Abstract
MicroRNAs (miRNAs) modulate gene expression and regulate many physiological and pathological conditions. However, their modulation and effect in osteoclastogenesis remain unknown. In this study, we investigated the role of miR-346-3p in regulating the osteoclast differentiation from RAW264.7 cells. We used the miRNA microarray assay, miR-346-3p mimic transfection, tartrate resistant acid phosphatase (TRAP) staining, bone resorption assay, qRT-PCR, and western blot. Our results showed that the expression of miR-346-3p was significantly upregulated during osteoclast differentiation. Further, by transfecting cells with miR-346-3p mimic, we observed an increased number of TRAP-positive multinucleated cells, increased pit area caused by bone resorption, and enhanced expression of osteoclast-specific genes and proteins. Conversely, miR-346-3p inhibition attenuated the osteoclast differentiation and function. Software-mediated prediction and validation using luciferase reporter assay showed that TRAF3, a negative regulator of osteoclast differentiation, was inhibited by miR-346-3p overexpression. Our results showed that miR-346-3p directly targeted TRAF3 mRNA via binding to its 3′-UTR and inhibited the expression of TRAF3 protein. Taken together, our results revealed that miR-346-3p promotes the regulation of osteoclastogenesis by suppressing the TRAF3 gene. In conclusion, miR-346-3p could be a novel therapeutic target for bone loss-related pathogenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ossification mediated by osteoblasts and bone resorption regulated by osteoclasts are the two predominant processes of bone remodeling that also maintain the bone metabolism homeostasis (Boyle et al. 2003). The delicate balance between bone formation and resorption is influenced by a complex network consisting of the immune, vascular, neuroendocrine, and musculoskeletal system (Okamoto et al. 2017). A loss of this balance may lead to enhanced bone resorption, which can adversely affect the bone structure and function and may lead to bone disorders, such as rheumatoid arthritis and osteoporosis.
Osteoclasts are multinucleated giant cells that originate from hematopoietic stem cells or monocyte/macrophage progenitor cells. Osteoclasts dissolve the mineralized bone matrix by secreting acid and proteinases, such as cathepsin K (Boyce 2013; Kitazawa et al. 2018). Osteoclast differentiation and functions are regulated by many cytokines, hormones, and signaling pathways, among which macrophage colony-stimulating factor (M-CSF) and receptor activator of nuclear factor-κB ligand (RANKL) play the key roles (Okamoto et al. 2017). The binding of M-CSF with colony-stimulating factor 1 receptor (CSF1R) initiates the signaling pathway responsible for the differentiation of osteoclast precursors (Lozano et al. 2019). Moreover, M-CSF also induces the expression of RANK, which in turn binds to its ligand RANKL and leads to the recruitment and activation of transcription factors belonging to the TNF receptor-associated factor (TRAF) protein family (Zhao et al. 2019). Among the TRAF protein family, TRAF6 is a crucial adaptor for RANKL-induced osteoclastogenesis, while TRAF2 and TRAF5 have been shown to play minor roles (Gravallese et al. 2001).
On the other hand, TRAF3, another member of the TRAF family has been reported to limit the activation of the RANKL-mediated signaling and inhibit osteoclastogenesis (Yao et al. 2009). Among the various transcription factors, the NF-κB family plays a crucial role in classical as well as alternate pathways of osteoclast differentiation. The NF-κB family is composed of several dimeric transcription molecules, such as NF-κB1 (the precursor protein of p50, also called as p105), NF-κB2 (the precursor protein of p52, also called as p100), RelA (p65), RelB, and c-Rel. A Rel homology domain (RHD), present in the N-terminal region of all the members, facilitates the homo- and heterodimerization among the NF-κB family members, as well as their sequence-specific binding to promoters of various genes. A C-terminal transcription activation domain (TAD) required for DNA binding is present in all NF-κB family members except p50 and p52. Therefore, p50 and p52 can regulate gene expression only when interacting with any of the three Rel proteins. While Rel A and c-Rel heterodimerize preferentially with p50, Rel B heterodimerizes with p100 and p52 (Ghosh and Karin 2002; Hayden and Ghosh 2008). The classical and alternative pathways of osteoclastogenesis are activated by RelA/p50 and RelB/p52, respectively (Zhao 2018). Further, NF-κB-inducing kinase (NIK) mediates non-canonical NF-κB signaling cascade in the alternative pathway, thereby leading to nuclear aggregation of active p52/RelB complexes. However, while the activity of NIK is regulated by ubiquitination and proteasomal degradation, TRAF3 is known to play a role in its inhibition (Yao et al. 2009; Yang et al. 2010). However, the precise mechanism by which TRAF3 downregulates osteoclastogenesis remains unknown.
MicroRNAs (miRNAs) are short (~22 nucleotides) single-stranded non-coding RNAs that regulate a wide variety of biological processes including cell survival, cell proliferation, differentiation, apoptosis, and autophagy. They bind to specific recognition sequences, which are mainly located in the 3′-untranslated regions (UTRs) or coding sequence (CDS) of target mRNAs and result in mRNA degradation or inhibition of its translation (Hrdlicka et al. 2019; Lozano et al. 2019). Emerging evidence indicates that miRNAs modulate osteoclast development and differentiation by regulating multiple signal transduction pathways and the expression of genes responsible for osteoclast differentiation and function. For example, in bone marrow-derived monocyte/macrophage precursors (BMMs), miR-21 regulates osteoclastogenesis via inhibiting programmed cell death protein 4 (PDCD4), while miR-223 targets nuclear factor IA (NFIA) and promotes osteoclastogenesis from the RAW264.7 cell line (Shibuya et al. 2013; Hu et al. 2017). On the other hand, miR-146a and miR-34a inhibit osteoclastogenesis by targeting TRAF6 and transforming growth factor-beta-induced factor 2 (TGIF2), respectively (Taganov et al. 2006; Krzeszinski et al. 2014). These results warrant further studies to elucidate the underlying molecular mechanisms behind microRNA-mediated regulation of osteoclast differentiation and function.
In the present study, we analyzed the profile of differentially expressed microRNAs during osteoclastogenesis using RAW264.7 cells. Our results showed that the expression of miR-346-3p was significantly modulated during osteoclastogenesis. Furthermore, the expression of TRAF3 was directly regulated by miR-346-3p.
Materials and methods
Differentiation of RAW264.7 cells into osteoclasts
The RAW264.7 cell line was obtained from the American Type Culture Collection (ATCC, Manassas, VA). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Gaithersburg, MD) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin-streptomycin (100 U/mL penicillin and 100 μg/ml streptomycin; Gibco) at 37°C in a humidified chamber with 5% CO2. For osteoclastogenesis, the cells were grown in alpha-minimal essential medium (α-MEM; Gibco) (3 × 103 cells/ml), seeded in 96-well plates, and incubated for 24 h. Subsequently, the culture medium was replaced with the fresh medium containing recombinant mouse-RANKL (30 ng/ml) and recombinant mouse-M-CSF (25 ng/ml) (R&D Systems, Minneapolis, MN), and cells were cultured for 5 d.
MicroRNA microarray assay
RAW264.7 cells were induced with or without M-CSF and RANKL as described in the previous section, and the total RNA was extracted. The miRNA microarray assay was designed by KangChen Bio-tech (Shanghai, China), and each sample was analyzed thrice. The RNA was labeled with miRCURY™ Array Power Labeling kit (Exiqon A/S, Vedbaek, Denmark), and the sample was hybridized with miRCURY™ LNA Array (v.19.0, Exiqon A/S, Denmark) chip according to the manufacturer’s instructions. The chip was scanned and the signal intensity was measured by the Axon GenePix 4000B chip scanner and analyzed by GenePix Pro 6.0. The differentially expressed miRNAs between the experimental and control groups were expressed as relative ratio. Finally, miRNAs with significant differential expression were clustered and the cluster diagram was made.
Total RNA isolation and qRT-PCR analysis
RAW264.7 cells were subjected to differentiation as described earlier, and total RNA was extracted from cells at designated time points (days 0, 1, and 3) by using Trizol reagent (Ambion, Austin, TX). Next, the total RNA was reverse transcribed into cDNA using the PrimeScript™ RT reagent kit (TaKaRa, Kusatsu, Japan). Amplification was detected using the SYBR Premix Ex Taq II kit (TaKaRa) on QuantStudio 6 Flex real-time PCR system (Thermo Fisher Scientific, Bartlesville, OK). Actin and U6 were used as the endogenous control for expression of genes and miRNAs, respectively. The relative mRNA expression of the target genes was calculated using the 2−ΔΔCT method. The primer sequences used to amplify the target genes are listed in Table 1.
Transfection of miR-346-3p mimic/inhibitors
miR-346-3p mimic, miR-346-3p inhibitor, non-specific control miR (NC), and miR-NC inhibitor were synthesized by Shanghai GenePharma (Shanghai, China). For transfection, RAW264.7 cells pretreated with M-CSF and RANKL were transfected with miR-346-3p mimic/inhibitor and their corresponding controls (control mimic/inhibitor, respectively) using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA) in the serum-free medium according to the manufacturer’s instructions. After 4 h of incubation, the cells were used for subsequent experiments as discussed below in the presence of fresh medium supplemented with M-CSF and RANKL.
TRAP staining
The differentiation of RAW264.7 into osteoclasts in the presence or absence of various miRNA mimics and their inhibitors was confirmed using TRAP staining using the TRAP staining kit (Sigma-Aldrich, St. Louis, MO) as per the manufacturer’s protocol. Briefly, the cells were fixed in 4% polyformaldehyde solution for 10 min at room temperature. After thorough rinsing in deionized water and addition of the TRAP staining fluid, the cells were incubated in dark at 37°C for 60 min. Next, the staining solution was removed and the cells were washed twice with distilled water. Images of TRAP-positive multinuclear cells (containing >3 nuclei) were captured using microscopy.
Bone resorption assay
Bone resorption activity was detected using the Corning Osteo Assay Surface (COAS; Corning, Corning, NY). Briefly, RAW264.7 cells (3000 cells/well) were seeded in plates in the presence of M-CSF and RANKL and cultured for 5 d. Subsequently, the culture media was removed and the wells were washed with distilled water. Adherent cells were removed by ultrasonication for 3 min. Subsequently, 10% sodium hypochlorite was added to the cells and incubated for 5 min at room temperature. Finally, the wells were washed thrice with deionized water, air-dried, and imaged randomly. The pit areas were calculated using Image-Pro Plus 6.0 software (Media Cybernetics Inc., Rockville, MD).
Western blot analysis
Total protein was isolated from experimental and control cells using RIPA lysis buffer (Beyotime, Wuhan, China). The protein was quantified using the BCA assay kit (Beyotime) by following the manufacturer’s protocol. Briefly, 30 μg of total protein from each sample were separated by SDS-PAGE at 110 V for 2 h and was electrically transferred onto PVDF membranes (Millipore, Burlington, MA). The membranes were blocked with 5% skimmed milk for 90 min at room temperature followed by overnight incubation with anti-TRAF3 antibody (1:1000; Abcam, Cambridge, UK), anti-TRAP antibody (1:1000; Abcam), and anti-GAPDH antibody (1:5000; Abcam) at 4°C. The next day, the membrane was washed to remove the unbound primary antibodies, and the membranes were incubated with goat anti-rabbit IgG-HRP secondary antibody (1:5000; Abcam) for 90 min at room temperature. The protein bands were visualized by chemiluminescent HRP substrate (Millipore) by using the Image Lab software (version 5.2, Bio-Rad, Santa Rosa, CA), and the grayscale values of the protein bands and the relative ratios of protein band intensity to GAPDH were quantified using ImageJ (National Institutes of Health, Bethesda, MD).
Dual-luciferase reporter assay
The 293T cell line was obtained from the ATCC. When 293T cells reach to 90% confluence, cells were washed, digested, and resuspended. Then a total of 1 × 105 293T cells/well were seeded in 24-well plates and co-transfected with wild-type or mutated pRL-TK TRAF3 3′-UTR and pRL-TK Renilla luciferase vector carrying miR-346-3p, miR-346-3p-mut, or NC using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Following 72 h of transfection, the cells were lysed and subjected to analysis by the dual-luciferase reporter assay system (Promega, San Luis Obispo, CA). The results were normalized against the ratio of the firefly/Renilla activity.
Statistical analysis
All experiments were performed at least in triplicates, and the data were shown as mean ± standard deviation (SD). Statistical differences among groups were analyzed by one-way ANOVA followed by Tukey’s post hoc tests using SPSS software version 20.0 (IBM, Armonk, NY). Results with P < 0.05 were considered as statistically significant.
Results
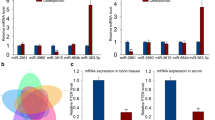
miR-346-3p was significantly upregulated during osteoclastogenesis
The miRNA microarray assay has been used to evaluate the miRNA expression profiles during osteoclastogenesis. Our results showed that the expression levels of miR-181a-5p, miR-146b-5p, and miR-322-5p were significantly downregulated during the osteoclastogenesis, while the expression levels of 5 microRNAs (miR-1958, miR-1931, miR-294-5p, miR-669d-3p, miR-346-3p) were significantly upregulated (Fig. 1, Fig. 2A). Since miR-346-3p was the most significantly upregulated among all, we investigated its role in osteoclastogenic using RAW264.7 cells. We monitored the expression of miR-346-3p on days 0, 1, and 3 after co-stimulation of RAW264.7 cells with M-CSF and RANKL; day 0 was regarded as the negative control. As expected, our results from the qRT-PCR showed that the miR-346-3p expression increased progressively during osteoclast differentiation (Fig. 2B). Further, these results were confirmed by the western blot which showed an increased expression of TRAP (a positive marker of mature osteoclasts) while the expression of TRAF3 was inhibited (Fig. 2C, D). These results suggested that miR-346-3p was overexpressed during osteoclast formation.
miR-346-3p was dramatically upregulated during osteoclastogenesis. (A) Relative miRNA levels were analyzed by qRT-PCR and normalized to U6. (B) miR-346-3p expression at designated time points (days 0, 1, and 3). (C, D) TRAP and TRAF3 protein expressions were detected by western blot and normalized to GAPDH. The data represented the mean ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001 compared with control.
miR-346-3p promoted osteoclastogenesis
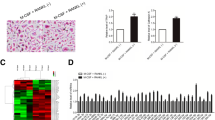
To evaluate the role of miR-346-3p in osteoclastogenesis, RAW264.7 cells were transfected with miR-346-3p mimic or its inhibitor before inducing osteoclast differentiation, while miR-NC and its inhibitor were used as control. Subsequently, RAW264.7 cells were cultured in the presence of M-CSF and RANKL for 6 d, and TRAP staining was performed to detect the number of mature osteoclasts. Our results showed that the number of TRAP-positive multinucleated giant cells increased in the experimental group treated with miR-346-3p mimic, while their numbers were significantly decreased in the group transfected with miR-346-3p inhibitor when compared with the control group (Fig. 3A, B).
miR-346-3p overexpression promoted osteoclastogenesis. (A) TRAP staining was performed in RAW 264.7 cells expressing miR-346-3p mimics/inhibitor, miR-NC, and miR-NC inhibitor. Osteoclasts were marked by black circles. Scale bars: 200 μm. (B) The number of TRAP-positive multinucleated cells was counted and shown. The data represented the mean ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001 compared with control.
miR-346-3p increased the bone resorptive ability of osteoclasts
Next, bone resorption activity assay was performed to assess the effect of transfection with miR-346-3p mimic on the bone resorptive capacity of osteoclasts. Our results showed that the areas of bone-resorbing pits significantly increased in the experimental group transfected with the miR-346-3p mimic, whereas the cells transfected with the miR-346-3p inhibitor showed significantly lower pit-formation ability. The group transfected with the miR-NC and its inhibitor exhibited similar baseline resorption ability (Fig. 4A). These results were consistent with the quantitative analysis of the area of absorption pits (Fig. 4B). Collectively, these results suggested that overexpression of miR-346-3p enhanced the bone resorption ability of osteoclasts.
miR-346-3p overexpression enhanced the bone resorption ability of osteoclasts. (A) The areas of bone-resorbing pits caused by osteoclast activity on the COAS bottom. Scale bars: 100 μm. (B) The quantitative analysis of the area of absorption pits. The data represented the mean ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001 compared with control.
miR-346-3p influenced the expression of osteoclastogenic genes and proteins
To understand the role of the miR-346-3p in osteoclastogenesis, we checked the expression levels of osteoclastogenesis associated genes by using qRT-PCR. Our results showed that in cells transfected with the miR-346-3p mimic, the expression of genes, such as NFATc1, TRAP, and cathepsin K were significantly upregulated. In contrast, cells transfected with the miR-346-3p inhibitor showed significant downregulation of these markers. Interestingly, the expression of TRAF3 was significantly downregulated in miR-346-3p mimic groups, while increased in miR-346-3p inhibitor groups (Fig. 5A–E). Further, these observations were confirmed by the western blot results as notably lower TRAF3 protein levels were seen in cells transfected with miR-346-3p mimic transfection, while increased TRAF3 expression was observed in cells subjected to miR-346-3p inhibitor transfection (Fig. 6A, B). Collectively, these results suggested that miR-346-3p promoted osteoclast differentiation via enhancing the expression of osteoclast-related genes and proteins.
miR-346-3p overexpression effected the expression of osteoclastogenic genes. Relative mRNA levels of osteoclast differentiation marker NFATc1 (A), TRAP (B), Ctsk (C), miR-346-3p (D), and TRAF3 (E) were determined by qRT-PCR and normalized to U6. The data represented the mean ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001 compared with control.
TRAF3 gene was a direct target of miR-346-3p
Since miRNAs mediate the degradation of target mRNAs by binding to the 3′-UTRs or CDSs of the target gene, we next sought to predict the potential target genes of miR-346-3p using TargetScan Release 7.1 (www.targetscan.org). Our analysis showed the presence of one conserved miR-346-3p target in the 3′-UTR of TRAF3 (Fig. 7A). To validate these observations, we co-transfected 293T cells with the luciferase expression vector including the 3′-UTR of TRAF3 along with the vector expressing miR-346-3p. Subsequently, we mutated the target site of miR-346-3p binding in TRAF3 and performed the dual-luciferase reporter assay. The results revealed that the luciferase activity was significantly downregulated in cells with wild-type (WT) TRAF3 but not in cells transfected with mutant TRAF3 (Fig. 7B). These findings confirmed that miR-346-3p directly targeted TRAF3.
miR-346-3p directly targeted TRAF3. (A) A schematic of the 3′-UTR region of wild-type (WT) TRAF3 mRNA illustrated the binding site was complementary paired with miR-346-3p. (B) The effect of miR-346-3p mimics/inhibitor, miR-NC, and miR-NC inhibitor on luciferase activity in 293T cells transfected with the TRAF3 3′-UTR WT/mutant reporter. The data represented the mean ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001 compared with control.
Discussion
The miRNAs have been identified as one of the pivotal regulators of gene expression during bone remodeling, homeostasis, and pathogenesis (Lian et al. 2012). There is an increasing interest in elucidating their role in osteoclast differentiation and function in the bone mass-related disorders. In this study, we explored the potential regulatory mechanism of miR-346-3p during osteoclast differentiation.
In the present study, our results showed that the expression of several microRNAs, such as miR-5622-5p, miR-1931, miR-294-5p, miR-669d-3p, and miR-346-3p was notably upregulated during osteoclastogenesis. Among these, the expression of miR-346-3p was the most significantly upregulated. Further, the overexpression of miR-346-3p increased the number of TRAP-positive multinuclear giant cells and promoted osteoclastogenesis. When osteoclastogenic cells were transfected with miR-346-3p mimic, it led to an increased expression of osteogenesis-related genes and proteins as confirmed by the results for qRT-PCR and western blot. In contrast, transfection with miR-346-3p inhibitor reduced the expression of these genes. Further, the dual-luciferase reporter assay and western blot assay revealed that TRAF3 was a potential target of miR-346-3p. Hence, our study demonstrated that miR-346-3p is one of the critical factors responsible for repressing TRAF3, an inhibitor of osteoclastogenesis, thereby promoting RANKL-induced osteoclastogenesis and osteoclastic bone resorption.
Osteoblasts and osteoclasts are the two important mediators of bone remodeling and metabolism. A growing number of studies have shown that an increase in osteoclastic bone resorption coupled with reduced osteoblastic bone formation leads to the pathogenesis of bone (Chen et al. 2018). Bone diseases that are characterized by a reduction in bone mass and low trauma fractures are a direct result of the increased osteoclastogenic activity. Hence, there is an imminent need to identify factors that are responsible for the imbalance observed in bone metabolism. Further, inhibition of excessive bone destruction could be an effective strategy to alleviate bone pathogenesis. Therefore, it is imperative to uncover the underlying mechanisms involved in the activation of osteoclastogenesis.
RANKL, a TNF-α superfamily cytokine, is widely expressed by several types of cells, such as osteoblasts, osteocytes, and bone marrow stromal cells among others. The binding between RANKL and RANK triggers the recruitment of adaptor molecules and induction of downstream signaling pathways (Lozano et al. 2019; van Dam et al. 2019). Extensive research using genetically modified mice has established RANK/RANKL signaling as the key molecular event during osteoclast differentiation (Okamoto et al. 2017). During the early stage of osteoclastogenesis, the activation of NF-κB and activator protein-1 (AP-1) by RANKL leads to a robust upregulation of nuclear factor of activated T cell cytoplasmic 1 (NFATc1) and its translocation to the nucleus where it modulates the transcription of NFATc1 target genes. NFATc1 is a crucial regulator during the terminal differentiation of osteoclasts and regulates the expression of TRAP, calcitonin receptor, β3 integrin, and cathepsin K (Park et al. 2017; Ono and Nakashima 2018).
miR-346 is widely expressed in different tissues and cells. Several studies have demonstrated that miR-346 regulates various pathophysiological processes by modulating the expression of different genes and promotes hepatocellular carcinoma occurrence, the biological function in breast cancer cells, and regulates the molecular mechanisms of cell death (Lin et al. 2015; Yang et al. 2017; Guo et al. 2018). However, the role of miR-346 in regulating osteoclastogenesis and functions of osteoclasts has not been reported.
Our previous study showed that miR-346-3p (one of mature miRNAs generated by miRNA precursor molecule, miR-346) was highly expressed during osteoclast differentiation (Niu et al. 2019). Our present work confirmed that miR-346-3p directly regulates osteoclastogenesis. Our results showed that the overexpression of miR-346-3p promoted osteoclastogenesis, elevated the number of TRAP-positive multinucleated giant cells, increased the expression levels of NFATc1, cathepsin K, and TRAP, and enhanced the bone resorption area. In contrast, inhibition of miR-346-3p attenuated osteoclastogenesis in RAW264.7 cells. These results confirmed that miR-346-3p has a negative regulatory effect during osteoclastogenesis. Our results further showed that TRAF3 might be a potential target of miR-346-3p. TRAF3, a death regulation signal, has been shown recently to serve as a pivotal multifunctional regulator in immune and other cells types (VanArsdale et al. 1997). Earlier studies have shown that TRAF3 blocked osteoclastogenesis in osteoclast precursors by inhibiting the activation of NIK-dependent alternative NF-κB signaling. Moreover, mice with TRAF3 conditionally deleted in myeloid cells, exhibited an early onset of osteoporosis, thereby indicating that TRAF3 was a major negative regulator of osteoclastogenesis (Li et al. 2019). In the present study, our results confirmed that miR-346-3p directly regulated TRAF3. Further observations such as enhanced osteoclastogenesis in response to overexpressed miR-346-3p validated these findings. Collectively, our results showed that miR-346-3p promoted osteoclastogenesis via repressing the expression of TRAF3. Nevertheless, the main limitation of this study is the lack of confirmation of our in vitro findings regarding the regulatory mechanisms of miR-346-3p in osteoclastogenesis in animal models. Thus, further in vivo investigation to fully understand its specific function is warranted.
Conclusions
To the best of our knowledge, the present study provides the first evidence of miR-346-3p-mediated regulation of osteoclastogenesis in RAW264.7 cells via targeting TRAF3 gene. Based on our results, we propose that miR-346-3p could be a potential therapeutic target for the treatment of osteoclast-related bone disorders.
References
Boyce BF (2013) Advances in osteoclast biology reveal potential new drug targets and new roles for osteoclasts. J Bone Miner Res 28:711–722
Boyle W, Simonet W, Lacey D (2003) Osteoclast differentiation and activation. Nature 423:337–342
Chen X, Wang Z, Duan N, Zhu G, Schwarz EM, Xie C (2018) Osteoblast-osteoclast interactions. Connect Tissue Res 59:99–107
Ghosh S, Karin M (2002) Missing pieces in the NF-kappaB puzzle. Cell 109:S81–S96
Gravallese E, Galson D, Goldring S, Auron P (2001) The role of TNF-receptor family members and other TRAF-dependent receptors in bone resorption. Arthritis Res 3:6–12
Guo Z, Li J, Sun J, Sun L, Zhou Y, Yu Z (2018) miR-346 promotes HCC progression by suppressing breast cancer metastasis suppressor 1 expression. Oncol Res 26:1073–1081
Hayden MS, Ghosh S (2008) Shared principles in NF-kappaB signaling. Cell 132:344–362
Hrdlicka HC, Lee SK, Delany AM (2019) MicroRNAs are critical regulators of osteoclast differentiation. Curr Mol Biol Rep 5:65–74
Hu CH, Sui BD, Du FY, Shuai Y, Zheng CX, Zhao P, Yu XR, Jin Y (2017) miR-21 deficiency inhibits osteoclast function and prevents bone loss in mice. Sci Rep 7:43191
Kitazawa R, Haraguchi R, Fukushima M, Kitazawa S (2018) Pathologic conditions of hard tissue: role of osteoclasts in osteolytic lesion. Histochem Cell Biol 149:405–415
Krzeszinski JY, Wei W, Huynh H, Jin Z, Wang X, Chang TC, Xie XJ, He L, Mangala LS, Lopez-Berestein G, Sood AK, Mendell JT, Wan Y (2014) miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature 512:431–435
Li J, Ayoub A, Xiu Y, Yin X, Sanders JO, Mesfin A, Xing L, Yao Z, Boyce BF (2019) TGFbeta-induced degradation of TRAF3 in mesenchymal progenitor cells causes age-related osteoporosis. Nat Commun 10:2795
Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, Zhang Y (2012) MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol 8:212–227
Lin CW, Jan MS, Kuo JH (2015) Exploring MicroRNA expression profiles related to the mTOR signaling pathway in mouse embryonic fibroblast cells treated with polyethylenimine. Mol Pharm 12:2858–2868
Lozano C, Duroux-Richard I, Firat H, Schordan E, Apparailly F (2019) MicroRNAs: key regulators to understand osteoclast differentiation? Front Immunol 10:375
Niu D, Gong Z, Sun X, Yuan J, Zheng T, Wang X, Fan X, Mao Y, Liu X, Tang B, Fu Y (2019) miR-338-3p regulates osteoclastogenesis via targeting IKKbeta gene. In Vitro Cell Dev Biol Anim 55:243–251
Okamoto K, Nakashima T, Shinohara M, Negishi-Koga T, Komatsu N, Terashima A, Sawa S, Nitta T, Takayanagi H (2017) Osteoimmunology: the conceptual framework unifying the immune and skeletal systems. Physiol Rev 97:1295–1349
Ono T, Nakashima T (2018) Recent advances in osteoclast biology. Histochem Cell Biol 149:325–341
Park JH, Lee NK, Lee SY (2017) Current understanding of RANK signaling in osteoclast differentiation and maturation. Mol Cells 40:706–713
Shibuya H, Nakasa T, Adachi N, Nagata Y, Ishikawa M, Deie M, Suzuki O, Ochi M (2013) Overexpression of microRNA-223 in rheumatoid arthritis synovium controls osteoclast differentiation. Mod Rheumatol 23:674–685
Taganov KD, Boldin MP, Chang KJ, Baltimore D (2006) NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A 103:12481–12486
van Dam PA, Verhoeven Y, Trinh XB, Wouters A, Lardon F, Prenen H, Smits E, Baldewijns M, Lammens M (2019) RANK/RANKL signaling inhibition may improve the effectiveness of checkpoint blockade in cancer treatment. Crit Rev Oncol Hematol 133:85–91
VanArsdale T, VanArsdale S, Force W, Walter B, Mosialos G, Kieff E, Reed J, Ware C (1997) Lymphotoxin-beta receptor signaling complex: role of tumor necrosis factor receptor-associated factor 3 recruitment in cell death and activation of nuclear factor kappaB. Proc Natl Acad Sci U S A 94:2460–2465
Yang C, McCoy K, Davis J, Schmidt-Supprian M, Sasaki Y, Faccio R, Novack D (2010) NIK stabilization in osteoclasts results in osteoporosis and enhanced inflammatory osteolysis. PLoS One 5:e15383
Yang F, Luo LJ, Zhang L, Wang DD, Yang SJ, Ding L, Li J, Chen D, Ma R, Wu JZ, Tang JH (2017) MiR-346 promotes the biological function of breast cancer cells by targeting SRCIN1 and reduces chemosensitivity to docetaxel. Gene 600:21–28
Yao Z, Xing L, Boyce BF (2009) NF-kappaB p100 limits TNF-induced bone resorption in mice by a TRAF3-dependent mechanism. J Clin Invest 119:3024–3034
Zhao B (2018) Does TNF promote or restrain osteoclastogenesis and inflammatory bone resorption? Crit Rev Immunol 38:253–261
Zhao K, Jia Y, Peng J, Pang C, Zhang T, Han W, Jiang J, Lu X, Zhu J, Qian Y (2019) Anacardic acid inhibits RANKL-induced osteoclastogenesis in vitro and prevents ovariectomy-induced bone loss in vivo. FASEB J 33:9100–9115
Acknowledgements
We would like to appreciate lab members for their support to our research and thank Editage (www.editage.cn) for English language editing.
Funding
This study was supported by grants from the National Natural Science Foundation of China (31700854), the Project of Natural Science Foundation of Anhui Province (1908085MH276), the Key Program of Anhui Educational Committee (KJ2018A1011 and KJ2019A0392), the Scientific Research Foundation of Bengbu Medical College (BYKY17118, BYKY18108, and BYKY1848ZD), the Translational Medicine Key Projects of Bengbu Medical College (BYTM2019006 and BYTM2019012), the Scientific Research Innovation Team of Bengbu Medical College (BYKC201910), the Postgraduate Research Innovation Project of Bengbu Medical College (Byycx1827, Byycx1954, Byycx1956, and Byycz1929), and the National Training Programs of Innovation and Entrepreneurship for Undergraduate (201810367013 and 201810367028).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no competing interests.
Additional information
Editor: Tetsuji Okamoto
Rights and permissions
About this article
Cite this article
Mao, Y., Chen, Y., Fu, Y. et al. miR-346-3p promotes osteoclastogenesis via inhibiting TRAF3 gene. In Vitro Cell.Dev.Biol.-Animal 56, 533–542 (2020). https://doi.org/10.1007/s11626-020-00479-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11626-020-00479-w