using statistical experimental design. J Appl Pharmaceut Sci 5(9):007–015. https://doi.org/10.7324/JAPS.2015.50902
Balaji L, Chittoor JT, Jayaraman G (2020) Optimization of extracellular lipase production by halotolerant Bacillus sp. VITL8 using factorial design and applicability of enzyme in pretreatment of food industry effluents. Prep Biochem Biotechnol 50(7):708–716. https://doi.org/10.1080/10826068.2020.1734936
Bharathi D, Rajalakshmi G, Komathi S (2019) Optimization and production of lipase enzyme from bacterial strains isolated from petrol spilled soil. J King Saud Univ-Sci 31(4):898–901. https://doi.org/10.1016/j.jksus.2017.12.018
Bhavani M, Chowdary GV, David M, Archana G (2012) Screening, isolation and biochemical characterization of novel lipase producing bacteria from soil samples. Int J Biol Eng 2(2):18–22. https://doi.org/10.5923/j.ijbe.20120202.03
Choo DW, Kurihara T, Suzuki T, Soda K, Esaki N (1998) A cold-adapted lipase of an Alaskan psychrotroph Pseudomonas sp. strain B11–1: gene cloning and enzyme purification and characterization. Appl Environ Microbiol 64(2):486–491. https://doi.org/10.1128/AEM.64.2.486-491.1998
Das S, Singha R, Rai C, Roy A (2014) Isolation and characterization of bacteria with spoilage potential from some refrigerated foods of West Bengal, India. Int J Curr Microbiol App Sci 3(9):630–639
De Pascale D, Cusano AM, Parrilli E, di Prisco G, Marino G, Tutino ML (2008) The cold-active Lip1 lipase from the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125 is a member of a new bacterial lipolytic enzyme family. Extremophiles 12(3):311–323. https://doi.org/10.1007/s00792-008-0163-9
Demirkan E, Çetinkaya AA, Abdou M (2021) Lipase from new isolate Bacillus cereus ATA179: optimization of production conditions, partial purification, characterization and its potential in the detergent industry. Turk J Biol 45(3):287–300. https://doi.org/10.3906/biy-2101-22
El-Bendary MA, Moharam ME, Mahmoud DA (2015) Economic production of polyethylene modifying lipase enzyme under solid state fermentation using banana peels and sand. BioTechnol: Indian J 11(3):94–101
Fact C, Chandra P, Singh R, Arora PK (2020) Microbial lipases and their industrial applications: a comprehensive review. BioMed Central 1–42. https://doi.org/10.1186/s12934-020-01428-8
Feller G, Thiry M, Gerday C (1991) Nucleotide sequence of the lipase gene lip2 from the Antarctic psychrotroph Moraxella TA 144 and site-specific mutagenesis of the conserved serine and histidine residues. DNA Cell Biol 10(5):381–388. https://doi.org/10.1089/dna.1991.10.381
Ghaima KK, Mohamed AI, Mohamed MM (2014) Effect of some factors on lipase production by Bacillus cereus isolated from diesel fuel polluted soil. Int J Sci Res Publ 4(8):416–420
Ghori MI, Iqbal MJ, Hameed A (2011) Characterization of a novel lipase from Bacillus sp isolated from tannery wastes. Braz J Microbiol 42(1):22–9. https://doi.org/10.1590/S1517-83822011000100003
Guncheva M, Zhiryakova D (2011) Catalytic properties and potential applications of Bacillus lipases. J Mol Catal B Enzym 68:1–2. https://doi.org/10.1016/j.molcatb.2010.09.002
Hassan SW, Abd El Latif HH, Ali SM (2018) Production of cold-active lipase by free and immobilized marine Bacillus cereus HSS: application in wastewater treatment. Front Microbiol 9:2377. https://doi.org/10.3389/fmicb.2018.02377
Ilesanmi OI, Adekunle AE, Omolaiye JA, Olorode EM, Ogunkanmi AL (2020) Isolation, optimization and molecular characterization of lipase producing bacteria from contaminated soil. Scientific African 8:e00279. https://doi.org/10.1016/j.sciaf.2020.e00279
IsiakaAdetunji A, OlufolahanOlaniran A (2018) Optimization of culture conditions for enhanced lipase production by an indigenous Bacillus aryabhattai SE3-PB using response surface methodology. Biotechnol Biotechnol Equipment 32(6):1514–1526. https://doi.org/10.1080/13102818.2018.1514985
Jadhav VV, Pote SS, Yadav A, Shouche YS, Bhadekar RK (2013) Extracellular cold active lipase from the psychrotrophic Halomonas sp. BRI 8 isolated from the Antarctic sea water. Songklanakarin J Sci Technol 35(6)
Javed S, Azeem F, Hussain S, Rasul I, Hussnain M, Riaz M, Afzal M (2018) Bacterial lipases : A review on purification and characterization. Progress in Biophysics and Molecular Biology 132: 23–34. Elsevier Ltd. https://doi.org/10.1016/j.pbiomolbio.2017.07.014
Joseph B, Upadhyaya S, Ramteke P (2011) Production of cold-active bacterial lipases through semisolid state fermentation using oil cakes. Enzyme research 2011. https://doi.org/10.4061/2011/796407
Katiyar P, Pratibha VH, Baghel VS (2017) Isolation, partial purification and characterization of a cold active lipase from Pseudomonas sp. isolated from Satopanth Glacier of Western Himalaya, India. Int J Sci Res Manag (IJSRM) 5(7):6106–6112. https://doi.org/10.18535/ijsrm/v5i7.37
Kuddus M (2018) Cold-active enzymes in food biotechnology : An updated mini-review. J Appl Biol Biotechnol 6:58–63. https://doi.org/10.7324/JABB.2018.60310
Kulakovaa L, Galkin A, Nakayama T, Nishino T, Esaki N (2004) Cold-active esterase from Psychrobacter sp. Ant300: gene cloning, characterization, and the effects of Gly→ Pro substitution near the active site on its catalytic activity and stability. Biochimica et Biophysica Acta (BBA)-Proteins Proteomics 1696(1):59–65. https://doi.org/10.1016/j.bbapap.2003.09.008
Lanka S, Latha JNL (2015) A Short Review on Various Screening Methods to Isolate Potential Lipase Producers : Lipases-the Present and Future Enzymes of Biotech Industry. Int J Biol Chem 9:207–219. https://doi.org/10.17311/ijbc.2015.207.219. (Science Alert)
Lee LP, Karbul HM, Citartan M, Gopinath SC, Lakshmipriya T, Tang TH (2015) Lipase-secreting Bacillus species in an oil-contaminated habitat: promising strains to alleviate oil pollution. BioMed Res Int 2015. https://doi.org/10.1155/2015/820575
Lindsay D, Brözel VS, Mostert JF, Von Holy A (2000) Physiology of dairy associated Bacillus spp. over a wide pH range. Int J Food Microbiol 54(1–2):49–62. https://doi.org/10.1016/S0168-1605(99)00178-6
Mazhar H, Abbas N, Ali S, Sohail A, Hussain Z, Ali SS (2017) Optimized production of lipase from Bacillus subtilis PCSIRNL-39. Afr J Biotech 16(19):1106–1115. https://doi.org/10.5897/AJB2017.15924
Melani NB, Tambourgi EB, Silveira E (2020) Lipases: from production to applications. Sep Purif Rev 49(2):143–158. https://doi.org/10.1080/15422119.2018.1564328
Mondal M, Biswas JK, Tsang YF, Sarkar B, Sarkar D, Rai M, Hooda PS (2019) A wastewater bacterium Bacillus sp. KUJM2 acts as an agent for remediation of potentially toxic elements and promoter of plant (Lens culinaris) growth. Chemosphere 232:439–452. https://doi.org/10.1016/j.chemosphere.2019.05.156
Mukhtar S, Rashid N, Haque MFU, Malik KA (2022) Metagenomic approach for the isolation of novel extremophiles. In Microbial Extremozymes (pp. 55–66). Academic Press. https://doi.org/10.1016/B978-0-12-822945-3.00010-5
RD Nadaf, SC Thimmappa (2020) Optimization of the media components by one factor at a time methodology to enhance lipase production by Bacillus Substilis KUBT4. Int J Sci Technol Res 9(01)
Neethu CS, Rahiman KM, Rosmine E, Saramma AV, Hatha AM (2015) Utilization of agro-industrial wastes for the production of lipase from Stenotrophomonas maltophilia isolated from Arctic and optimization of physical parameters. Biocatal Agric Biotechnol 4(4):703–709. https://doi.org/10.1016/j.bcab.2015.09.002
Novototskaya-Vlasova KA, Petrovskaya LE, Rivkina EM, Dolgikh DA, Kirpichnikov MP (2013) Characterization of a cold-active lipase from Psychrobacter cryohalolentis K5T and its deletion mutants. Biochem Mosc 78(4):385–394. https://doi.org/10.1134/S000629791304007X
Patel GB, Shah KR, Shindhal T, Rakholiya P, Varjani S (2021) Process parameter studies by central composite design of response surface methodology for lipase activity of newly obtained actinomycete. Environ Technol Innov 23:101724. https://doi.org/10.1016/j.eti.2021.101724
Pham VHT, Kim J, Chang S, Chung W (2021) Investigation of Lipolytic-Secreting Bacteria from an Artificially Polluted Soil Using a Modified Culture Method and Optimization of Their Lipase Production. Microorganisms 9(12):2590. https://doi.org/10.3390/microorganisms9122590
Prasad MP, Sethi R (2013) Comparative Studies on the production of Lipase by Bacillus species under various growth parameters. Int J Curr Microbiol Appl Sci 2(11):179–185. https://doi.org/10.12691/jaem-6-1-2
Pulicherla KK, Ghosh M, Kumar PS, Rao KRSS (2011) Psychrozymes-the next generation industrial enzymes. J Marine Sci Res Dev 1(2). https://doi.org/10.4172/2155-9910.1000102
Ramle Z, Rahim RA (2016) Psychrophilic lipase from Arctic bacterium. Trop Life Sci Res 27(supp1):151. https://doi.org/10.21315/2Ftlsr2016.27.3.21
Rampelotto PH (2014) Polar microbiology: Recent advances and future perspectives. Biology 3(1):81–84. https://doi.org/10.3390/biology3010081
Rasol R, Rashidah AR, Nazuha RSN, Smykla J, Maznah WW, Alias SA (2014) Psychrotrophic lipase producers from Arctic soil and sediment samples. Pol J Microbiol 63(1):75
Remonatto D, Miotti RH Jr, Monti R, Bassan JC, de Paula AV (2022) Applications of immobilized lipases in enzymatic reactors: A review. Process Biochem 114:1–20. https://doi.org/10.1016/j.procbio.2022.01.004
Saleena SK, Johnson JI, Joseph JK, Padinchati KK, Abdulla MH (2022) Production and optimization of l-asparaginase by Streptomyces koyangensis SK4 isolated from Arctic sediment. J Basic Microbiol 63(3–4):417–426. https://doi.org/10.1002/jobm.202200116
Salwoom L, Raja Abd Rahman RNZ, Salleh AB, MohdShariff F, Convey P, Pearce D, Mohamad Ali MS (2019) Isolation, characterisation, and lipase production of a cold-adapted bacterial strain Pseudomonas sp. LSK25 isolated from Signy Island Antarctica. Molecules 24(4):715. https://doi.org/10.3390/molecules24040715
Saraswat R, Bhushan I, Gupta P, Kumar V, Verma V (2018) Production and purification of an alkaline lipase from Bacillus sp. for enantioselective resolution of (±)-Ketoprofen butyl ester. 3 Biotech 8:1–12. 10.1007/2Fs13205–018–1506–6
Shaheen M, Ullah I, Rafiq M, Maqsood Ur Rehman M, Shah AA, Hasan F (2020) Purification and characterization of lipase from psychrophilic bacteria Pseudomonas Mandelii htb2 from Batura glacier Pakistan. Appl Ecol Environ Res 18(3):4103. https://doi.org/10.15666/aeer/1803_41034114
Sharma AK, Sharma V, Saxena J (2016) A review on applications of microbial lipases. Int J Biotech Trends Technol 19(1):1–5. https://doi.org/10.14445/22490183/IJBTT-V19P601
Siddiqui KS (2015) Some like it hot, some like it cold: Temperature dependent biotechnological applications and improvements in extremophilic enzymes. Biotechnol Adv 33(8):1912–1922. https://doi.org/10.1016/j.biotechadv.2015.11.001
Sirisha E, Rajasekar N, Narasu ML (2010) Isolation and optimization of lipase producing bacteria from oil contaminated soils. Adv Biol Res 4(5):249–252
Stoytcheva M, Montero G, Zlatev RA, Leon J, Gochev V (2012) Analytical methods for lipases activity determination: a review. Curr Anal Chem 8(3):400–407
Verma S, Meghwanshi GK, Kumar R (2021) Current perspectives for microbial lipases from extremophiles and metagenomics. Biochimie 182:23–36. https://doi.org/10.1016/j.biochi.2020.12.027
Wang J, Li K, He Y, Wang Y, Han X, Yan Y (2019) Enhanced performance of lipase immobilized onto Co2+-chelated magnetic nanoparticles and its application in biodiesel production. Fuel 255:115794. https://doi.org/10.1016/j.fuel.2019.115794
Wi AR, Jeon SJ, Kim S, Park HJ, Kim D, Han SJ, Yim JH, Kim HW (2014) Characterization and a point mutational approach of a psychrophilic lipase from an arctic bacterium. Bacillus Pumilus Biotechnol Lett 36(6):1295–1302. https://doi.org/10.1007/s10529-014-1475-8
Willerding AL, Oliveira LAD, Moreira FW, Germano MG, Chagas AF (2011) Lipase activity among bacteria isolated from Amazonian soils. Enzyme Research 2011. https://doi.org/10.4061/2011/720194
Yasemin S, Arabac N, Güvenmez HK (2017) Production of Cold Active Lipase from Bacillus sp. J Appl Biol Sci 11(2):24–27
Zhang J, Lin S, Zeng R (2007) Cloning, expression, and characterization of a cold-adapted lipase gene from an Antarctic deep-sea psychrotrophic bacterium, psychrobacter sp. 7195. J Microbiol Biotechnol 17(4):604–610
Zhu B, Connolly PJ, Zhang YM, McDonnell ME, Bian H, Lin SC, Macielag MJ (2020) The discovery of azetidine-piperazine di-amides as potent, selective and reversible monoacylglycerol lipase (MAGL) inhibitors. Bioorg Med Chem Lett 30(14):127243. https://doi.org/10.1016/j.bmcl.2020.127243
Acknowledgment
The first author is grateful to the Council of Scientific and Industrial Research (CSIR) for the awarded fellowship. The authors wish to thank the Department of Marine Biology, Microbiology & Biochemistry, Cochin University of Science and Technology for providing the necessary facilities to carry out the work. The authors are thankful to the Director, National Centre for Antarctic and Ocean Research, Ministry of Earth Sciences, for logistic support and facilities for sample, collection, and analysis.
Funding
The Council of Scientific and Industrial Research (CSIR) supported the first author with grant no.09/239(0534)2018-EMR-1.
Author information
Authors and Affiliations
Contributions
HK designed and performed the experiments, analyzed the data, and prepared the manuscript. VS helped to perform the experiments. SK helped in reviewing and finalizing the manuscript. AP helped to analyze the data and in reviewing the manuscript. KKP helped with logistical support and funding. MH conceptualized the project, designed the experiments, and reviewed and finalized the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declared that they have no competing interests.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kattatheyil, H., Sajeela, V., Kabeer, S.S. et al. Screening, optimization, and molecular characterization of cold-active lipase producing Bacillus cereus I13 from Arctic sediments. Biologia 79, 1041–1055 (2024). https://doi.org/10.1007/s11756-024-01610-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11756-024-01610-y