Abstract
Purpose
African animal trypanosomiasis (AAT) is a disease affecting livestock in sub-Saharan Africa. The use of trypanocidal agents is common practice to control AAT. This study aimed to identify drug-resistant Trypanosoma congolense in Lambwe, Kenya, and assess if molecular test backed with mice tests is reliable in detecting drug sensitivity.
Methods
Blood samples were collected from cattle, in Lambwe, subjected to buffy coat extraction and Trypanosoma spp. detected under a microscope. Field and archived isolates were subjected to molecular characterization. Species-specific T. congolense and TcoAde2 genes were amplified using PCR to detect polymorphisms. Phylogenetic analysis were performed. Four T. congolense isolates were evaluated individually in 24 test mice per isolate. Test mice were then grouped (n=6) per treatement with diminazene, homidium, isometamidium, and controls. Mice were subsequently assessed for packed cell volume (PCV) and relapses using microscopy.
Results
Of 454 samples, microscopy detected 11 T. congolense spp, eight had TcoAde2 gene, six showed polymorphisms in molecular assay. Phylogenetic analysis grouped isolates into five. Two archived isolates were homidium resistant, one was also diminazene resistant in mice. Two additional isolates were sensitive to all the drugs. Interestingly, one sensitive isolate lacked polymorphisms, while the second lacked TcoAde2, indicating the gene is not involved in drug sensitivity. Decline in PCV was pronounced in relapsed isolates.
Conclusion
T. congolense associated with homidium and diminazene resistance exist in Lambwe. The impact can be their spread and AAT increase. Polymorphisms are present in Lambwe strains. TcoAde2 is unlikely involved in drug sensitivity. Molecular combined with mice tests is reliable drug sensitivity test and can be applied to other genes. Decline in PCV in infected-treated host could suggest drug resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
African animal trypanosomiasis (AAT) is a serious disease that affects livestock in most sub-Sahara African countries. The disease is known to cause tremendous losses in the livestock production industry, thus causing food insecurity [1, 2]. The disease is caused by protozoan organisms of the genus Trypanosoma. Trypanosoma congolense and T. vivax are among the species that are pathogenic in cattle [3]. The pathogens are transmitted by tsetse fly, of the genus Glossina. Tsetse flies are found in many regions in Africa including Tanzania, Ethiopia, Uganda, and Kenya [4,5,6,7]. In Kenya, six different species have been identified with Glossina pallidipes and Glossina brevipalpis being the most common vector species. AAT prevalence of 3.2% in domestic animals from four regions based on pathogen detection by microscopy has recently been reported in Kenya [8].
AAT is widely distributed in sub-Saharan African countries, including Ethiopia, Uganda, Tanzania, Kenya, Zambia, Nigeria, and Cameroon among others [8,9,10,11,12,13,14,15]. Several risk factors have been associated with the prevalence and spread of AAT. These include re-emergence of tsetse flies [6, 16], short-lived control programs that lack regularity on a large scale [17], emergence and spread of drug-resistant parasites [18], breeds of domestic animals [11], variation in seasons across the period of a year which could be associated to climate change [4], and lack of enough funds channeled toward research and eradication of AAT. This has led to huge economic losses of approximately US$ 4.5 billion in the agricultural industry in sub-Saharan Africa [19]. In Kenya, AAT endemic areas include Teso, Suba of Busia county, and Lambwe valley in Western Kenya [20], Kwale county in Coastal region [21,22,23], Mwea National Reserve in Central Kenya [8, 24], and Trasmara area in the Rift-Valley region [25].
Signs and symptoms of AAT in livestock include fever, anemia, weight loss, enlarged lymph nodes, and possible death if not treated [26, 27]. Anemia is associated with low packed cell volume (PCV) and it is indicated to be one of the signs of Trypanosoma infection progression in animals [28]. Use of trypanocidal drugs has been a common practice in the control AAT in most endemic regions in sub-Saharan Africa [1]. The major drugs used are isometamidium chloride, diminazene diaceturate, and homidium [2, 29,30,31,32,33]. Although homidium is no longer recommended for use due to its mutagenic nature [34], it is still being used in some AAT endemic regions in sub-Saharan Africa, including Kenya.
Diminazene is used to treat sheep, goats, and cattle that are infected with T. vivax and T. congolense. Its mode of action involves an interaction with the AT rich region at the minor groove of kinetoplast DNA (kDNA), thus preventing replication and formation of the DNA [35, 36]. On the other hand, isometamidium is used for both prophylaxis and curing livestock infected with T. vivax, T. congolense, and some selective T. evansi strains. Its mode of action is reported to be a high interaction with kDNA, thus causing possible linearization of the DNA in some species of Trypanosoma [36,37,38]. Homidium salts are also commonly used to treat T. vivax and T. congolense infections in goats, sheep, and cattle, despite being implicated as being a potential cancer-causing agent [34]. It is also known to have some prophylactic activity but not to the extent of isometamidium [29]. Its mode of action is an interaction with both the nucleus and kDNA [38], thus interrupting genome activity. In regions with high use of the trypanocidal drugs, there have been reports of drug-resistant parasites emerging [32, 39].
Drug resistance can be defined as lack of or reduction of sensitivity of trypanosomes to trypanocidal drugs at a dosage prescribed by the manufacturer and taken according to veterinary doctor’s directives. Easy acquisition of trypanocidal drugs by farmers due to privatization of veterinary offices in Africa has led to overfrequent use, and under-dose usage of trypanocidal drugs by livestock farmers. In addition, the use of poor quality drugs could cause drug-resistant trypanosomes [30, 40, 41]. Trypanocidal drug resistance has been reported in Nigeria, Niger, Ghana, Kenya, Uganda, Tanzania, Somalia, Sudan, Chad, Malawi, Zimbabwe, and Mozambique among others [42, 43]. Drug resistance in T. congolense isolates has been linked to point mutations/polymorphisms in P2 purine transporter gene TcoAT1, with polymorphisms in the TcoAde2 region of the transporter gene, linked to diminazene resistance [44, 45]. Several studies have been done using mouse test and polymerase chain reaction-restriction fragment length polymorphism (PCR–RFLP) for detection of diminazene and isometamidium resistance [42, 46, 47]. These mutations are said to spread with more exposure to trypanocidal drugs [48], thus causing possible increase in prevalence rate in some animals. However, this was later debunked from a study that showed a high level of polymorphism associated with diminazene resistance without exposure of isolates to trypanocidal drugs [43]. In addition, a more recent study confirmed through genetic, biochemical, and molecular methods that TcoAT1 has no ability to bind and transport diminazene, but that it could be a transporter for purine nucleosides [49]. However, these molecular, biochemical, and genetic experiments did not confirm whether the presence or absence of the TcoAT1 genes had an effect on drug sensitivity using single-dose mice experiments, which has been one of the common and reliable approaches to confirm drug-resistant Trypanosoma isolates from the field [50]. This is so since Trypanosoma-Swiss white mouse model closely resembles physiological conditions of trypanosome-infected cattle, thus providing accurate data on therapeutic and chemoprophylactic efficacy of trypanocidal drugs for management of animal trypanosomiasis. Since the immune system is an important factor for drug efficacy in in vivo systems, live animals are also useful than in vitro testing systems in this type of studies [51]. Further, findings of drug sensitivity studies using mice can easily be extrapolated in livestock [50, 52].
In Kenya, T. congolense and T. vivax have been identified as the most common species affecting cattle [53]. Here also, there have been cases of parasites resistant to the three major trypanocidal drugs used in the Coastal region of Kenya and in parts of Western Kenya [54]. However, regions, such as Lambwe valley in Homa Bay in Western Kenya, which is endemic to AAT, lack studies on drug resistance and molecular characterization of the Trypanosoma parasites, despite being endemic to AAT [21]. Drug resistance could be one of the major causes of high mortality rate in cattle. Our previous study reported an AAT prevalence of 15.63% in cattle, and T. congolense was one of the most common species in Lambwe villages [55]. Trypanosoma congolense is also known to propagate in mice. This was a follow-up study aimed at identifying and characterizing T. congolense isolates from field and archived isolates of Lambwe region. In addition, this study aimed to assess whether molecular assays, backed up with in vivo mice experiments, are a reliable approach to detect drug sensitivity. Data from such a study can help epidemiologists determine whether trypanocidal drug resistance could be contributing to the high amount of AAT cases reported in endemic regions, such as Lambwe. In a previous study, sub-Saharan countries reporting high trypanocidal drug resistance were also shown to report high AAT prevalence [56]. Moreover, it could create awareness in drug applications, and prevent possible consumption of remnant drug particles by humans consuming the meat products, due to lack of drug uptake by the parasites.
Materials and Methods
Study Area
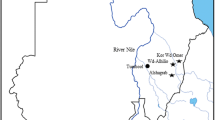
This study was carried out in villages in the Lambwe region of Homa Bay, western Kenya, located between 000 5’ S and 34° 12’ E (Fig. 1). The area lies in the Lambwe river valley that rises about 1,100 m above the sea level. This region is densely dominated with grassland areas, with different species of trees forming forested areas. Mixed farming is a common practice in the area, with most farmers planting cereals and keeping cattle [57]. Ruma National Park forms part of Lambwe region, with G. pallidipes infesting the area and affecting livestock productivity in the area through reported cases of AAT. The study location was chosen based on our previous study in the region, which reported a high prevalence of cattle trypanosomiasis, and that there is poor control practices and use of trypanocidal drugs by livestock keepers in the region [55]. Hence, we wanted to see if the presence of drug-resistant parasites could also be one of the contributing factors to the high prevalence seen.
Study Design, Sample Size Determination, and Sampling Procedure
Sample analysis was conducted to determine presence of drug-resistant T. congolense species from Lambwe. A cross-sectional study was performed to determine drug sensitivity, using molecular approach followed by in vivo drug sensitivity test.
A 9.2% prevalence (P) from a previous study was used to calculate sample size [21]. A confidence interval (Z) of 1.96 value, an absolute desired precision (d) of 0.05, number of cows (m) sampled in each village cluster being 15, and an intercluster correlation coefficient (ρ) of 0.15 were used [58]. The formula for cluster sampling design was then applied to determine sample size [59].
Approximately 110 cattle were blood sampled each from four villages (Kamato, Gendo, Kigoto, and Nyatoto) within Lambwe region, close to Ruma National Park. No consideration was made on type of breeds (all farmers had local breeds), age, sex, or body state of the animal. Every single household with cattle were included in the study. Animals that had received trypanocidal drug treatment in the preceding 90 days were excluded from this study.
Data Collection
Trypanosoma Collection, Detection, and Preservation
For detection of live Trypanosoma parasites at the field level, a volume of 5 ml blood was collected from jugular vein of the cattle in May 2021 and subjected to buffy coat extraction for Trypanosoma pathogen detection [60]. The parasite isolates were then cryopreserved in capillary tubes containing 20% glycerol in Phosphate Saline Glucose (PSG) pH 8. This was then stored in liquid nitrogen for shipment to the molecular, and pharmacology laboratory at Kenya Agricultural and Livestock Research Organization (KALRO), Nairobi, Kenya. Additionally, three archived T. congolense isolates originally from Lambwe region, available via the KALRO-BioRI cryobank, Kenya Trypanosomiasis Research Institute (KETRI) 3696 isolated in 1962, KETRI 3735 isolated in 1980, and KETRI 2940 isolated in 1984, were also included in this study to check for the status of drug resistance from isolates that were collected before, and also have more isolates to confirm whether molecular gene detection, backed up with in vivo assays, is a sure approach to detect drug sensitivity.
Laboratory Experiments
Molecular Assays–DNA Extraction and Amplification
Molecular assays were performed to confirm Trypanosoma species and sub-species collected from the field and the KETRI isolates, and to confirm the presence of TcoAde2 gene of the transporter gene TcoAT1. Isolates were first subjected to DNA extraction using a QIAamp DNA blood mini kit (Qiagen, Valencia, California, USA). PCR was then performed in a 25 μl reaction containing, 1X buffer (Bioline, UK), 1 μM forward and 1 μM reverse primers, 0.8 U/μl MyTaq (Bioline, UK), 12.6 μl Dnase-free water and 5 μl of template DNA. Primers used were ITS1 primers ITS1-F: 5′ CCG GAA GTT CAC CGA TAT TG 3’ and ITS1-R: 5′ TTG CTG CGT TCT TCA ACG AA 3′. ITS1-F binds to 18 S, while ITS1-R binds to 5.8 s regions of rDNA [61, 62]. These produce different band sizes in different Trypanosoma species, based on their internal transcribed spacer regions. All ITS1 positive for T. vivax and T. congolense were confirmed by band size of 250 bp and 700 bp, respectively, after gel electrophoresis. All ITS1 positive for T. congolense were then subjected to species-specific amplification of T. congolense Savannah (TCS). Primers TCS1: CGAGAACGGGCACTTGCGA and TCS2: GGACAAACAAATCCCGCACA were used [63]. The expected band size for TCS was 316 bp, found in position 293 of the genome [63]. Once confirmed positive for TCS, TcoAde2 primers: Ade2F-ATAATCAAAGCTGCCATGGATGAAG and Ade2R-GATGACTAACAATATGCGGGCAAAG for the T. congolense P2 putative transporter gene (TcoAT1) were used to confirm the presence of the Ade2 gene [44]. These primers bind at position 522 bp and 1145 bp from start codon of the TcoAT1 genome. Expected band size for TcoAde2 amplicon was 648 bp after running in 1.5% agarose gel. All positive TcoAde2 bands were purified using GeneJET PCR Purification kit, according to the manufacturer’s instructions and sent for DNA Sanger sequencing at Inqaba Biotech (South Africa). Sequencing was performed to confirm the presence of common polymorphisms that were also detected from other countries of sub-Saharan Africa, such as Zambia, Ethiopia, Zimbabwe, and Cameroon [42, 43, 46, 47], and evaluate if the polymorphisms were also present in isolates from Kenya. Schematic overview of the procedure is shown in Fig. 2.
Mice Work
Ethics Statement
During the meeting of the Institutional Animal Care and Use Committee (IACUC) of the KALRO–Biotechnology Research Institute on June 2, 2021 held at BioRI-Muguga, the proposal titled “Epidemiology, Drug sensitivity pattern and control practice of African Animal Trypanosomes in Western Kenya” in which the proposed study was to carry out in vivo drug sensitivity experiments at the Institute was reviewed. The committee further received the revised animal protocol and confirmed that all issues raised by IACUC reviewers were adequately addressed. The committee was also convinced that the study addressed an important research topic that would contribute in the management of animal trypanosomiasis in the region. The committee therefore resolved to support the study and hold the person involved responsible to ensure high standards of animal welfare were observed. The committee made impromptu visits to ensure compliance with its regulations.
In vivo Mice Experiments
Molecular experiments were followed up with mice experiments for all T. congolense positive isolates. This was done to detect if the absence or presence of TcoAde2 gene has an effect on drug sensitivity in mice model, and also detect isolates resistant to trypanocidal drugs in the mice experiment. All T. vivax positive isolates were excluded from the mice experiments as they do not propagate in a murine model. Swiss white female mice (25-30 g; 6–8 weeks old) were used for in vivo drug sensitivity experiments. A total of 11 T. congolense isolates (eight field and three KETRI isolates), two donor mice for in vivo propagation of each isolate, and 24 test mice (for each T. congolense stabilates collected from donor mice) were used. Treatment was then done per group (n=6) in each isolate. The mice were acquired from KALRO-BioRI small animal unit. The mice were laboratory inbred mice, housed in groups of six per cage, and kept in medium cages, with dry wood shavings used as bedding material changed twice a week. They were provided with mice pellets sourced from Unga Feeds Kenya Limited, and chlorinated tap water was also provided ad libitum [64]. The experimental mice were allowed to acclimatize for 14 days, during which physical and visual examination of skin for signs of mite infestation (mange) and fecal examination were done and treatment with 20 mg/kg body weight (BW) of ivermectin was administered once intraperitoneally as a cover for endo- and ectoparasites [65].
Single-dose mice experiments were applied as previously described [50], with some adjustments. Live body weight was assessed once before infection using digital balance, to determine the amount of drug to be administered per mouse. For in vivo propagation assessment of parasitemia and packed cell volume (PCV) determination, not more than 10% of total blood volume in mice (77–80 ml/kg) was taken. This was 2.5 µl of peripheral blood samples. Packed cell volume was determined once a week after infection and treatment for the test mice and also for the control mice [28]. This was done to assess whether there were any signs of anemia progression after mice received treatment with a trypanocidal drug, to determine if the drug was working and whether isolate infection were susceptible or resistant to the drug treatment. Anemia is low PCV, hematocrit and hemoglobin, and it is indicated to be one of the signs of Trypanosoma infection progression in animals [28].
Mice were screened by microscopy for any trypanosome infection prior to inoculation with the isolates for propagation of the trypanosomes. For in vivo propagation in donor mice, two capillary tubes of the relevant isolate were put into 0.4 ml of ESG buffer, and 0.2 ml infected in each donor mice for propagation. A blood sample of 2.5 µl was taken each day for approximately seven days by the tail tip of each mouse, and viewed under microscope, until a parasitemia count of 3.2 × 107 trypanosomes/ml was reached.
Blood trypanosomes from donor mice were then diluted in ESG buffer until a count of 1 × 105 blood trypanosomes was reached. This concentration is considered adequate for inoculation in mice as per previous single-dose mice experiments documented [50]. This was then used for intraperitoneal infection in test mice. Each of the T. congolense isolates was infected in 24 mice: six then treated with isometamidium chloride (69,007-LYON France TFP 711A), six with diminazene diaceturate (Batch 1022A1-Libourne France), six with homidium bromide (batch 22,600 Loudeac, France), and six used as negative control. Prior to treatment, each drug was dissolved in distilled water to make the final concentrations. Recommended single-dose concentration of 1 mg/kg of isometamidium chloride, 20 mg/kg for diminazene diaceturate, and 1 mg/kg homidium bromide intraperitoneal treatment was used 24 h after infection [50]. Negative control groups were infected, but were treated with distilled water only. Blood samples of 2.5 µl were then taken each day by tail tip from each mouse for 14 days, and twice a week after 14 days until 60 days post-treatment for detection of relapses under a light microscope. A resistant isolate was confirmed when more than one mouse had a relapse within the 60 days post-treatment. A sensitive isolate was confirmed when at least five mice showed no relapse and were cured within the 60 days [50]. A schematic overview of the procedure is shown in Fig. 2.
The procedure performed in mice were not invasive; therefore, there was no need for pain management. However, all mice were euthanized at the end of the experiment using concentrated carbon dioxide.
Data Analysis
DNA sequences from the field collected and KETRI isolates were edited and merged using Bioedit software [66, 67]. The Blastn tool in NCBI was then used to identify homologous sequences in Genbank to the query sequences. The sequences, including the identified drug-sensitive species in GenBank, were translated to protein sequences using Expasy translation tool. Both the DNA and protein sequences were deposited in Genbank with Accession Numbers: OK424593-OK424594, OK137195-OK137198, and OK545528. These were sequences from isolates C7, C3, C4, C5, C6, C8, and C10 from Gendo and Kamato in Lambwe, and KETRI 3735 from Lambwe too. Clustal W multiple alignment in MEGA11 was then applied to compare protein sequences from these study isolates from Lambwe to those of T. congolense-sensitive species in Genbank [68]. The evolutionary history was inferred by using the Maximum Likelihood method and JTT matrix-based model, and 1000 bootstraps applied [69]. Evolutionary analyses were conducted in MEGA11 [69]. The average mean weekly PCV was determined for each trypanocidal drug in each of the four isolates for the eight weeks.
Results
Ex vivo Parasitological and Molecular Results
A total of 454 blood samples were collected. Out of these, morphological microscopic identification of eight T. congolense from Gendo and Kamato villages in Lambwe Valley and seven T. vivax live parasites from Kamato, Gendo, and Kigoto was made under a light microscope. There were no live parasites detected from Nyatoto village and no co-infections detected microscopically. Thus, an overall prevalence of 1.76% for T. congolense and 1.54% for T. vivax was detected. Additionally, three archived KETRI isolates from Lambwe that were pre-detected microscopically were used. Out of the 18 Trypanosoma isolates considered in this study (seven T. vivax and eight T. congolense field isolates and additional three archived T. congolense isolates), all eleven T. congolense isolates gave amplification products for T. congolense Savannah Fig. 3. Of the eleven T. congolense Savannah positive isolates, eight (six field and two KETRI isolates) gave amplification products with TcoAde2 gene Fig. 4. A total of eight sequences re-amplified, after sequencing PCR-purified TcoAde2 products. A total of seven sequences achieved submission acceptance in GenBank (Table 1).
Gel image showing TcoAde2 gene bands (a) Row 1: Lane 1: 100bp ladder; lane 2, 4, and 6: indicate positive Ade2 gene; lane 10: shows a negative control; lane 3, 5, 7, 8 and 9: were negative for Ade2 gene; arrow on the right shows expected position of the band (b) Row 2: Lane 1: 100bp ladder; lane 2, 3 and 7: show negative result for Ade2; Lanes 4, 5, 6, 8, 9: were positive to Ade2 gene; Lane 10, positive control; arrow on the right shows expected position of the band
Multiple sequence alignment analysis of the TcoAde2 region revealed polymorphisms in 6/7 Valine to an Isoleucine at the conserved region position 132, 5/7 Alanine to a Glycine at the position 88 and 3/7 Alanine to a Glycine at position 30 of the T. congolense isolates. These were isolates C3-OK137196, C6-OK424593, C10- OK137195, C5-OK137198, and C4-OK137197 from Gendo and Kamato in Lambwe, and KETRI 3735-OK545528 from Lambwe South (Table 1). This was based on comparisons of the isolates, to that of drug-sensitive species in the GenBank Accession #CCC92853.1-IL3000. Only the sequence from T. congolense isolate C7-OK424594 did not show any polymorphism at positions 132, 88, and 30, C6-OK424593 at positions 88 and 30, and C4-OK137197 and KETRI 3735-OK545528 at position 30.
The phylogenetic tree of the highest log likelihood (-667.11) is shown in Fig. 5, indicating five branches of the T. congolense isolates. The percentage of trees in which the associated taxa clustered together is shown next to the branches. There were a total of 216 positions in the final dataset. Isolates, C7-OK424594 and C6-OK424593, were very closely and closely related to the drug-sensitive isolate IL3000-CCC92853 from GenBank, respectively. These were the isolates that lacked polymorphism at positions 132, 88, and 30. The rest of the isolates, C3-OK137196, C6-OK424593, C10- OK137195, C5-OK137198, and C4-OK137197, from Gendo and Kamato, as well as KETRI 3735-OK545528 from Lambwe South, were distantly related to IL3000-CCC92853.
In vivo Mice Results
Assessment of Relapses
Out of the 11 Trypanosoma isolates considered in this study, three field isolates (C3, C6, and C7) and three biobank isolates (KETRI 3696, KETRI 3735, and KETRI 2940) propagated successfully in donor mice. Out of these six isolates, only four isolates (C7 and the three KETRI isolates) propagated in drug test mice (Table 1). One isolate (C7) was sensitive to all three trypanocidal drugs used (Table 2), had TcoAde2 gene, and did not show polymorphisms in the molecular assay (Fig. 6). KETRI 3696 lacked TcoAde2 gene but was sensitive to all three trypanocidal drugs in mice assay. For infection with KETRI 2940, three mice showed relapse 10–12 days post-treatment with homidium; with KETRI 3735, four mice showed relapse 15, 21, 22, and 25 days post-treatment with homidium, and two mice relapsed on 8 and 15 days post-treatment with diminazene (Table 2). Thus, giving an overall drug resistance prevalence of 0.44% for T. congolense isolates.
Multiple alignment showing polymorphic regions of field and archived isolates. Isolates C3, C4, C5, C6, KETRI 3735 & C10 compared to drug-sensitive species (IL3000), showed polymorphisms at conserved regions position 132, 88 and 30, fromValine to Isoleucine and from Alanine to Glycine. Arrows show regions
PCV Results
Mean weekly PCV varied after infection and treatment with (homidium, isometamidium, and diminazene) for the four isolates, in addition to the control groups (Fig. 7, Supplementary files 1, 2 & 3). The control groups in all the isolates had more pronounced drop in mean PCV over the 8 weeks, C7––56.85 ± 2.08 to 0.00, KETRI 3696––55.86 ± 5.95 to 0.00, KETRI 2940––54.45 ± 7.05 to 0.00, and KETRI 3735––55.86 ± 5.95 to 0.00. Drop in PCV was more in control KETRI 3696, followed by KETRI 2940, KETRI 3735, and finally C7 isolates. PCV for isolate KETRI 3735 homidium-treated mice was observed to decline considerably on the 4th, 5th, and 7th weeks from 57.00 ± 5.58 to 46.00 ± 5.11 compared to other isolates. On the other hand, PCV for isolate KETRI 2940 homidium-treated mice was also seen to have a noticeable drop on 6th week from 50.50 ± 2.68 to 47.00 ± 6.90 compared to the rest. Of important to note is that relapses were seen earlier from 3rd to 4th weeks, and the 2nd week for the two isolates KETRI 3735 and 2940, respectively. For diminazene treatment, KETRI 3735 isolate showed a big drop in PCV on the 5th week from 54.16 ± 5.38 to 50.50 ± 2.68. Relapses were, however, seen on the 2nd week. Isolates treated with isometamidium did not show much decline over the weeks, but for KETRI 3735 there was a noticeable decline on the 4th to 5th weeks. Overall mean PCV for the 8 weeks for isolate KETRI 3735 isometamidium-treated mice was higher, 55.48 ± 2.08, than its mean PCV for diminazene and homidium treatment, 53.89 ± 2.76 and 53.13 ± 5.44 respectively. Similarly, overall mean PCV for the 8 weeks in isometamidium-treated mice in isolate KETRI 2940 54.52 ± 1.68 was higher than in homidium-treated mice 52.50 ± 2.26. However, the difference was not that much between isolate KETRI 3735 and KETRI 2940 isometamidium-treated mice (Fig. 7, Supplementary file 1, 2 & 3).
Discussion
This study aimed at assessing the current state of T. congolense drug sensitivity to the three main trypanocidal drugs (isometamidium chloride, diminazene, and homidium) used in AAT endemic regions in Kenya, characterizing resistant species and assessing whether molecular test backed up with in vivo drug test is a reliable approach in detecting drug sensitivity. Molecular assays using TcoAde2 gene were conducted, backed up with in vivo single-dose drug sensitivity mice test using the three main trypanocidal drugs. In molecular test, all 11 fields and KETRI biobanked isolates considered were confirmed to be positive for T. congolense Savannah; however, not all TcoAde2 gene could be amplified. This could be attributed to lack of the TcoAde2 gene in some isolates. Similar findings have been reported from a study in Ethiopia [47]. In isolates that gave amplification products for TcoAde2 gene, polymorphism in the Valine to Isoleucine and Alanine to Glycine amino acids (positions 132, 88 and 30 – conserved regions) indicated the presence of polymorphisms from Kamato and Gendo villages in Lambwe, and in one of the KETRI isolates from Lambwe South. These polymorphism identified could be linked to a shift in codon, GTC to ATC at the molecular level. This confirms that the common polymorphism identified in some T. congolense isolates from other sub-Saharan countries, such as Zambia, Ethiopia, Zimbabwe, South Africa, and Cameroon [42, 43, 46, 47], is also present in Kenyan T. congolense isolates region from Lambwe.
For the low levels of species associated with drug resistance, we suggest that it is due to the assessment being based on T. congolense species that were detected by microscopy, confirmed by molecular assay, and inoculated in mice models. It could also be due to the fact that most isolates, especially from 2021 could not propagate well in mice model for detection of drug resistance. We suggest this could be due to the fact that parasites from recent years are more susceptible to immune system of the host. Another justification for the low drug resistance could be that the area has more drug-sensitive species. However, the implication can be a spread of the resistant parasites with more exposure or poor use of the trypanocidal drugs [30, 70]. Similar reports of low levels of drug-resistant Trypanosoma species have been reported elsewhere in Kenya [70].
A phylogenetic tree of the sequenced isolates showed the existence of genetic variations in the T. congolense isolates formed five branches of the species. Similar reports on genetic variation in T. congolense have been reported in West, South, and East Africa [71, 72].
In the mice experiment described here, four (C7, KETRI 3735, KETRI 3696, and KETRI 2940) of the 11 T. congolense isolates grew successfully in mice. This could be due to the fact that not all T. congolense species are known to grow well in mice [42]. In addition, the immune system of the mice models may vary hence making some mice immunotolerant to the isolates. The trypanocidal drugs that showed relapse in mice experiments were homidium and diminazene. Homidium together with isometamidium belong to phenanthridine group of trypanocidals, which work by inhibiting gene expression and replication of kinetoplast DNA (kDNA) [29, 73, 74]. Resistance to these drugs and diamidines are caused by mutations or loss of subunits of vacuolar ATPase and transport protein or polymorphism in mitochondrial F-1 ATPase subunit. This is known to cause contact between the ATPases, thus causing the parasite to get rid of the kDNA hence leading to drug resistance [73, 75, 76]. Trypanosoma congolense resistance to homidium has also been reported in Ethiopia and Nigeria [77, 78]. In addition, comparable results on possible existance of T. congolense-resistant isolates to trypanocidal drugs have been reported from Transmara region of Kenya [25]. Resistance to diminazene and multi-drug resistance to diminazene, homidium, and isometamidium have also been identified in Ethiopia and Burkina Faso [79,80,81]. Isolate KETRI 3696 that had shown lack of TcoAde2 gene in molecular assay and was sensitive to all three trypanocidal drugs proved that indeed this transporter gene is not involved in the drug uptake. This correlates with the findings from a different study, but did not involve mice tests as a confirmatory test [49]. This also correlates to a recent study that proved that diminazene uptake in T. congolense was due to low processes of transporters, such as folate transporters and resistance, may be due to lower/decreased mitochondrial membrane potential [82]. The correlation from our study shows that, indeed molecular assays, backed up with in vivo mice test, are a sure approach to detect drug sensitivity in isolates. However, the search or finding for other genes and new reliable and easy methods to evaluate drug resistance of Trypanosoma species are needed. There were no isometamidium-resistant isolates. This is probably due to the fact that immune system of the mice was not compromised. It has been documented from previous studies that immune system of host animal plays a role in preventing isometamidium resistance in T. congolense isolates [83], and that the drug is only involved in preventing replication of the parasite, while immune system is involved in killing the parasite.
Average PCV values were seen to decline in most Trypanosoma-treated mice. However, the drop in PCV being more in control groups than in experimental groups was because the mice did not receive any trypanocidal treatment. The drop in mean PCV in the control groups went to 0.00 as most of the control mice did not reach the end of the experiment. The different level of drop of PCV in the different isolate control groups could be attributed to the difference in the virulence state of the isolates, in which the most recent isolate was less virulent than isolates collected from years before. General decline in mean PCV could be attributed to anemia progression in infected mice [28]. This is despite receiving treatment, which could also indicate the virulence state of the different isolates and that it required considerable time for the drug to effectively circulate, and reduce the parasitemia load. Similar findings have been reported from elsewhere in Kenya [84]. Consistent results were also seen in T. evansi-infected mice. The decline was more in homidium and followed by diminazene-treated mice in two isolates. Homidium was one of the drugs showing more relapses in two of the isolates, followed by diminazene in one of the isolates, thus explaining their noticeable decline in average PCV. The preceding time period for relapses in most isolates occurring prior to the considerable drop in average PCV could be explained by the fact that anemia, which is linked to low PCV, normally occurs at a later stage after an infection with the animal trypanosomes.
Limitations
The authors accept that not all isolates from recent years could propagate well in mice. Hence, more studies need to be done with greater number of isolates that can propagate well in mice to confirm cases of drug-resistant species.
Conclusion
Potential T. congolense species that are resistant to homidium and diminazene exist in the Lambwe region of Kenya. If not controlled, the impact can be a spread and increase of the resistant parasites, and eventual increase in AAT prevalence. This may affect efforts toward managing the disease. Polymorphisms that are present in TcoAde2 gene of T. congolense isolates from other sub-Saharan countries are also present in isolates from Lambwe region of Kenya. Molecular assays combined with in vivo mice test is a sure approach to detect drug sensitivity and can be applied to other genes or parasites for drug sensitivity tests. Nonetheless, using new reliable and easier to use techniques would be more appropriate. Decline in PCV in trypanocidal-treated host species that are infected with animal trypanosomes could indicate resistant Trypanosoma species to the used drugs.
Data Availability
The datasets generated and/or analyzed during the current study are available in the GenBank repository, some are included in this published article [and its supplementary information files].
References
Kristjanson PM, Swallow BM, Rowlands GJ, Kruska RL, De Leeuw PN (1999) Measuring the costs of African animal trypanosomosis, the potential benefits of control and returns to research. Agric Syst 59:79–98. https://doi.org/10.1016/S0308-521X(98)00086-9
Shaw A (2004) The trypanosomiases;Economics of african trypanosomiasis. Wallingford. https://doi.org/10.1079/9780851994758.0369
Tchamdja E, Kulo AE, Vitouley HS, Batawui K, Bankolé AA, Adomefa K et al (2017) Cattle breeding, trypanosomosis prevalence and drug resistance in Northern Togo. Vet Parasitol 236:86–92. https://doi.org/10.1016/j.vetpar.2017.02.008
Simwango M, Ngonyoka A, Nnko HJ, Salekwa LP, Ole-Neselle M, Kimera SI et al (2017) Molecular prevalence of trypanosome infections in cattle and tsetse flies in the maasai Steppe, northern Tanzania. Parasit Vectors 10:1–11. https://doi.org/10.1186/s13071-017-2411-2
Rodrigues CMF, Garcia HA, Sheferaw D, Rodrigues AC, Pereira CL, Camargo EP et al (2019) Genetic diversity of trypanosomes pathogenic to livestock in tsetse flies from the Nech Sar National Park in Ethiopia: A concern for tsetse suppressed area in Southern Rift Valley. Infect Genet Evol 69:38–47. https://doi.org/10.1016/j.meegid.2019.01.010
Waiswa C, Picozzi K, Katunguka-rwakishaya E, Olaho-mukani W (2006) Glossina fuscipes fuscipes in the trypanosomiasis endemic areas of south eastern Uganda: Apparent density, trypanosome infection rates and host feeding preferences. Acta Trop 99:23–29. https://doi.org/10.1016/j.actatropica.2006.06.005
KENTTEC (2017) Tsetse and Trypanosomiasis Problem in Kenya. http://www.kenttec.go.ke/tsetse-and-trypanosomiasis-problem-in-kenya/. Accessed 12 Dec 2019
Ngari NN, Gamba DO, Olet PA, Zhao W, Paone M, Cecchi G (2020) Developing a national atlas to support the progressive control of tsetse-transmitted animal trypanosomosis in Kenya. Parasit Vectors 13:1–12. https://doi.org/10.1186/s13071-020-04156-5
Dagnachew S, Tsegaye B, Awukew A, Tilahun M, Ashenafi H, Rowan T et al (2017) Prevalence of bovine trypanosomosis and assessment of trypanocidal drug resistance in tsetse infested and non-tsetse infested areas of Northwest Ethiopia. Parasite Epidemiol Control 2:40–49. https://doi.org/10.1016/j.parepi.2017.02.002
Weny G, Okwee-Acai J, Okech SG, Tumwine G, Ndyanabo S, Abigaba S et al (2017) Prevalence and risk factors associated with hemoparasites in cattle and goats at the edge of kibale national park, Western Uganda. J Parasitol 103:69–74. https://doi.org/10.1645/16-33
Nhamitambo NL (2017) Molecular identification of trypanosome species in cattle of the mikumi human/livestock/wildlife interface areas. J Infect Dis Epidemiol, Tanzania. https://doi.org/10.23937/2474-3658/1510029
Kivali V, Kiyong AN, Fyfe J, Toye P, Fèvre EM, Cook EAJ (2020) Spatial Distribution of trypanosomes in cattle from Western Kenya. Front Vet Sci 7:1–6. https://doi.org/10.3389/fvets.2020.00554
Mbewe NJ, Namangala B, Sitali L, Vorster I, Michelo C (2015) Prevalence of pathogenic trypanosomes in anaemic cattle from trypanosomosis challenged areas of Itezhi-tezhi district in central Zambia. Parasit Vectors 15:4–9. https://doi.org/10.1186/s13071-015-1260-0
Balogun JB, Chechet GD, Ndams IS, Okubanjo O, Mamman M (2021) Molecular Prevalence of Trypanosome infections in kachia grazing reserve of kaduna state nigeria. Niger J Parasitol 42:6–9. https://doi.org/10.4314/njpar.v42i1.7
Ngomtcho SCH, Weber JS, Ngo Bum E, Gbem TT, Kelm S, Achukwi MD (2017) Molecular screening of tsetse flies and cattle reveal different trypanosoma species including T. grayi and T. theileri in northern cameroon. Parasit Vect Parasit Vect 10:1–16. https://doi.org/10.1186/s13071-017-2540-7
Magona JW, Walubengo J, Odiit M, Okedi LA (2005) Implications of the re-invasion of Southeast Uganda by Glossina pallidipes on the epidemiology of bovine trypanosomosis. Vet Parasitol 128:1–9. https://doi.org/10.1016/j.vetpar.2004.10.020
Adungo F, Mokaya T, Makwaga O, Mwau M (2020) Tsetse distribution, trypanosome infection rates, and small-holder livestock producers’ capacity enhancement for sustainable tsetse and trypanosomiasis control in Busia, Kenya. Trop Med Heal 48:1–8. https://doi.org/10.1186/s41182-020-00249-0
Mekuria S, Gadissa F (2011) Survey on bovine trypanosomosis and its vector in metekel and Awi zones of Northwest Ethiopia. Acta Trop 117:146–151. https://doi.org/10.1016/j.actatropica.2010.11.009
Holt HR, Selby R, Mumba C, Napier GB, Guitian J (2016) Assessment of animal African trypanosomiasis (AAT) vulnerability in cattle-owning communities of sub-Saharan Africa the LCNTDR Collection: Advances in scientific research for NTD control. Parasit Vectors 9:1–12. https://doi.org/10.1186/s13071-016-1336-5
Thumbi SM, Jung JO, Mosi RO, Mcodimba FA (2010) Spatial distribution of african animal trypanosomiasis in suba and teso districts in western kenya. BMC Res Notes 3:2–6. https://doi.org/10.1186/1756-0500-3-6
Magak G, Okoth WO, Michael OG, Kennedy O, Thedeus OO, Awino B et al (2019) Human and Animal trypanosomiasis in lambwe valley foci, kenya – current situation and latent trypanotolerance. Asian J Res Anim Vet Sci 3:1–12
Muraguri GR, McLeod A, McDermott JJ, Taylor N (2005) The incidence of calf morbidity and mortality due to vector-borne infections in smallholder dairy farms in Kwale district, Kenya. Vet Parasitol 130:305–315. https://doi.org/10.1016/j.vetpar.2004.11.026
Mbahin N, Affognon H, Andoke J, Tiberius M, Mbuvi D, Otieno J et al (2013) Parasitological prevalence of bovine trypanosomosis in kubo division of kwale county of coastal: baseline survey. Am J Anim Vet Sci 8:28–36. https://doi.org/10.3844/ajavssp.2013.28.36
Shaw APM, Cecchi G, Wint GRW, Mattioli RC, Robinson TP (2014) Mapping the economic benefits to livestock keepers from intervening against bovine trypanosomosis in Eastern Africa. Prev Vet Med 113:197–210. https://doi.org/10.1016/j.prevetmed.2013.10.024
Mapenay IM, Maichamo MW (2008) Epidemiology of trypanocidal drug resistance in the transmara district of kenya. Kenya Vet 30:57–61. https://doi.org/10.4314/kenvet.v30i2.39624
Bekele EE (2016) The Current situation and diagnostic approach of nagana in africa : a review the current situation and diagnostic approach of nagana in africa : a review. J Nat Sci Res 5:119–120
Fineile P, Murray M, Barry JD, Morrison WI, Williams RO, Hirumi H, Rovis L (1983) African animal trypanosomiasis. World Anim Rev. FAO Animal Production and Health Paper 37
Naessens J, Kitani H, Nakamura Y, Yagi Y, Sekikawa K, Iraqi F (2005) TNF-α mediates the development of anaemia in a murine Trypanosoma brucei rhodesiense infection, but not the anaemia associated with a murine Trypanosoma congolense infection. Clin Exp Immunol 139:405–410. https://doi.org/10.1111/j.1365-2249.2004.02717.x
Giordani F, Morrison LJ, Rowan TG, De Koning HP, Barrett MP (2016) The animal trypanosomiases and their chemotherapy: A review. Parasitology 143:1862–1889. https://doi.org/10.1017/S0031182016001268
Delespaux V, Geerts S, Brandt J, Elyn R, Eisler MC (2002) Monitoring the correct use of isometamidium by farmers and veterinary assistants in Eastern Province of Zambia using the isometamidium-ELISA. Vet Parasitol 110:117–122. https://doi.org/10.1016/S0304-4017(02)00316-3
Jamal S, Sigauque I, Macuamule C, Neves L, Penzhorn BL, Marcotty T et al (2005) The susceptibility of Trypanosoma congolense isolated in zambézia province, mozambique, to isometamidium chloride, diminazene aceturate and homidium chloride. Onderstepoort J Vet Res 72:333–338. https://doi.org/10.4102/OJVR.V72I4.190
Mekonnen G, Mohammed EF, Kidane W, Nesibu A, Yohannes H, Van Reet N et al (2018) Isometamidium chloride and homidium chloride fail to cure mice infected with Ethiopian Trypanosoma evansi type A and B. PLoS Negl Trop Dis 12:1–12. https://doi.org/10.1371/journal.pntd.0006790
Mugunieri GL, Murilla GA (2003) Resistance to trypanocidal drugs - Suggestions from field survey on drug use in Kwale district, Kenya. Onderstepoort J Vet Res 70:29–36
Sutcliffe OB, Skellern GG, Araya F, Cannavan A, Sasanya JJ, Dungu B et al (2014) Animal trypanosomosis: making quality control of trypanocidal drugs possible. OIE Rev Sci Tech. 33:813–820. https://doi.org/10.20506/rst.33.3.2320
Wilson WD, Tanious FA, Mathis A, Tevis D, Hall JE, Boykin DW (2008) Antiparasitic compounds that target DNA. Biochimie 90:999–1014. https://doi.org/10.1016/j.biochi.2008.02.017
Shapiro TA, Englund PT (1990) Selective cleavage of kinetoplast DNA minicircles promoted by antitrypanosomal drugs. Proc Natl Acad Sci U S A 87:950–954
Dougherty G, Waring MJ (1982) The interaction between prothidium dibromide and DNA at the molecular level. Biophys Chem 15:27–40. https://doi.org/10.1016/0301-4622(82)87014-2
Boibessot I, Turner CMR, Watson DG, Goldie E, Connel G, McIntosh A et al (2002) Metabolism and distribution of phenanthridine trypanocides in Trypanosoma brucei. Acta Trop 84:219–228. https://doi.org/10.1016/s0001-706x(02)00188-2
Sinyangwe L, Delespaux V, Brandt J, Geerts S, Mubanga J, Machila N et al (2004) Trypanocidal drug resistance in eastern province of zambia. Vet Parasitol 119:125–135. https://doi.org/10.1016/j.vetpar.2003.11.007
Van Den Bossche P, Doran M, Connor RJ (2000) An analysis of trypanocidal drug use in the eastern province of zambia. Acta Trop 75:247–258. https://doi.org/10.1016/S0001-706X(00)00059-0
KARI E-Mifugo, Cattle (2014) KARI E-Mifugo (Livestock) Clinic. https://kalro.org/emimi/sites/default/files/E-Mifugo Animal Trypanosomiasis final 28/. Accessed 20 June 2020
Delespaux V, Geysen D, Van den Bossche P, Geerts S (2008) Molecular tools for the rapid detection of drug resistance in animal trypanosomes. Trends Parasitol 24:236–242. https://doi.org/10.1016/j.pt.2008.02.006
Chitanga S, Marcotty T, Namangala B, Van den Bossche P, Van Den Abbeele J, Delespaux V (2011) High prevalence of drug resistance in animal trypanosomes without a history of drug exposure. PLoS negl trop dis Public Library Sci 5:14–54. https://doi.org/10.1371/journal.pntd.0001454
Delespaux V, Chitanga S, Geysen D, Goethals A, Van den Bossche P, Geerts S (2006) SSCP analysis of the P2 purine transporter TcoAT1 gene of Trypanosoma congolense leads to a simple PCR-RFLP test allowing the rapid identification of diminazene resistant stocks. Acta Trop 100:96–102. https://doi.org/10.1016/j.actatropica.2006.10.001
Vitouley HS, Mungube EO, Allegye-Cudjoe E, Diall O, Bocoum Z, Diarra B et al (2011) Improved pcr-rflp for the detection of diminazene resistance in Trypanosoma congolense under field conditions using filter papers for sample storage. PLoS Negl Trop Dis 5:7–10. https://doi.org/10.1371/journal.pntd.0001223
Mamoudou A, Delespaux V, Chepnda V, Hachimou Z, Andrikaye JP, Zoli A et al (2008) Assessment of the occurrence of trypanocidal drug resistance in trypanosomes of naturally infected cattle in the adamaoua region of cameroon using the standard mouse test and molecular tools. Acta Trop 106:115–118. https://doi.org/10.1016/j.actatropica.2008.02.003
Moti YF, Abbeele VDJ, Van den Bossche DLPB, Delespaux V (2012) Ghibe river basin in Ethiopia: Present situation of trypanocidal drug resistance in Trypanosoma congolense using tests in mice and PCR-RFLP. Vet Parasitol 189:197–203. https://doi.org/10.1016/j.vetpar.2012.04.022
O’Meara WP, Smith DL, McKenzie FE (2006) Potential impact of intermittent preventive treatment (IPT) on spread of drug-resistant malaria. PLoS Med 3:633–642. https://doi.org/10.1371/journal.pmed.0030141
Munday JC, Rojas KE, Eze AA, Delespaux V, Van Den AJ, Rowan T et al (2013) Drugs and drug resistance functional expression of tcoat1 reveals it to be a p1-type nucleoside transporter with no capacity for diminazene uptake. Int J Parasitol Drugs Drug Resist 3:69–76. https://doi.org/10.1016/j.ijpddr.2013.01.004
Eisler MC, Brandt J, Bauer B, Clausen P, Delespaux V, Holmes PH et al (2001) Standardised tests in mice and cattle for the detection of drug resistance in tsetse-transmitted trypanosomes of African domestic cattle. Vet Parasitol. https://doi.org/10.1016/s0304-4017(01)00415-0
Nefertiti ASG, Batista MM, Da Silva PB, Batista DGJ, Da Silva CF, Peres RB et al (2018) In vitro and in vivo studies of the trypanocidal effect of novel quinolines. Antimicrob Agents Chemother 62:1–11. https://doi.org/10.1128/AAC.01936-17
Peregrine AS, Knowles G, Ibitayo AI, Scott JR, Moloo SK, Murphy NB (1991) Variation in resistance to isometamidium chloride and diminazene aceturate by clones derived from a stock of Trypanosoma congolense. Parasitology 102:93–100. https://doi.org/10.1017/s0031182000060388
Wangwe II, Wamwenje SA, Mirieri C, Masila NM, Wambua L, Kulohoma BW (2019) Modelling appropriate use of trypanocides to restrict wide-spread multi-drug resistance during chemotherapy of animal african trypanosomiasis. Parasitology 17:774–780. https://doi.org/10.1017/S0031182018002093
Geerts SI, Holmes PH, Eisler MCDO (2001) African bovine trypanosomiasis: the problem of drug resistance. Trends Parasitol 111:25–28. https://doi.org/10.1016/S1471-4922(00)01827-4
Okello I, Mafie E, Eastwood G, Nzalawahe J, Mboera LEG (2022) Prevalence and associated risk factors of african animal trypanosomiasis in cattle in lambwe, kenya. J Parasitol Res 22:1–12. https://doi.org/10.1155/2022/5984376
Okello I, Mafie E, Eastwood G, Nzalawahe J, Mboera LEG (2022) African animal trypanosomiasis: a systematic review on prevalence, risk factors and drug resistance in sub-saharan Africa. J Med Entomol 59:1099–1143. https://doi.org/10.1093/jme/tjac018
University B (2013) The Ruma / Lambwe Valley Research Activities Location Climate and land use change impacts on ecosystem processes and stability in the Lambwe savanna. Available from https://www.bayceer.uni-bayreuth.de/CREATE/en/forschung/114937/120964/The_Lambwe_valley_Research_Activities. Accessed 28 Jan 2013
Otte MJ, Gumm ID (1997) Intra-cluster correlation coefficients of 20 infections calculated from the results of cluster-sample surveys. Prev Vet Med 9:147–150. https://doi.org/10.1016/S0167-5877(96)01108-7
Thrusfield M (2005) Veterinary epidemiology. blackwell science, Third Edit
Chagas CRF, Binkienė R, Ilgūnas M, Iezhova T, Valkiūnas G (2020) The buffy coat method: A tool for detection of blood parasites without staining procedures. Parasit Vectors 13:1–12. https://doi.org/10.1186/s13071-020-3984-8
Njiru ZK, Constantine CC, Guya S, Crowther J, Kiragu JM, Thompson RCA et al (2005) The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitol Res 95:186–192. https://doi.org/10.1007/s00436-004-1267-5
Daniel KM, Audra JS, Paul HTJ, WC, (1992) Sensitive detection of trypanosomes in tsetse flies by DNA amplification. Int J Parasitol 22:909–918. https://doi.org/10.1016/0020-7519(92)90047-O
Marc DB, Gerald M, Adrien ZAM (2015) Detection and identification of Trypanosoma of African livestock through a single PCR based on internal transcribed spacer 1 of rDNA. Int J Parasitol 31:610–614. https://doi.org/10.1016/S0020-7519(01)00161-8
Mukabane DK, Shivairo RS, Mdachi RE, Orenge CO, Mulama DK (2014) Effect of Aflatoxin B-1 on transmissibility of trypanosoma congolense in mice. J Biol Agric Healthc 24:28–37
Gitonga PK, Ndungu K, Murilla GA, Thande PC, Wamwiri FN, Auma JE et al (2017) Differential virulence and tsetse fly transmissibility of Trypanosoma congolense and Trypanosoma brucei strains. Onderstepoort J Vet Res 84:1–10. https://doi.org/10.4102/ojvr.v84i1.1412
Tipman H-F (2004) Software review analysis for free: comparing programs for sequence analysis. Brief Bioinform 5:82–87
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. https://doi.org/10.1093/molbev/msr121
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices. Bioinformatics 8:275–282. https://doi.org/10.1093/bioinformatics/8.3.275
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Kulohoma BW, Wamwenje SAO, Wangwe II, Masila N, Mirieri CK, Wambua L (2020) Prevalence of trypanosomes associated with drug resistance in Shimba Hills, Kwale County, Kenya. BMC Res Notes 13:1–6. https://doi.org/10.1186/s13104-020-05077-3
Baker RD, Godfrey DG (2009) Trypanosoma congolense: the distribution of enzymic variants in east and West Africa. Parasitology 96:475–486. https://doi.org/10.1017/S0031182000080112
Eze AA, Gould MK, Munday JC, Tagoe DNA, Stelmanis V, Schnaufer A et al (2016) Reduced mitochondrial membrane potential is a late adaptation of trypanosoma brucei brucei to isometamidium preceded by mutations in the γ Subunit of the F1Fo-ATPase. PLoS Negl Trop Dis 10:1–21. https://doi.org/10.1371/journal.pntd.0004791
Chowdhury AR, Bakshi R, Wang J, Yildirir G, Liu B, Pappas-Brown V et al (2010) The killing of african trypanosomes by ethidium bromide. PLoS Pathog 6:5–12. https://doi.org/10.1371/10.1371/journal.ppat.1001226
Baker N, Hamilton G, Wilkes JM, Hutchinson S, Barrett MP, Horn D (2005) Vacuolar ATPase depletion affects mitochondrial ATPase function, kinetoplast dependency, and drug sensitivity in trypanosomes. Proc Natl Acad Sci U S A 112:9112–9117. https://doi.org/10.1073/pnas.1505411112
Gould MK, Schnaufer A (2014) Independence from kinetoplast DNA maintenance and expression is associated with multidrug resistance in Trypanosoma brucei In vitro. Antimicrob Agents Chemother 58:2925–2928. https://doi.org/10.1128/AAC.00122-14
Jones-davies WJ, Folkers C (1968) The prevalence of homidium-resistant strains of trypanosomes in cattle in Northern Nigeria. Bull Epizoot Dis Africa 14:65–72
Scott JM, Pegram RG (1974) A high incidence of Trypanosoma congolense strains resistant to homidium bromide in Ethiopia. Trop Anim Heal Prod 6:215–221. https://doi.org/10.1007/bf02383280
Clausen PH, Sidibe I, Kaboré I, Bauer B (1992) Development of multiple drug resistance of Trypanosoma congolense in Zebu cattle under high natural tsetse fly challenge in the pastoral zone of samorogouan, burkina faso. Acta Trop 51:229–236. https://doi.org/10.1016/0001-706x(92)90041-u
Codjia V, Mulatu W, Majiwa PAO, Leak SGA, Rowlands GJ, Authié E et al (1993) Epidemiology of bovine trypanosomiasis in the Ghibe valley, southwest Ethiopia. Occurrence of populations of Trypanosoma congolense resistant to diminazene, isometamidium and homidium. Acta Trop 53:151–163. https://doi.org/10.1016/0001-706X(93)90026-8
Mulugeta W, Wilkes J, Mulatu W, Majiwa PAO, Masake R, Peregrine AS (1997) Long-term occurrence of Trypanosoma congolense resistant to diminazene, isometamidium and homidium in cattle at Ghibe, Ethiopia. Acta Trop 64:205–217. https://doi.org/10.1016/s0001-706x(96)00645-6
Carruthers LV, Munday JC, Ebiloma GU, Steketee P, Jayaraman S, Campagnaro GD et al (2021) Diminazene resistance in Trypanosoma congolense is not caused by reduced transport capacity but associated with reduced mitochondrial membrane potential. Mol Microbiol 116:564–588. https://doi.org/10.1111/mmi.14733
Tihon E, Imamura H, Van den Broeck F, Vermeiren L, Dujardin JC, Van Den Abbeele J (2017) Genomic analysis of Isometamidium Chloride resistance in Trypanosoma congolense. Int J Parasitol Drugs Drug Resist 7:350–361. https://doi.org/10.1016/j.ijpddr.2017.10.002
Ndung’u K, Murilla GA, Thuita JK, Ngae GN, Auma JE, Gitonga PK et al (2020) Differential virulence of Trypanosoma brucei rhodesiense isolates does not influence the outcome of treatment with anti-trypanosomal drugs in the mouse model. PLoS ONE 15:1–16. https://doi.org/10.1371/journal.pone.0229060
Christine MK, Joanna A, Paul OM, Kariuki N, Rosemary B, Richard K, Collins O, Serap A, GM (2018) Differential virulence of camel Trypanosoma evansi isolates in mice. Parasitology 176:139–148. https://doi.org/10.1017/S0031182017002359
Acknowledgements
The authors wish to thank the IACUC review committee and staff at KALRO-BioRI Pharmacology and Molecular Laboratory, KALRO-BioRI’s former Director Dr. Raymond Mdachi, livestock farmers whose cattle were used for collection of Trypanosoma isolates, and the Kenya Tsetse and Trypanosomiasis Eradication Council (KENTTEC) staff.
Funding
This study was funded by Partnership for Skills in Applied Sciences, Engineering and Technology-Regional Scholarship Innovative Fund. The funders had no role in developing the study design, collection, analysis, interpretation of the results, and writing the manuscript.
Author information
Authors and Affiliations
Contributions
IO conceptualized the study and performed molecular assays. IO and KALRO-BioRI’s pharmacology laboratory staff performed in vivo drug test experiments. IO and JNH performed the sequence analysis. IO and KO performed PCV analysis. IO wrote original draft of the manuscript, IO, EM, GE, LEGM, and JN reviewed and edited the manuscript. All the authors approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interests.
Ethical Approval
This study was approved by the Institutional Animal Care and Use Committee (IACUC) of Kenya Agricultural and Livestock Research Organization-Biotechnology Research Institute (protocol code No. (4/325/III/9). An informed consent was solicited from the owners of the cattle whose blood samples were collected from. The animal study was approved by Director of Veterinary Services in Kenya Ref MOALF/SDL/DVS/DS/RES/77 and Sokoine University of Agriculture provided research clearance permit Ref SUA/ADM/R.1.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Okello, I., Mafie, E., Nzalawahe, J. et al. Trypanosoma Congolense Resistant to Trypanocidal Drugs Homidium and Diminazene and their Molecular Characterization in Lambwe, Kenya. Acta Parasit. 68, 130–144 (2023). https://doi.org/10.1007/s11686-022-00640-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11686-022-00640-3