Abstract
The undesirable effects of silver nanoparticles (AgNPs) on soil environment have caused much concern. The previous studies, however, focused on sandy soil, with little known on others. In present study, the effects of polyvinylpyrrolidone-coated AgNPs (0, 1, 10, and 100 mg kg− 1 soil) on enzyme activities (urease and dehydrogenase), ammonia-oxidizing bacteria (AOB) and archaea (AOA), bacterial and archaeal communities, and microbial function profile in a yellow-brown loam soil were investigated. The significant dose-response inhibitions of AgNPs on enzyme activities were observed, with dehydrogenase more susceptible to AgNPs. Both of bacterial and archaeal amoA genes were reduced by AgNPs above 10 mg kg− 1, with AOB more susceptible to AgNPs than AOA. AgNPs at 100 mg kg− 1 caused reductions on the dominant Nitrosospira and Nitrosomonas, and even disappearance on Nitrosovibrio, while increase on Nitrososphaera significantly. AgNPs also changed bacterial and archaeal community structure. Exposure to AgNPs at 100 mg kg− 1 caused significant increases by 186.79% and 44.89% for Bacteroidetes and Proteobacteria, while decreases by 47.82%, 44.09%, 43.67%, and 80.44% for Actinobacteria, Chloroflexi, Planctomycetes, and Verrucomicrobia, respectively. Moreover, three dominant archaeal phyla (Thaumarchaeota, Euryarchaeota, and Parvarchaeota) were also reduced in the presence of AgNPs, especially Thaumarchaeota with the significant reduction of 13.71%. PICRUSt prediction revealed that AgNPs indeed had the potential to change soil microbial community’s functional contributions. It must be cautious on the interference of AgNPs to soil ecological functions in the future.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Silver nanoparticles (AgNPs) as antibacterial agent are being widely used in diverse fields (e.g., medical products (Tolaymat et al. 2010), textiles (Button et al. 2016), and household appliances (Baranwal et al. 2018)). However, AgNPs could enter into environment during their life span (Benn and Westerhoff 2008; Auvinen et al. 2016) and further cause questions on environmental health and sanitation (Farid et al. 2018). The great concern is safety of soil ecosystem which is regarded as a main sink for nanoparticles. Surface runoff, agricultural application of sewage sludge, and effluent irrigation are main release sources of AgNPs into soil environment. It was reported that 145 μg L− 1 of AgNPs was detected during initial runoff due to release of outdoor paints (Kaegi et al. 2010), and the predicted incremental increases of AgNPs in sludge-amended soil were 110 ng kg− 1· per year (Sun et al. 2014). Therefore, it is urgent to evaluate the ecological effects of AgNPs on soil ecosystem.
The shifts in microbial community could well indicate the response of soil ecosystem to invasion of exotic pollutants, further mirroring soil health (Holden et al. 2014). AgNPs in soils will inevitably interact with microbes which are responsible for biogeochemical cycle (e.g., carbon, nitrogen, phosphorus, and sulfur cycle) and waste degradation. Morones et al. (2005) introduced that AgNPs toxicity to Gram-negative bacteria (S. typhus, E. coli, P. aeruginosa, and V. cholera) was size dependent. Guo et al. (2016) reported that inhibition extent of AgNPs on Phanerochaete chrysosporium could be changed by sulfide. AgNPs properties (e.g., size, shape, and coating) (Arnaout and Gunsch 2012), concentrations (Sheng and Liu 2017), and dosing regimens (Zeng et al. 2019) could determine the interactions between AgNPs and organisms. AgNPs exhibit toxicity to organisms via (1) attaching to cell membranes, resulting in the changes of membrane permeability, redox cycling in the cytosol, accumulation of intracellular radicals, and dissipation of the proton motive force for ATP synthesis, (2) entering bacterial cell directly to further cause damage by interfering with DNA and protein synthesis (Lok et al. 2006; Morones et al. 2005; Nel et al. 2006; Reidy et al. 2013). Compared to pure culture, the behaviors of AgNPs in complex environment need to be more caution.
It could be seen from Table 1 that AgNPs had negative effects on microbial biomass, enzyme activities, functional microbes, and even microbial community, which resulted from AgNPs themselves and released silver ions (He et al. 2016; Peyrot et al. 2014; Shin et al. 2012). AOB and AOA with amoA gene could convert ammonia to nitrite in the first step of nitrification, which had high sensitivity to AgNPs (Beddow et al. 2017). Archaea, an important prokaryote, which is different from bacteria and eukaryotes, often composes of extremophiles or exists in the extreme conditions (Woese and Fox 1977). They also play important roles in nutrient cycle as well as bacteria, such as AOA (Thion et al. 2016) and methanogens (Marti et al. 2015). Research on archaeal community exposed to AgNPs is still lag behind compared to bacterial community. McGee et al. (2018) reported that AgNPs reduced archaeal amoA gene copy numbers, while the other archaea were not assessed.
Soil properties and compositions (i.e., pH, organic matter content, soil texture, and ionic strength) could affect the fate and bioavailability of nanoparticles, further changing biological toxicity of nanoparticles (Tiede et al. 2009; Cornelis et al. 2014; Garcia-Gomez et al. 2018). Simonin and Richaume (2015) pointed out that soil type should be taken into account when evaluating ecotoxicity of AgNPs. A study by Rahmatpour et al. (2017) demonstrated that soils with lower clay content and ionic strength could cause greater inhibitions of AgNPs on microbial and enzyme activities. While extended to other soil types, the above-reported information still needs to be unraveled due to the diverse compositions in different soil type.
This study was conducted in a yellow-brown loam soil ecosystem which was common in Nanjing of China. The overall goals of present study were to (1) explore the impacts of AgNPs on soil enzyme activities of dehydrogenase and urease (dehydrogenase was an indicator of microbial activity, and urease was chosen as a biological indicator of nitrogen cycle (Li et al. 2018)); (2) clarify the changes of bacterial and archaeal community structure besides AOB and AOA in soils exposed to AgNPs; and (3) reveal the functional profile variations of soil microbial community.
Materials and methods
Characterization of AgNPs
PVP-AgNPs solution was provided by Shanghai Huzheng Nano Technology Co., Ltd. PVP-AgNPs were considered to be lower biological toxicity versus AgNPs with gum Arabic or citrate (Arnaout and Gunsch 2012). A transmission electron microscope (TEM) was used to characterize the morphology of AgNPs. AgNPs with diameter 10–40 nm had fine dispersion and uniform (Fig. S1). The more detailed descriptions on AgNPs have been described in the study of Huang et al. (2018).
Test soil
The surface soils (0–10 cm depth) were collected from natural field at Nanjing Agricultural University (Nanjing (32.03 N, 118.83E), Jiangsu Province, China) in April 2015. In order to reflect the whole properties of soil well, soils were collected from 5 different positions and fully mixed, and then removed bulk materials (i.e., stones, plant roots, and litter), followed by over 4 mm sieve. The prepared test soils were stored at 4 °C for further analytical purpose. The properties of soil were yellow-brown loam, 7.13 of pH value, 12.5 g organic matter kg− 1 soil, 0.91 g N kg− 1 soil, and 313.4 g water kg− 1 soil of maximum water holding capacity.
Experimental design
The experiment was performed in 250 mL sterilized plastic jar with flat bottom (Fisher, Pittsburgh, USA) (Fig. S2). The experimental design was detailed in the study of Huang et al. (2018). Briefly, each of jar as a test unit contained 60 g dry soil with 40% of the field capacity. After pre-cultured at 25 °C for 4 days, AgNPs solution was added into soils with micropipette while stirring soils slightly with spoon to obtain final silver concentrations of 1, 10, and 100 mg kg− 1, respectively. Meanwhile, control groups only received deionized water. Exactly 1 mg kg− 1 AgNPs was as environmentally relevant concentration according to the study of Gottschalk et al. (2009), and high AgNPs concentrations of 10 and 100 mg kg− 1 were chosen because of increasing consumption of AgNPs, and based on our preliminary experiment that AgNPs at 100 mg kg− 1 could evidently inhibit soil enzyme activities. After addition of AgNPs, the moisture content of soils was adjusted to 60% of field capacity. All test units were sealed with breathable parafilm (PM 996, Parafilm M®, US) and then incubated at 25 °C in a dark environment for 1, 4, 9, 16, 23, 30, and 37 days according to sampling interval, respectively. In addition, the test units without AgNPs on day 0 were used to investigate the initial enzyme activities and microbial community. Each treatment in the different sampling time was independent test unit and prepared in triplicate, thus totaling 87 test units constructed in the experiment. The analysis of total Ag concentration in soils was conducted using an inductively coupled plasma mass spectrometer (ICP-MS) after acid digestion according to US EPA 3050B method. The measured Ag concentrations corresponding to the nominal concentrations were 1.131 ± 0.213, 10.582 ± 0.657, and 97.235 ± 3.097 mg kg− 1, respectively.
Analysis of enzyme activities
Soil enzyme activities were measured using colorimetric methods on days 1, 4, 9, 16, 23, 30, and 37, respectively. The dehydrogenase activities were analyzed in accordance with the method of Gong (1997), and urease activities of Kandeler and Gerber (1988). Briefly, soils (dehydrogenase for 2.5 g, and urease for 1.0 g) were collected into a glass tube with stopper and suspended in buffered substrate solutions. For buffer solutions and substrates, Tris buffer (0.05 M, pH 7.6) and TTC (1%) were used for dehydrogenase, borate buffer (0.05 M, pH 10.0) and urea (5%) for urease. Soil samples were thoroughly mixed on a vortex shaker before placed in a constant temperature incubator at 37 °C for 24 h in the dark environment.
Real-time fluorescence quantitative analysis and 454-pyrosequencing
The soil DNA was extracted and purified using PowerSoil DNA Isolation Kit (Mo Bio Laboratories, Carlsbad, USA). The bacterial and archaeal amoA genes were quantified using real-time fluorescence quantitative analysis (Huang et al. 2018). AOB, AOA, bacterial, and archaeal communities were conducted using 454-pyrosequencing by Personal Biotechnology Co., Ltd. (Shanghai, China) as described in text S1 of Supporting Information (SI). Pyrosequencing reads were available in the NCBI Sequence Read Archive (SRA) database under the accession number SRP218979.
To identify the functional profile shifts in soil microbes exposed to AgNPs, KEEG functional annotations were achieved via PICRUSt (phylogenetic investigation of communities by reconstruction of unobserved states) prediction. PICRUSt predicted metagenomes using established evolutionary model from 16S rRNA data and reference genome database. The differences of 16S rRNA gene copy number among different species were also considered, and raw data of species abundance were corrected to make the predictive results more accurate and reliable. The technique with more cost-effective was often used to provide functional insights when there was only 16S data available in samples (Langille et al. 2013).
Statistical analysis
The enzyme activities were presented as means ± standard deviations. The statistical differences in soil enzyme activities were analyzed through one-way ANOVA at 95% of significance level (p < 0.05). Pearson correlation analysis was used to characterize AgNPs concentration-dependent effect on soil enzyme activities. The value of semi-inhibitory concentration (IC50) is often considered as an estimate of toxicant effects on specific organism. In present study, IC50 value presented 50% of inhibitions on enzyme activities and was calculated as the previous study (Huang et al. 2018).
Results
Response of soil enzyme activities to AgNPs
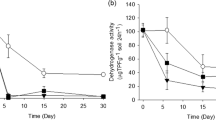
Before dosing AgNPs, the tested dehydrogenase activity was 85.13 mg (g·d)− 1, and urease activity was 512.79 mg N (g·d)− 1. Soil enzyme activities, especially dehydrogenase activities, were found to be affected by AgNPs (Fig. 1). The dehydrogenase activity at 1 mg kg− 1 AgNPs showed the significant increase on day 1 and then reduction from 4th day onwards (p < 0.05), while variations in urease activities were not observed at the same AgNPs level. When AgNPs concentration reached ≥ 10 mg kg− 1, two enzyme activities showed the significant decrease (p < 0.05). The dehydrogenase activity on day 1 was reduced by 92% at 100 mg kg− 1 of AgNPs, and barely detectable after 37 days. Correspondingly, the urease activity decreased by 75% and 87% on day 1 and 37, respectively.
Moreover, two enzyme activities in all AgNPs treatment groups kept relatively stable and did not recover to the level of control group at the end of experiment, indicating that inhibitions of AgNPs on enzyme activities were persistent. Pearson analysis showed the significantly negative correlation between enzyme activities and AgNPs concentrations (urease, r = − 0.928, p < 0.05; dehydrogenase r = −0.737, p < 0.05). These results revealed that the influences of AgNPs on enzyme activities were dose-dependent, with the higher correlation between urease activities and AgNPs levels. It could be seen from Table 2 that variations in IC50 were also obvious. The IC50 of dehydrogenase and urease decreased from 12.134 to 5.446 mg kg− 1 and 32.644 to 14.976 mg kg− 1 with time, respectively, which meant that exposure time could evidently affect AgNPs ecotoxicity. The lower IC50 of dehydrogenase indicated that the dehydrogenase was more sensitive to AgNPs than urease in the study.
Response of ammonia-oxidizing microorganisms to AgNPs
AOB on day 37 was more abundant than AOA, approximately 4 to 5 times (Fig.2). Bacterial and archaeal amoA genes at 1 mg kg− 1 AgNPs were higher than those in control group, indicating that 1 mg kg− 1 AgNPs did not affect ammonia-oxidizing microorganism, even with slight stimulations. While AgNPs were above 10 mg kg− 1, the inhibitions were observed. Compared to control group, exposure to 100 mg kg− 1 AgNPs caused decreases of 39% and 21% for bacterial and archaeal amoA genes, respectively.
AgNPs at 100 mg kg−1 lowered the richness and diversity both of AOB and AOA communities assessed by Chao1, ACE, and Shannon indexes (Tables S1 and S2). UPGMA analysis showed high similarity of AOB community between control group on days 0 and 37, while AOA between control group and AgNPs treatment group on day 37 (Fig. S3). These results indicated that AOB had higher sensitivities to AgNPs than AOA. PCA analysis showed the significant separation of AOB community with the principal components 1 and 2 explaining 97.42% and 2.58% for total variations, and AOA community with 99.87% and 0.13%, respectively (Fig. S4).
Before soils were treated with AgNPs, the most dominant AOB was Nitrosospira, followed by Nitrosomonas and Nitrosovibrio (Fig. 3 (a)). Exposure to AgNPs for 37 days changed AOB distribution. AgNPs caused reductions of Nitrosospira from 5.44% to 4.08% and Nitrosomonas from 1.73% to 1.65%, even the disappearance of Nitrosovibrio. However, AOA exposed to AgNPs showed the different variations from AOB. Nitrososphaera was the only indentified genus, and their relative abundance in the presence of AgNPs was 8 times higher than that in control group on day 37. These results demonstrated that AgNPs did change nitrogen cycle pathway in yellow-brown loam soil.
Response of microbial community structure to AgNPs
As shown in Table S3 and S4, bacterial Chao1, ACE, and Shannon indexes exposed to AgNPs reduced, indicating that AgNPs exposure could lead to a low richness and diversity of bacterial community. However, archaeal richness and diversity under AgNPs exposure increased. In addition, a comparison between days 0 and 37 on control group suggested that durable culture could also lead to an obvious reduction in microbial richness and diversity.
As show in Fig. S5, the high similarity among bacterial community was still found between control group on days 0 and 37, while archaeal community between control group and AgNPs treatment group on day 37. PCA analysis via principal components 1 and 2 explained 92.32% and 7.68% for bacterial community variations, and 95.42% and 4.58% for archaeal community variations, respectively (Fig. S6). The difference and similarity in microbial communities were also presented using Venn diagram with common and unique OTUs (Fig. S7). The number of common OTUs accounted for 3.69% and 10.24% of the total observed OTUs of bacteria and archaea, respectively. It suggested that both of bacteria and archaea had tolerance to AgNPs. The common bacteria OTUs at phylum level belonged to Proteobacteria (29.68%), Actinobacteria (23.44%), Bacteroidetes (8.20%), and Chloroflexi (7.85%), while the common archaea OTUs mostly belonged to Thaumarchaeota (79.78%).
In addition, the microbial community compositions were compared at phylum, class, and family level, respectively. As shown in Fig. 4(a), the relative abundance of phylum Proteobacteria (25.02%–39.09%) was the highest in samples, followed by Actinobacteria (6.34%–14.50%), Acidobacteria (11.94%–13.85%), Chloroflexi (5.11%–9.14%), Planctomycetes (3.07%–5.45%), Bacteroidetes (4.39%–12.59%), Nitrospirae (2.80%–2.88%), Gemmatimonadetes (2.59%–3.64%), and Verrucomicrobia (0.53%–2.71%). Metastats analysis revealed the significant differences in some phyla between AgNPs and control group on day 37. The presence of AgNPs significantly decreased the relative abundances of dominant phyla Actinobacteria (47.82%), Chloroflexi (44.09%), Planctomycetes (43.67%), and Verrucomicrobia (80.44%), while increasing the relative abundances of phyla Bacteroidetes (186.79%) and Proteobacteria (44.89%) (p < 0.05). Moreover, comparing control group on days 0 and 37, the difference caused by duration time was found only for Actinobacteria (p < 0.05).
At class level (Fig. 4b), after 37-day AgNPs exposure, the relative abundances of Betaproteobacteria and Cytophagia increased, while Acidmicrobial and Actinobacteria decreased. Further study on family level (Fig. 4c) showed that the relative abundance of Methylophilaceae presented the most significant rise from 0.05% to 14.3%. The relative abundance of family Cytophagaceae also increased obviously. However, there was no obvious change in the relative abundance of Nitrospirae.
As shown in Fig.5 (a), three dominant archaeal phyla were observed in soils. Compared with bacteria, the archaeal classification was more singular. The phylum Thaumarchaeota (73.0%–84.6%) was the most dominant in three samples. All archaea at phylum level decreased at 100 mg kg− 1 AgNPs, especially Thaumarchaeota with the significant reduction of 13.71%, indicating that AgNPs exposure had the negative influence on the archaeal community structure.
To further study the effect of AgNPs on archaeal community, the analysis was carried out on class and order level. Three classes in phylum Euryarchaeota were observed to be Methanobacteria, Methanomicrobia, and Thermoplasmata (Fig. 5b). Six dominant orders were observed and compared (Fig. 5c). Nitrososphaerales was the most abundant orders. After 37-day AgNPs exposure, the relative abundance of order Nitrososphaerales decreased from 79.8% to 72.9%. Among four dominant archaeal orders in phylum Euryarchaeota, Methanocellales, and Methanosarcinales increased from 0.02% and 0.15% to 0.22% and 0.43%, while Methanobacteriales and Methanomicrobiales decreased from 0.43% and 0.15% to 0.08% and 0%, respectively.
Response of predictive functional profile to AgNPs
The microbial community’s predictive functional contributions were summarized into six categories based on KEEG annotations (Fig. 6). The reads related to metabolism were 59.5%~59.9%, followed by genetic information processing of 18.4%~19.6%, and environmental information processing of 15.0%~15.2%. There were no obvious shifts in the functional classification between control group on days 0 and 37. However, six metabolic pathways occurred among the different variations after 37-day exposure to AgNPs.
It was found that the relative abundances related to cellular processes, human diseases, and genetic information increased, while others decreased. In metabolism categories, most of secondary metabolism pathways exposed to AgNPs were lower than control group on day 37, and xenobiotics biodegradation and metabolism with the greatest reduction. In environmental information processing categories, signal transduction increased, while membrane transport and signaling molecules and interaction decreased. In cellular processes categories, cell motility on day 37 showed a slight increase.
Discussion
The slight increase of dehydrogenase activity at lower AgNPs level of 1 mg kg− 1 on day 1 may be due to enhanced microbial activity at lower toxic stress (Gu et al. 2014), which needs further to be explored in yellow-brown loam soil. The inhibitions of AgNPs on dehydrogenase could be explained that the main sites of producing dehydrogenase were located in the plasma membrane of bacteria and the mitochondrial membrane of fungi (Dick 1994; Sinsabaugh 1994). AgNPs could absorb to cell membrane and then directly destroy its permeability (Lok et al. 2006; Nel et al. 2006). The previous studies have demonstrated that smaller particles (< 10 nm) could even enter cell directly and cause more damages, such as interference with DNA and protein synthesis, redox process, and function of organelle (Morones et al. 2005; Choi and Hu 2008). These mechanisms could lead to a reduction in soil dehydrogenase activity.
The negative effects of AgNPs on soil enzyme activities, especially on dehydrogenase activities, were accordance with the study of McGee et al. (2017). McGee et al. (2017) reported that soil dehydrogenase activity declined immediately and was significantly lower than control group (p ≤ 0.0001) at 50 mg kg− 1 AgNPs, but urease activity declining significantly on day 3 (p ≤ 0.0001). Samarajeewa et al. (2017) also claimed that dehydrogenase activities had the greatest sensitivity in sand soil treated with PVP-AgNPs, and PVP showed no effects on soil enzyme activities. IC50 values of dehydrogenase in our study ranged from 5.446 mg kg− 1 to 12.134 mg kg− 1, lower than those reported by other studies, such as 14-day IC50 of 19.9 mg kg− 1 PVP-AgNPs by Samarajeewa et al. (2017) and 7-day IC50 of 107.98 mg kg− 1 citrate-AgNPs by Shin et al. (2012). These differences were largely attributed to soil texture and AgNPs coating. Arnaout and Gunsch (2012) reported that citrate-AgNPs showed higher toxicity to microorganisms than AgNPs with PVP or gum Arabic coating.
For inhibitions of AgNPs on soil urease activities, AgNPs might inhibit the microbial activity related to urease (Unine et al. 2012). It has been widely known that urease is more vulnerable to Ag+, but Ag+ concentration from AgNPs in soils was less than the predicted effect concentration (Shin et al. 2012). Urease was found to be less susceptible to PVP-AgNPs than dehydrogenase in our study. However, Shin et al. (2012) reported urease with the highest sensitivity to citrate-AgNPs in sand. A 37-day IC50 of 14.976 mg kg− 1 PVP-AgNPs in our study was also comparable to 7-day IC50 of 14.20 mg kg− 1 citrate-AgNPs in the study of Shin et al. (2012). Rahmatpour et al. (2017) reported that PVP-AgNPs below 1 mg kg− 1 could inhibit urease activities in the typical Torriorthents and Haplocalcids soil, while PVP-AgNPs at 1 mg kg− 1 in present study did not affect urease activities. These results indicated that PVP-AgNPs in yellow-brown loam soil had the lower toxicity to urease. Meanwhile, the response of enzyme activities to AgNPs was dose-dependent, which was in accordance with the study of Rahmatpour et al. (2017).
Both of AOB and AOA play the important roles in ammonia oxidation process. The dominant AOB in present study was mainly due to be incubated at 25 °C. AOB generally had a wide range of temperature adaption (4–37 °C), while AOA was more active under warmer conditions (37 °C) (Wu et al. 2013). The presence of AgNPs above 10 mg kg− 1 decreased amoA gene copy numbers of AOB and AOA, indicating that AgNPs indeed interfered with nitrogen cycling. AOB in present study had higher sensitivity to AgNPs than AOA, which was accordance with the study of Beddow et al. (2017), while McGee et al. (2017) reported a contrary result. The different response of AOB and AOA to PVP-AgNPs might be due to the different predicted structures of archaeal and bacterial enzyme ammonia monooxygenase (Walker et al. 2010).
The high percentage of unclassified in ammonia-oxidizing microorganism community structure might be due to low portion of ammonia oxidizing microbes in overall microbial community and low specificity of primers used in the experiment. Nitrosospira and Nitrosomonas were dominant in the presence of AgNPs, which was consistent with the study of Zhang et al. (2014). The disappearance of Nitrosovibrio revealed that Nitrosovibrio was the most susceptible to AgNPs among AOB community. Nitrososphaera was reported to retrieve in all soils except acidic soil (Pester et al. 2012); this could explain the presence of Nitrososphaera in yellow-brown loam soil with 7.13 of pH value.
The reduction of microbial alpha diversity in control group with time might be due to prolonged poverty of nutrient input. Based on analysis of beta diversity, AgNPs indeed brought about variations in microbial communities. AgNPs exposure changed the bacterial community structure. AgNPs caused significant reduction on phylum Verrucomicrobia while increase on phylum Proteobacteria in present study, and these results were accordance with the study of McGee et al. (2017). Dunfield et al. (2007) reported that phylum Verrucomicrobia had the function of oxidizing methane under anaerobic conditions and reducing the escape of methane in soil ecosystem, which was important for controlling greenhouse effect. It meant that soils contaminated with AgNPs have the potential to increase greenhouse gas emissions, which must be cautious. The high resistance of Proteobacteria to AgNPs in present study might be due to their defense mechanisms to toxic pollutants. Some bacteria belonged to Proteobacteria possessed various metal-resistance integrons and secreted extracellular polymers (Nemergut et al. 2004; Nilgiriwala et al. 2008).
Acidobacteria was linked to organic carbon transformation (Ward et al. 2009). Acidobacteria in soil often accounted for 20% of bacterial community (Naether et al. 2012). McGee et al. (2017) reported that Acidobacteria on day 30 decreased from 14% to 6.5% at 50 mg kg− 1 of AgNPs. Grün and Emmerling (2018) reported that Acidobacteria at 0.01 mg kg− 1 of polyacrylate–AgNPs in loamy soil significantly increased under short exposure and diminished after 1 year, while no variations were found at 0.1–1 mg kg− 1 AgNPs. However, no significant variations in Acidobacteria were observed in our study. It was reported that Acidobacteria had the potential to excrete extracellular slime and siderophores and possessed genes encoding a range of ion channels, resistance-nodulation-cell division transporters, and drug transporters (Ward et al. 2009).
One explanation for reduction on Chloroflexi was lack of a lipid outer membrane and specialized secretion systems (Sutcliffe 2011). Meanwhile, Actinobacteria on day 37 decreased significantly (from 12.15% to 6.34%) in the presence of AgNPs, while McGee et al. (2017) reported no effects of AgNPs on Actinobacteria. Most of phylum Actinobacteria were heterotrophic and aerobic, which played an important role in organic matter degradation (Chater et al. 2010). It could be concluded that AgNPs had an inhibition on carbon cycle in soil environment, even causing a more profound impact on the earth’s environment. Kalyuhznaya et al. (2009) reported that Methylophilaceae linked methanol oxidation to denitrification. The relative abundance of family Methylophilaceae belonged to phylum Proteobacteria increased at 100 mg kg− 1 AgNPs, which would interfere with the carbon and nitrogen cycle. Family Cytophagaceae also increased obviously, and McBride et al. (2014) reported that most members of this family digested macromolecules such as polysaccharides or proteins.
For archaeal community, some shifts were also observed after AgNPs exposure. According to the variations of Chao1, ACE, and Shannon indexes, archaeal community showed high tolerance to AgNPs compared to bacterial community in the current study. The AgNPs decreased the most abundant order Nitrososphaerales, which was an AOA from soil and played an essential role in nitrogen cycle on earth (Tourna et al. 2011). Zhang et al. (2012) reported that AOA had more important roles in ammonia oxidation than AOB in the strongly acidic soils. These meant that nitrogen cycle in soil could be interfered after AgNPs exposure. In addition, AgNPs also had negative effects on Methanomicrobia and Methanobacteria in the study. Both of Methanomicrobia and Methanobacteria are the important Methanogens which is the only known microorganism producing methane with small molecule carbohydrates and hydrogen (Zhao et al. 2018). Hence, AgNPs exposure might have an impact on carbon cycling.
PICRUSt prediction provided needed insights that AgNPs had a potential to change soil microbial community’s functional contributions, which need further evidence. Carbohydrate metabolism and amino acid metabolism could be reduced by nanoparticles, such as AgNPs and graphene oxides (Li et al. 2019). Liu et al. (2019) reported that PVP-AgNPs at 50 mg L− 1 could significantly inhibit 31% of amino acid transport and metabolism pathways. The inhibitions of AgNPs on amino acid metabolism besides the decline of AOB and AOA abundance revealed AgNPs at 100 mg kg− 1 indeed affected the nitrogen cycle. The increase on cell motility in the presence of AgNPs was also accordance with the study of Li et al. (2019).
Conclusions
The present study investigated the effects of AgNPs (1–100 mg kg− 1) on enzyme activities, bacterial community, archaeal community, and microbial function profile in a yellow-brown loam soil. It was found that the inhibitions of AgNPs had a clear dose-response between enzyme activities and AgNPs levels. Meanwhile, AgNPs exposure time also had negative effects on enzyme activities. Soil dehydrogenase activities were observed to be more susceptible to AgNPs than urease activities. The variations in microbial richness and diversity exposed to AgNPs demonstrated that bacteria and archaea had different response to AgNPs. The community structures of bacteria and archaea were also evidently influenced by AgNPs, with some functional microorganisms especially sensitive to AgNPs. PICRUSt prediction further provided insights into effects of AgNPs on microbial function. Therefore, the effects of AgNPs on soil health need to be cautious.
References
Arnaout CL, Gunsch CK (2012) Impacts of silver nanoparticle coating on the nitrification potential of Nitrosomonas europaea. Environ Sci Technol 46(10):5387–5395
Auvinen H, Sepúlveda VV, Rousseau DPL, Du Laing G (2016) Substrate -and plant-mediated removal of citrate-coated silver nanoparticles in constructed wetlands. Environ Sci Pollut Res 23:21920–21926
Baranwal A, Srivastava A, Kumar P, Bajpai VK, Maurya PK, Chandra P (2018) Prospects of nanostructure materials and their composites as antimicrobial agents. Front Microbiol 9
Beddow J, Stolpe B, Cole PA, Lead JR, Sapp M, Lyons BP, Colbeck I, Whitby C (2017) Nanosilver inhibits nitrification and reduces ammonia-oxidising bacterial but not archaeal amoA gene abundance in estuarine sediments. Environ Microbiol 19:500–510
Benn TM, Westerhoff P (2008) Nanoparticle silver released into water from commercially available sock fabrics. Environ Sci Technol 42:4133–4139
Button M, Auvinen H, Van Koetsem F, Hosseinkhani B, Rousseau D, Weber KP, Du Laing G (2016) Susceptibility of constructed wetland microbial communities to silver nanoparticles: a microcosm study. Ecol Eng 97:476–485
Chater KF, Biro S, Lee KJ, Palmer T, Schrempf H (2010) The complex extracellular biology of Streptomyces. FEMS Microbiol Rev 34:171–198
Choi O, Hu Z (2008) Size dependent and reactive oxygen species related nanosilver toxicity to nitrifying bacteria. Environ Sci Technol 42:4583–4588
Cornelis G, Hund-Rinke K, Kuhlbusch T, Van Den Brink N, Nickel C (2014) Fate and bioavailability of engineered nanoparticles in soils: a review. Crit Rev Environ Sci Technol 44:2720–2764
Dick RP (1994) Soil enzyme activities as indicators of soil quality. Soil Sci Soc Am J 58:107–124
Dunfield PF, Yurgey AQ, Senin P, Smirnova AV, Stott MB, Hou S, Ly B, Saw JH, Zhou Z, Ren Y, Wang J, Mountain BW, Crowe MA, Weatherby TM, Bodelier PLE, Liesack W, Feng L, Wang L, Alam M (2007) Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450:879–882
Eivazi F, Afrasiabi Z, Jose E (2018) Effects of silver nanoparticles on the activities of soil enzymes involved in carbon and nutrient cycling. Pedosphere 28:209–214
Farid M, Ali S, Zubair M, Saeed R, Rizwan M, Sallah-Ud-Din R, Azam A, Ashraf R, Ashraf W (2018) Glutamic acid assisted phyto-management of silver-contaminated soils through sunflower; physiological and biochemical response. Environ Sci Pollut Res 25:25390–25400
Garcia-Gomez C, Fernandez MD, Garcia S, Obrador AF, Leton M, Babin M (2018) Soil pH effects on the toxicity of zinc oxide nanoparticles to soil microbial community. Environ Sci Pollut Res 25:28140–28152
Gong P (1997) Dehydrogenase activity in soil: a comparison between the TTC and INT assay under their optimum conditions. Soil Biol Biochem 29:211–214
Gottschalk F, Sonderer T, Scholz RW, Nowack B (2009) Modeled environmental concentrations of engineered nanomaterials (TiO2, ZnO, Ag, CNT, fullerenes) for different regions. Environ Sci Technol 43:9216–9222
Grün A, Emmerling C (2018) Long-term effects of environmentally relevant concentrations of silver nanoparticles on major soil bacterial phyla of a loamy soil. Environ Sci Eur 30:31
Grün A, Straskraba S, Schulz S, Schloter M, Emmerling C (2018) Long-term effects of environmentally relevant concentrations of silver nanoparticles on microbial biomass, enzyme activity, and functional genes involved in the nitrogen cycle of loamy soil. J Environ Sci 69:12–22
Gu L, Li Q, Quan X, Cen Y, Jiang X (2014) Comparison of nanosilver removal by flocculent and granular sludge and short- and long-term inhibition impacts. Water Res 58:62–70
Guo Z, Chen G, Liu L, Zeng G, Huang Z, Chen A, Hu L (2016) Activity variation of Phanerochaete chrysosporium under nanosilver exposure by controlling of different sulfide sources. Sci Rep 6:20813
Hänsch M, Emmerling C (2010) Effects of silver nanoparticles on the microbiota and enzyme activity in soil. J Plant Nutr Soil Sci 173:554–558
He S, Feng Y, Ni J, Sun Y, Xue L, Feng Y, Yu Y, Lin X, Yang L (2016) Different responses of soil microbial metabolic activity to silver and iron oxide nanoparticles. Chemosphere 147:195–202
Holden PA, Schimel JP, Godwin HA (2014) Five reasons to use bacteria when assessing manufactured nanomaterial environmental hazards and fates. Curr Opin Biotech 27:73–78
Huang J, Cao C, Li RQ, Guan WZ (2018) Effects of silver nanoparticles on soil ammonia-oxidizing microorganisms under temperatures of 25 and 5 °C. Pedosphere 28:607–616
Kaegi R, Sinnet B, Zuleeg S, Hagendorfer H, Mueller E, Vonbank R, Boller M, Burkhardt M (2010) Release of silver nanoparticles from outdoor facades. Environ Pollut 158(9):2900–2905
Kalyuhznaya MG, Martens-Habbena W, Wang T, Hackett M, Stolyar SM, Stahl DA, Lidstrom ME, Chistoserdova L (2009) Methylophilaceae link methanol oxidation to denitrification in freshwater lake sediment as suggested by stable isotope probing and pure culture analysis. Environ Microbiol Rep 1:385–392
Kandeler E, Gerber H (1988) Short-term assay of soil urease activity using colorimetric determination of ammonium. Biol Fertil Soils 6:68–72
Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821
Li M, Liang Z, Callier MD, d'Orbcastel ER, Ma X, Sun L, Li X, Wang S, Song X, Liu Y (2018) Nitrogen and organic matter removal and enzyme activities in constructed wetlands operated under different hydraulic operating regimes. Aquaculture 496:247–254
Li H, Chi Z, Yan B (2019) Long-term impacts of graphene oxide and Ag nanoparticles on anammox process: performance, microbial community and toxic mechanism. J Environ Sci 79:239–247
Liu X, Yang X, Hu X, He Q, Zhai J, Chen Y, Xiong Q, Vymazal J (2019) Comprehensive metagenomic analysis reveals the effects of silver nanoparticles on nitrogen transformation in constructed wetlands. Chem Eng J 358:1552–1560
Lok CN, Ho CM, Chen R, He QY, Yu WY, Sun HZ, Tam P, Chiu JF, Che CM (2006) Proteomic analysis of the mode of antibacterial action of silver nanoparticles. J Proteome Res 5:916–924
Marti M, Juottonen H, Robroek B, Yrjala K, Danielsson A, Lindgren PE, Svensson BH (2015) Nitrogen and methanogen community composition within and among three Sphagnum dominated peatlands in Scandinavia. Soil Biol Biochem 81:204–211
McBride MJ, Liu W, Lu X, Zhu Y, Zhang W (2014) The family Cytophagaceae, 4th edn. Springer Reference, London, pp 577–593
McGee CF, Storey S, Clipson N, Doyle E (2017) Soil microbial community responses to contamination with silver, aluminium oxide and silicon dioxide nanoparticles. Ecotoxicology 26:449–458
McGee CF, Storey S, Clipson N, Doyle E (2018) Concentration-dependent responses of soil bacterial, fungal and nitrifying communities to silver nano and micron particles. Environ Sci Pollut Res 25:18693–18704
Morones JR, Elechiguerra JL, Camacho A, Holt K, Kouri JB, Ramírez JT, Yacaman MJ (2005) The bactericidal effect of silver nanoparticles. Nanotechnology 16:2346–2353
Naether A, Foesel BU, Naegele V, Wüst PK, Weinert J, Bonkowski M, Alt F, Oelmann Y, Polle A, Lohaus G, Gockel S, Hemp A, Kalko EKV, Linsenmair KE, Pfeiffer S, Renner S, Schöning I, Weisser WW, Wells K, Fischer M, Overmann J, Friedrich MW (2012) Environmental factors affect Acidobacterial communities below the subgroup level in grassland and forest soils. Appl Environ Microbiol 78:7398–7406
Nel A, Xia T, Madler L, Li N (2006) Toxic potential of materials at the nanolevel. Science 311:622–627
Nemergut DR, Martin AP, Schmidt SK (2004) Integron diversity in heavy-metal-contaminated mine tailings and inferences about integron evolution. Appl Environ Microbiol 70(2):1160–1168
Nilgiriwala KS, Alahari A, Rao AS, Apte SK (2008) Cloning and overexpression of alkaline phosphatase PhoK from Sphingomonas sp. strain BSAR-1 for bioprecipitation of uranium from alkaline solutions. Appl Environ Microbiol 74(17):5516–5523
Pester M, Rattei T, Flechl S, Grongroft A, Richter A, Overmann J, Reinhold-Hurek B, Loy A, Wagner M (2012) amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ Microbiol 14:525–539
Peyrot C, Wilkinson KJ, Desrosiers M, Sauvé S (2014) Effects of silver nanoparticles on soil enzyme activities with and without added organic matter. Environ Toxicol Chem 33:115–125
Rahmatpour S, Shirvani M, Mosaddeghi MR, Nourbakhsh F, Bazarganipour M (2017) Dose-response effects of silver nanoparticles and silver nitrate on microbial and enzyme activities in calcareous soils. Geoderma 285:313–322
Reidy B, Haase A, Luch A, Dawson KA, Lynch I (2013) Mechanisms of silver nanoparticle release, transformation and toxicity: a critical review of current knowledge and recommendations for future studies and applications. Materials 6:2295–2350
Samarajeewa AD, Velicogna JR, Princz JI, Subasinghe RM, Scroggins RP, Beaudette LA (2017) Effect of silver nano-particles on soil microbial growth, activity and community diversity in a sandy loam soil. Environ Pollut 220:504–513
Sheng Z, Liu Y (2017) Potential impacts of silver nanoparticles on bacteria in the aquatic environment. J Environ Manag 191:290–296
Shin Y, Kwak JI, An Y (2012) Evidence for the inhibitory effects of silver nanoparticles on the activities of soil exoenzymes. Chemosphere 88:524–529
Simonin M, Richaume A (2015) Impact of engineered nanoparticles on the activity, abundance, and diversity of soil microbial communities: a review. Environ Sci Pollut Res 22:13710–13723
Sinsabaugh RS (1994) Enzymic analysis of microbial pattern and process. Biol Fertil Soils 17:69–74
Sun TY, Gottschalk F, Hungerbühler K, Nowack B (2014) Comprehensive probabilistic modelling of environmental emissions of engineered nanomaterials. Environ Pollut 185:69–76
Sutcliffe IC (2011) Cell envelope architecture in the Chloroflexi: a shifting frontline in a phylogenetic turf war. Environ Microbiol 13(2):279–282
Thion CE, Poirel JD, Cornulier T, De Vries FT, Bardgett RD, Prosser JI (2016) Plant nitrogen-use strategy as a driver of rhizosphere archaeal and bacterial ammonia oxidiser abundance. FEMS Microbiol Ecol 92(7):w91
Tiede K, Hassellov M, Breitbarth E, Chaudhry Q, Boxall A (2009) Considerations for environmental fate and ecotoxicity testing to support environmental risk assessments for engineered nanoparticles. J Chromatogr A 1216:503–509
Tolaymat TM, El Badawy AM, Genaidy A, Scheckel KG, Luxton TP, Suidan M (2010) An evidence-based environmental perspective of manufactured silver nanoparticle in syntheses and applications: a systematic review and critical appraisal of peer-reviewed scientific papers. Sci Total Environ 408:999–1006
Tourna M, Stieglmeier M, Spang A, Konneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C (2011) Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci U S A 108:8420–8425
Unine JM, Colman BP, Bone AJ, Gondikas AP, Matson CW (2012) Biotic and abiotic interactions in aquatic microcosms determine fate and toxicity of Ag nanoparticles. Part 1. Aggregation and dissolution. Environ Sci Technol 46:6915–6924
Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, Brochier-Armanet C, Chain PSG, Chan PP, Gollabgir A, Hemp J, Hügler M, Karr EA, Könneke M, Shin M, Lawton TJ, Lowe T, MartensHabbena W, Sayavedra-Soto LA, Lang D, Sievert SM, Rosenzweig AC, Manning G, Stahl DA (2010) Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci U S A 107:8818–8823
Ward NL, Challacombe JF, Janssen PH, Henrissat B, Coutinho PM, Wu M, Xie G, Haft DH, Sait M, Badger J, Barabote RD, Bradley B, Brettin TS, Brinkac LM, Bruce D, Creasy T, Daugherty SC, Davidsen TM, Deboy RT, Detter JC, Dodson RJ, Durkin AS, Ganapathy A, GwinnGiglio M, Han CS, Khouri H, Kiss H, Kothari SP, Madupu R, Nelson KE, Nelson WC, Paulsen I, Penn K, Ren QH, Rosovitz MJ, Selengut JD, Shrivastava S, Sullivan SA, Tapia R, Thompson LS, Watkins KL, Yang Q, Yu CH, Zafar N, Zhou LW, Kuske CR (2009) Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl Environ Microbiol 75:2046–2056
Woese CR, Fox GE (1977) Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Nat Acad Sci U S A 74:5088–5090
Wu Y, Ke X, Hernández M, Wang B, Dumont MG, Jia Z, Conrad R (2013) Autotrophic growth of bacterial and archaeal ammonia oxidizers in freshwater sediment microcosms incubated at different temperatures. Appl Environ Microbiol 79:3076–3084
Zeng JW, Xu P, Chen GQ, Zeng GM, Chen AW, Hu L, Huang ZZ, He K, Guo Z, Liu WW, Wu J, Shi JB (2019) Effects of silver nanoparticles with different dosing regimens and exposure media on artificial ecosystem. J Environ Sci 75:181–192
Zhang LM, Hu HW, Shen JP, He JZ (2012) Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. Isme J 6:1032
Zhang C, Liang Z, Hu Z (2014) Bacterial response to a continuous long-term exposure of silver nanoparticles at sub-ppm silver concentrations in a membrane bioreactor activated sludge system. Water Res 50:350–358
Zhao J, Li YQ, Pan S, Tu QQ, Dang WY, Wang Z, Zhu HT (2018) Effects of magnesium chloride on the anaerobic digestion and the implication on forward osmosis membrane bioreactor for sludge anaerobic digestion. Bioresour Technol 268:700–707
Funding
This work was funded by the National Natural Science Foundation of China (No. 51479034 and No.51650410657), Fundamental Research Funds for the Central Universities (No.2242019 K40064), and Postgraduate Research & Practice Innovation Program of Jiangsu Province (No. KYCX19_0096).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Thomas D. Bucheli
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 655 kb)
Rights and permissions
About this article
Cite this article
Yan, C., Huang, J., Cao, C. et al. Effects of PVP-coated silver nanoparticles on enzyme activity, bacterial and archaeal community structure and function in a yellow-brown loam soil. Environ Sci Pollut Res 27, 8058–8070 (2020). https://doi.org/10.1007/s11356-019-07347-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-019-07347-5